Efficient Production of 9,22-Dihydroxy-23,24-bisnorchol-4-ene-3-one from Phytosterols by Modifying Multiple Genes in Mycobacterium fortuitum
Abstract
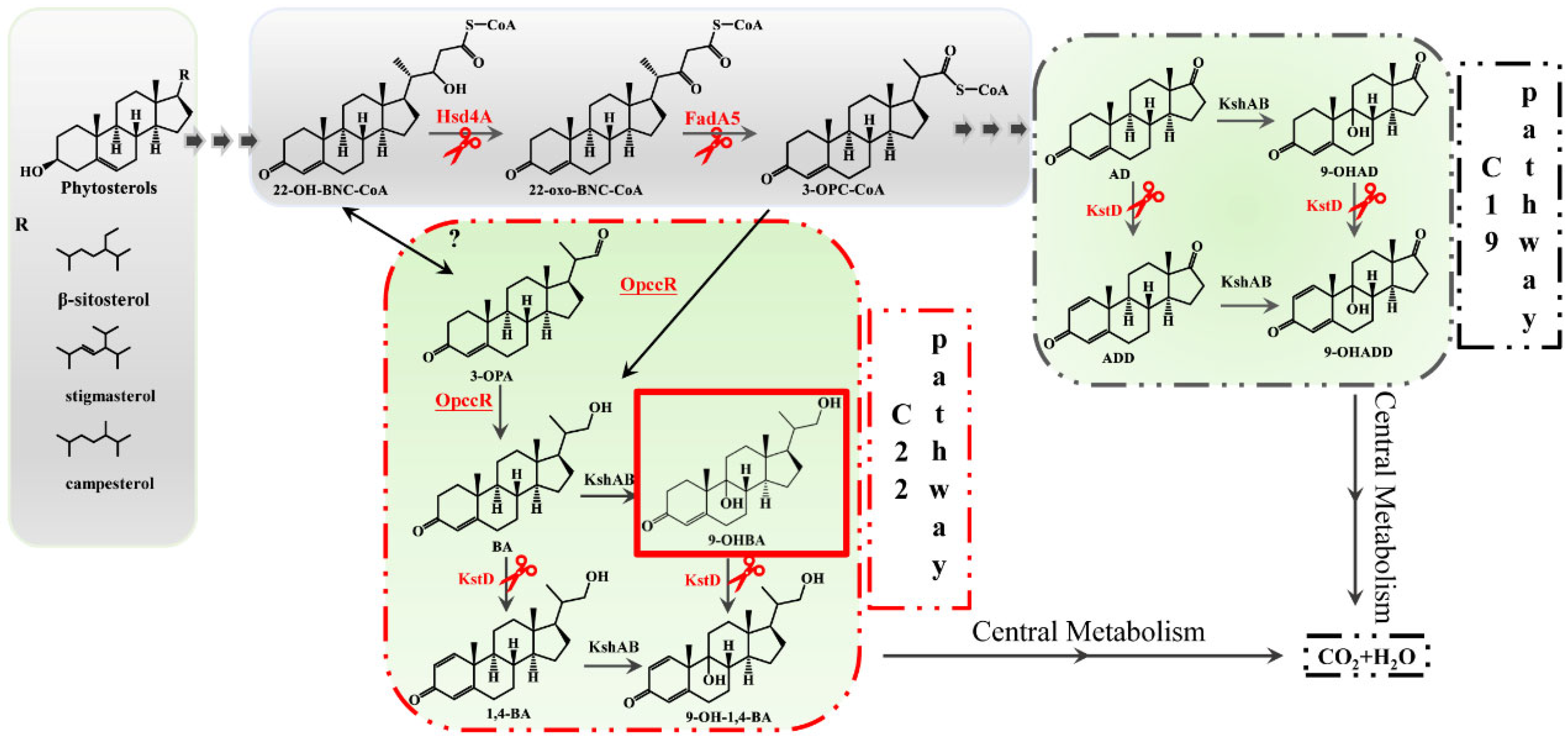
1. Introduction
2. Results
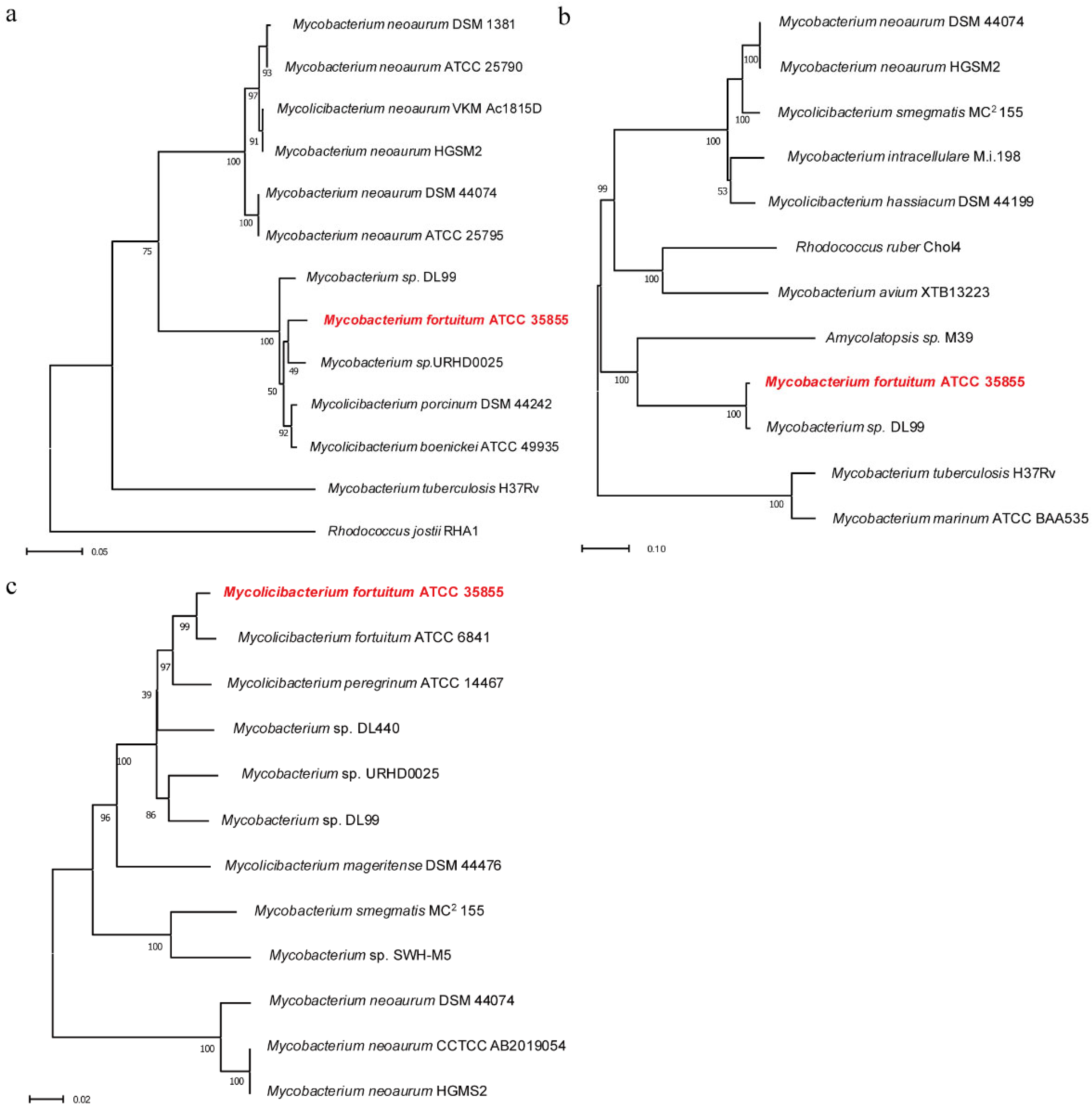
2.1. Construction of Phylogenetic Tree of Hsd4A, FadA5, and OpccR
2.2. Hsd4A—The Key Enzyme in the C19 Pathway
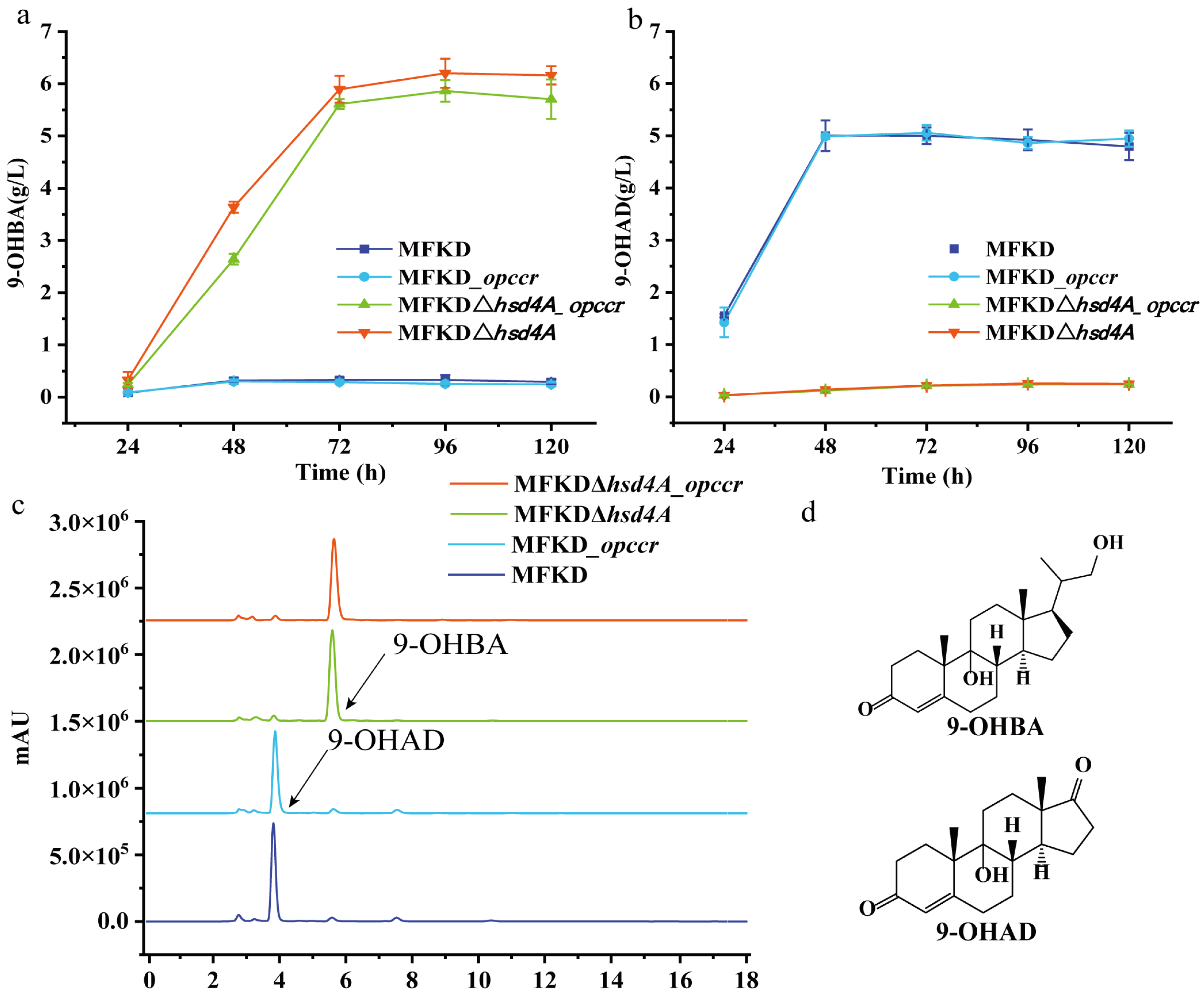
2.3. Deletion of fadA5 Improves the Selectivity of 9-OHBA by Eliminating 9-OHAD
2.4. Evaluation of the 9-OHBA-Producing Strain
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, Reagents, and Culture Conditions
4.2. Bioinformatic Analysis
4.3. Construction of Mutant Strains
| Name | Description | Source |
|---|---|---|
| Strains | ||
| Escherichia coli | E. coli DH5α | Vazyme Biotech Co., Ltd., Nanjing, China |
| MFKD | 9-OHAD producer, kstD1&2&3&4&5 deletion mutant of ATCC 35855 | Our lab |
| MFKD_opccR | ATCC 35855 opccR overexpression in MFKD via p40-opccR | This study |
| MFKDΔhsd4A | hsd4A deletion mutant of MFKD | This study |
| MFKDΔhsd4A_ opccR | ATCC 35855 opccR overexpression in MFKDΔhsd4A via p40-opccR | This study |
| MFKDΔhsd4AΔfadA5 | hsd4A and fadA5 double-deletion mutant of MFKD | This study |
| Plasmids | ||
| pKADel | Plasmid for allelic exchange, Pag85-lacZ Phsp60-sacB, AprR, KanR | [19] |
| pKADelΔhsd4A | pKADel carrying two homologous arms of hsd4A | This study |
| pKADelΔfadA5 | pKADel carrying two homologous arms of fadA5 | This study |
| p40 | pMV306 with Psmyc promoter, KanR | [19] |
| p40-opccR | p40 possessing opccR from M. fortuitum ATCC 35855 | This study |
4.4. Bioconversion and Analytical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Donova, M.V. Steroid Bioconversions. Methods Mol. Biol. 2017, 1645, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rugutt, J.K.; Rugutt, K.J. Antimycobacterial activity of steroids, long-chain alcohols and lytic peptides. Nat. Prod. Res. 2012, 26, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Seth, D.; Kamat, D. Intranasal Steroid Therapy for Allergic Rhinitis. Pediatr. Ann. 2019, 48, e43–e48. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Ji, W.T.; Song, L.; Tao, X.Y.; Zhao, M.; Gao, B.; Meng, H.; Wang, F.Q.; Wei, D.Z. Transformation of phytosterols into pregnatetraenedione by a combined microbial and chemical process. Green Chem. 2022, 24, 3759–3771. [Google Scholar] [CrossRef]
- Peng, H.; Wang, Y.; Jiang, K.; Chen, X.; Zhang, W.; Zhang, Y.; Deng, Z.; Qu, X. A Dual Role Reductase from Phytosterols Catabolism Enables the Efficient Production of Valuable Steroid Precursors. Angew. Chem. Int. Ed. Engl. 2021, 60, 5414–5420. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Zhang, X.; Li, Y.; Wang, Z.; Lv, Y.; Liu, J.; Alam, M.A.; Xiong, W.; Xu, J. Mycolicibacterium cell factory for the production of steroid-based drug intermediates. Biotechnol. Adv. 2021, 53, 107860. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V. Transformation of steroids by actinobacteria: A review. Appl. Biochem. Microbiol. 2007, 43, 1–14. [Google Scholar] [CrossRef]
- Almeida, C.A.S.; Baggio, S.R.; Mariutti, L.R.B.; Bragagnolo, N. One-step rapid extraction of phytosterols from vegetable oils. Food Res. Int. 2020, 130, 108891. [Google Scholar] [CrossRef]
- Andryushina, V.A.; Rodina, N.V.; Stytsenko, T.S.; Huy, L.D.; Druzhinina, A.V.; Yaderetz, V.V.; Voishvillo, N.E. Conversion of soybean sterols into 3,17-diketosteroids using actinobacteria Mycobacterium neoaurum, Pimelobacter simplex, and Rhodococcus erythropolis. Appl. Biochem. Microbiol. 2011, 47, 270–273. [Google Scholar] [CrossRef]
- Fernandez-Cabezon, L.; Galan, B.; Garcia, J.L. New Insights on Steroid Biotechnology. Front. Microbiol. 2018, 9, 958. [Google Scholar] [CrossRef]
- McLeod, M.P.; Warren, R.L.; Hsiao, W.W.L.; Araki, N.; Myhre, M.; Fernandes, C.; Miyazawa, D.; Wong, W.; Lillquist, A.L.; Wang, D.; et al. The complete genome of Rhodococcus sp RHA1 provides insights into a catabolic powerhouse. Proc. Natl. Acad. Sci. USA 2006, 103, 15582–15587. [Google Scholar] [CrossRef]
- Schaefer, C.M.; Lu, R.; Nesbitt, N.M.; Schiebel, J.; Sampson, N.S.; Kisker, C. FadA5 a thiolase from Mycobacterium tuberculosis: A steroid-binding pocket reveals the potential for drug development against tuberculosis. Structure 2015, 23, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Wipperman, M.F.; Sampson, N.S.; Thomas, S.T. Pathogen roid rage: Cholesterol utilization by Mycobacterium tuberculosis. Crit. Rev. Biochem. Mol. 2014, 49, 269–293. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Wang, F.Q.; Zhang, H.C.; Wei, D.Z. Identification and engineering of cholesterol oxidases involved in the initial step of sterols catabolism in Mycobacterium neoaurum. Metab. Eng. 2013, 15, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Donova, M.V.; Egorova, O.V. Microbial steroid transformations: Current state and prospects. Appl. Microbiol. Biotechnol. 2012, 94, 1423–1447. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.C.; He, B.R.; Zhang, J.X.; Yuan, C.Y.; Han, S.W.; Du, G.L.; Shi, J.P.; Sun, J.S.; Zhang, B.G. Phytosterol conversion into C9 non-hydroxylated derivatives through gene regulation in. Appl. Microbiol. Biotechnol. 2023, 107, 7635–7646. [Google Scholar] [CrossRef]
- Xu, L.Q.; Liu, Y.J.; Yao, K.; Liu, H.H.; Tao, X.Y.; Wang, F.Q.; Wei, D.Z. Unraveling and engineering the production of 23,24-bisnorcholenic steroids in sterol metabolism. Sci. Rep. 2016, 6, 21928. [Google Scholar] [CrossRef]
- Yuan, C.Y.; Ma, Z.G.; Zhang, J.X.; Liu, X.C.; Du, G.L.; Sun, J.S.; Shi, J.P.; Zhang, B.G. Production of 9,21-dihydroxy-20-methyl-pregna-4-en-3-one from phytosterols in Mycobacterium neoaurum by modifying multiple genes and improving the intracellular environment. Microb. Cell Fact. 2021, 20, 229. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Yuan, C.; Du, G.; Han, S.; Shi, J.; Sun, J.; Zhang, B. Improving the production of 9alpha-hydroxy-4-androstene-3,17-dione from phytosterols by 3-ketosteroid-Delta(1)-dehydrogenase deletions and multiple genetic modifications in Mycobacterium fortuitum. Microb. Cell Fact. 2023, 22, 53. [Google Scholar] [CrossRef]
- Song, S.; He, J.; Gao, M.; Huang, Y.; Cheng, X.; Su, Z. Loop pathways are responsible for tuning the accumulation of C19- and C22-sterol intermediates in the mycobacterial phytosterol degradation pathway. Microb. Cell Fact. 2023, 22, 19. [Google Scholar] [CrossRef]
- Zhang, J.X.; Zhang, R.J.; Song, S.K.; Su, Z.D.; Shi, J.P.; Cao, H.J.; Zhang, B.G. Whole-Genome Analysis of Mycobacterium neoaurum DSM 1381 and the Validation of Two Key Enzymes Affecting C22 Steroid Intermediates in Sterol Metabolism. Int. J. Mol. Sci. 2023, 24, 6148. [Google Scholar] [CrossRef] [PubMed]
- Lu, R.; Schaefer, C.M.; Neshitt, N.M.; Kuper, J.; Kisker, C.; Sampson, N.S. Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis Is Controlled by a Redox-Sensitive Thiol Switch. ACS Infect. Dis. 2017, 3, 666–675. [Google Scholar] [CrossRef]
- Turk, M.; Lietzow, R. Stabilized nanoparticles of phytosterol by rapid expansion from supercritical solution into aqueous solution. AAPS PharmSciTech 2004, 5, e56. [Google Scholar] [CrossRef] [PubMed]
- Mancilla, R.A.; Little, C.; Amoroso, A. Efficient Bioconversion of High Concentration Phytosterol Microdispersion to 4-Androstene-3,17-Dione (AD) by Mycobacterium sp B3805. Appl. Biochem. Biotechnol. 2018, 185, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Huang, Y.L.; Rathman, J.F.; Yang, S.T. Lecithin-enhanced biotransforation of cholesterol to androsta-1,4-diene-3,17-dione and androsta-4-ene-3,1 7-dione. J. Chem. Technol. Biotechnol. 2002, 77, 1349–1357. [Google Scholar] [CrossRef]
- Zhou, L.F.; Li, H.; Xu, Y.A.; Liu, W.; Zhang, X.M.; Gong, J.S.; Xu, Z.H.; Shi, J.S. Effects of a nonionic surfactant TX-40 on 9 alpha-hydroxyandrost-4-ene-3,17-dione biosynthesis and physiological properties of Mycobacterium sp. LY-1. Process Biochem. 2019, 87, 89–94. [Google Scholar] [CrossRef]
- Su, L.Q.; Xu, S.P.; Shen, Y.B.; Xia, M.L.; Ren, X.X.; Wang, L.F.; Shang, Z.H.; Wang, M. The Sterol Carrier Hydroxypropyl-beta-Cyclodextrin Enhances the Metabolism of Phytosterols by Mycobacterium neoaurum. Appl. Environ. Microbiol. 2020, 86, e00441-20. [Google Scholar] [CrossRef]
- Parish, T. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 2000, 146, 1969–1975. [Google Scholar] [CrossRef]


| Strain | Relative Selectivity (%) | |||
|---|---|---|---|---|
| 9-OHAD | 9-OHBA | AD | BA | |
| MFKD | 78.09 ± 0.21 | 4.21 ± 0.08 | 0.82 ± 0.01 | 0.09 ± 0.01 |
| MFKD_opccR | 77.98 ± 2.06 | 4.76 ± 0.31 | 0.87 ± 0.16 | 0.12 ± 0.03 |
| MFKDΔhsd4A | 4.07 ± 0.12 | 81.47 ± 0.04 | 0.35 ± 0.01 | 0.21 ± 0.01 |
| MFKDΔhsd4A_ opccR | 4,67 ± 0.13 | 81.22 ± 0.37 | 0.63 ± 0.22 | 0.19 ± 0.01 |
| MFKDΔhsd4AΔfadA5 | 0.90 ± 0.08 | 95.13 ± 0.46 | 0.37 ± 0.08 | 0.19 ± 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Liu, X.; He, B.; Zhai, X.; Yuan, C.; Li, Y.; Lin, W.; Wang, H.; Zhang, B. Efficient Production of 9,22-Dihydroxy-23,24-bisnorchol-4-ene-3-one from Phytosterols by Modifying Multiple Genes in Mycobacterium fortuitum. Int. J. Mol. Sci. 2024, 25, 3579. https://doi.org/10.3390/ijms25073579
Han S, Liu X, He B, Zhai X, Yuan C, Li Y, Lin W, Wang H, Zhang B. Efficient Production of 9,22-Dihydroxy-23,24-bisnorchol-4-ene-3-one from Phytosterols by Modifying Multiple Genes in Mycobacterium fortuitum. International Journal of Molecular Sciences. 2024; 25(7):3579. https://doi.org/10.3390/ijms25073579
Chicago/Turabian StyleHan, Suwan, Xiangcen Liu, Beiru He, Xinghui Zhai, Chenyang Yuan, Yixin Li, Weichao Lin, Haoyu Wang, and Baoguo Zhang. 2024. "Efficient Production of 9,22-Dihydroxy-23,24-bisnorchol-4-ene-3-one from Phytosterols by Modifying Multiple Genes in Mycobacterium fortuitum" International Journal of Molecular Sciences 25, no. 7: 3579. https://doi.org/10.3390/ijms25073579
APA StyleHan, S., Liu, X., He, B., Zhai, X., Yuan, C., Li, Y., Lin, W., Wang, H., & Zhang, B. (2024). Efficient Production of 9,22-Dihydroxy-23,24-bisnorchol-4-ene-3-one from Phytosterols by Modifying Multiple Genes in Mycobacterium fortuitum. International Journal of Molecular Sciences, 25(7), 3579. https://doi.org/10.3390/ijms25073579






