Comparison of Extracellular Vesicles from Induced Pluripotent Stem Cell-Derived Brain Cells
Abstract
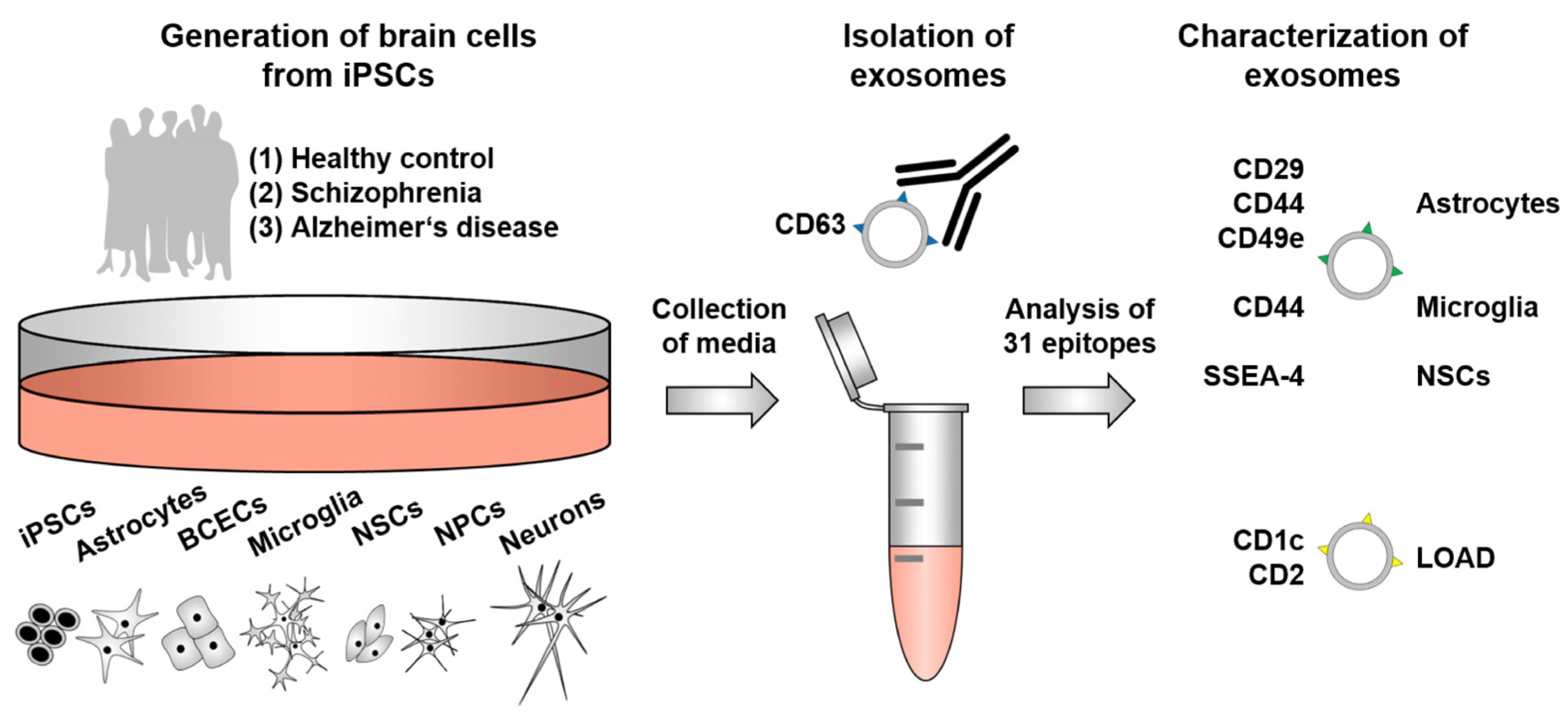
1. Introduction
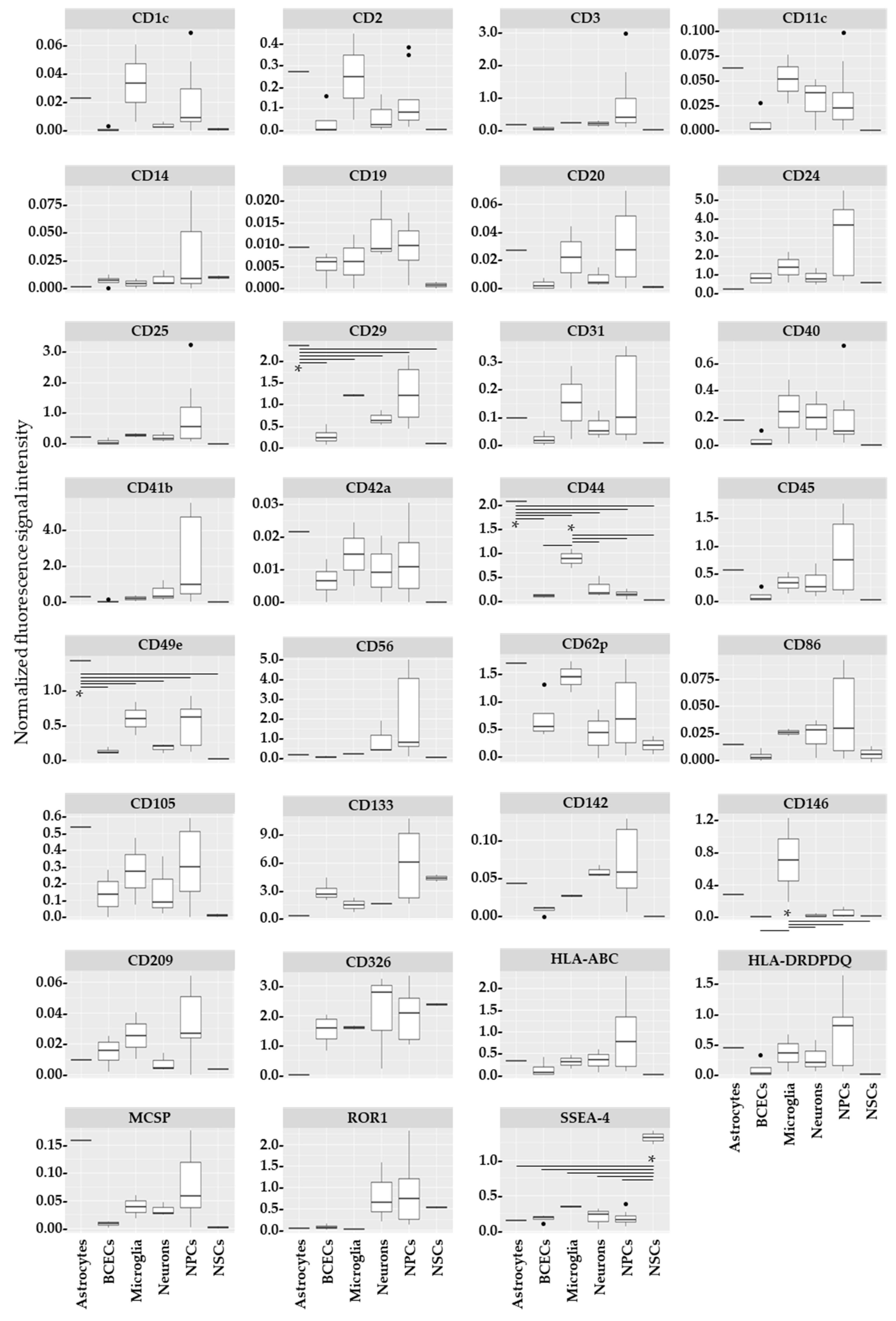
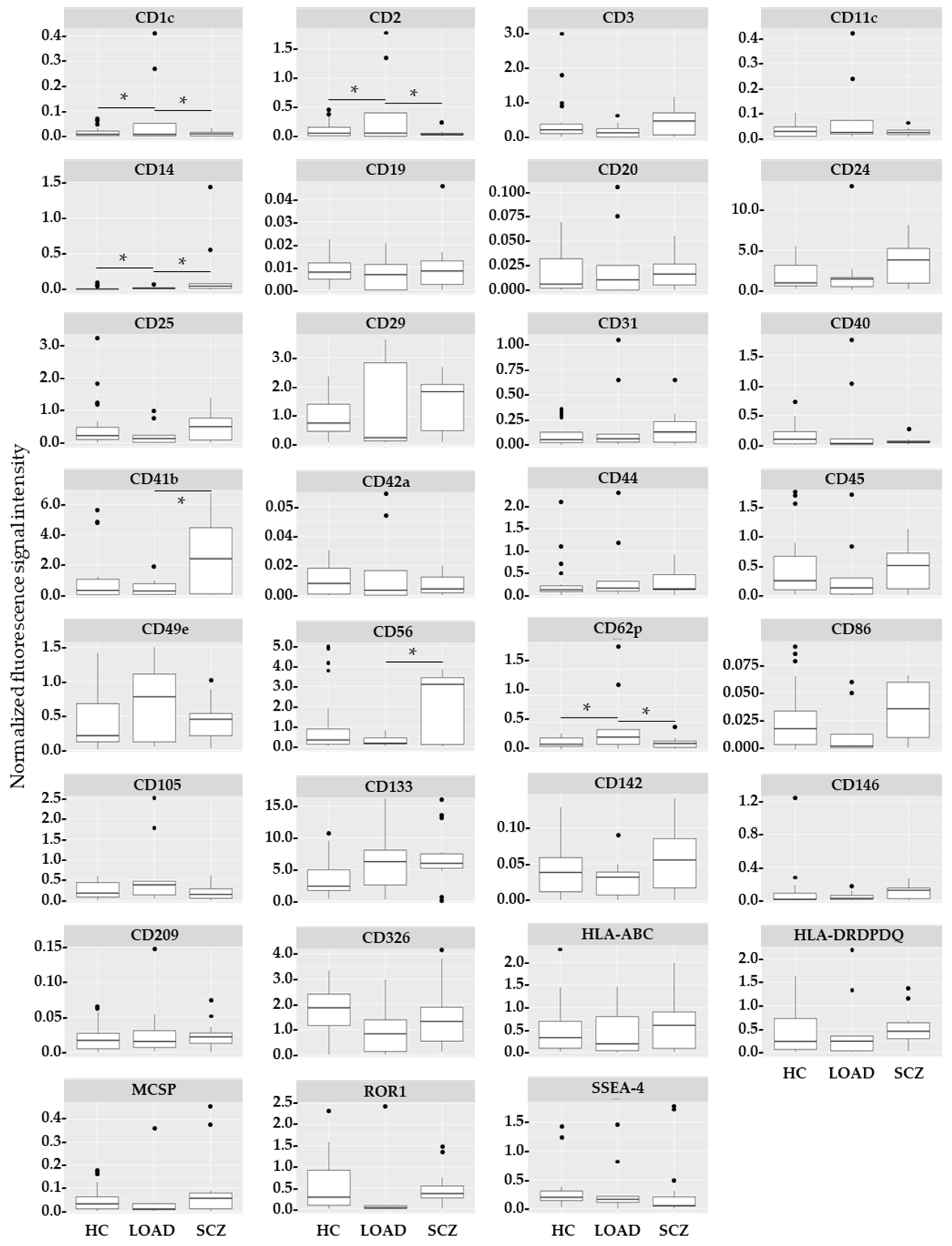
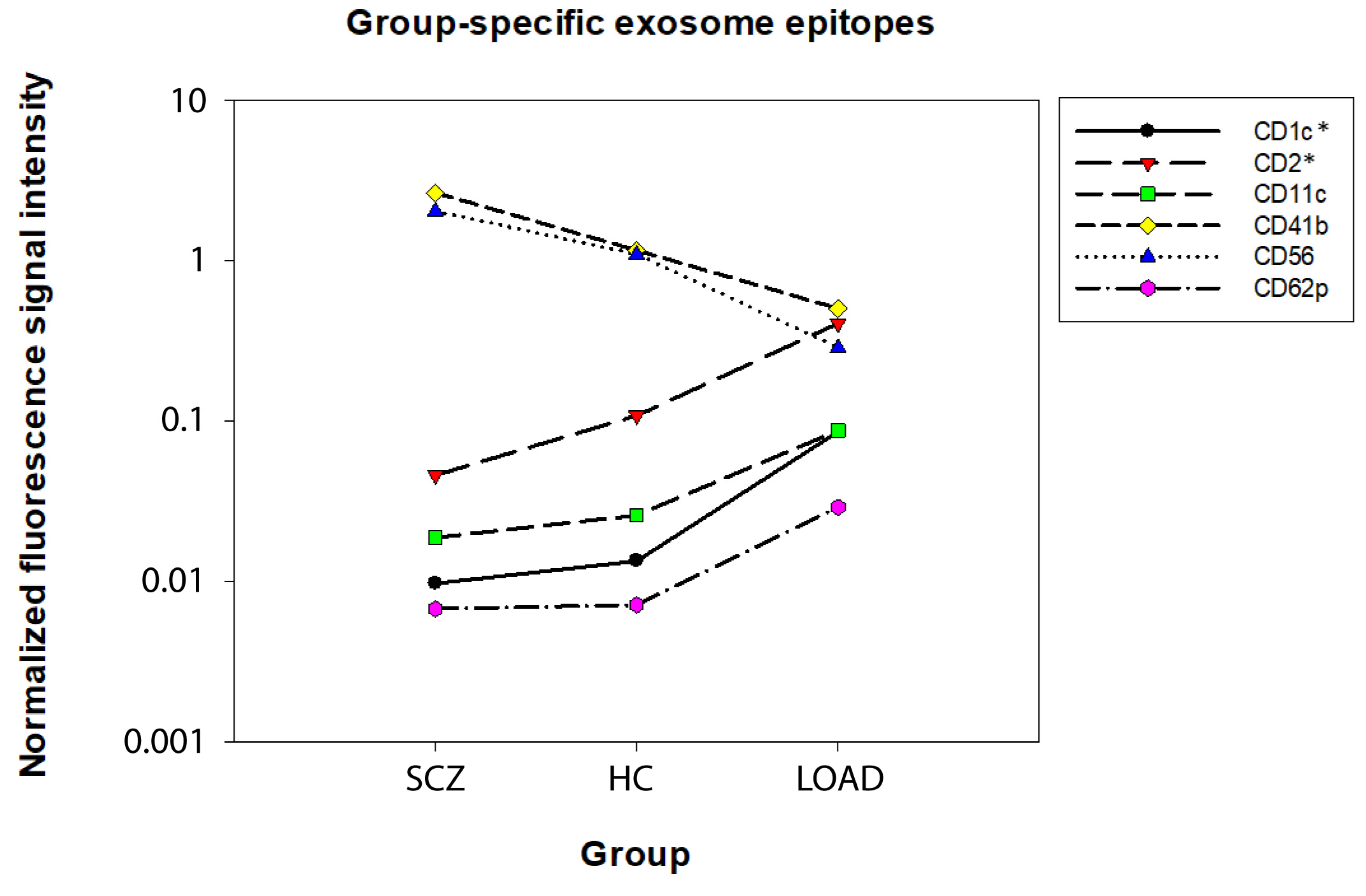
2. Results
3. Discussion
4. Materials and Methods
4.1. Origin and Characterization of iPSCs
4.2. Cultivation of Human iPSCs
4.3. Generation of NSCs, NPCs, and Neurons
4.4. Generation of Astrocytes
4.5. Generation of MGCs
4.6. Generation of BCECs
4.7. Isolation of EVs and Analysis of Exosome Epitopes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Saeedi, S.; Israel, S.; Nagy, C.; Turecki, G. The emerging role of exosomes in mental disorders. Transl. Psychiatry 2019, 9, 122. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Appelt-Menzel, A.; Oerter, S.; Mathew, S.; Haferkamp, U.; Hartmann, C.; Jung, M.; Neuhaus, W.; Pless, O. Human iPSC-Derived Blood-Brain Barrier Models: Valuable Tools for Preclinical Drug Discovery and Development? Curr. Protoc. Stem Cell Biol. 2020, 55, e122. [Google Scholar] [CrossRef]
- Jung, M.; Rujescu, D. Translational medicine: From disease- and patient-specific stem cell research to clinical trials and back again. Eur. Arch. Psychiatry Clin. Neurosci. 2016, 266, 679–680. [Google Scholar] [CrossRef][Green Version]
- Skene, N.G.; Bryois, J.; Bakken, T.E.; Breen, G.; Crowley, J.J.; Gaspar, H.A.; Giusti-Rodriguez, P.; Hodge, R.D.; Miller, J.A.; Muñoz-Manchado, A.B.; et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 2018, 50, 825–833. [Google Scholar] [CrossRef]
- Blumenfeld, J.; Yip, O.; Kim, M.J.; Huang, Y. Cell type-specific roles of APOE4 in Alzheimer disease. Nat. Rev. Neurosci. 2024, 25, 91–110. [Google Scholar] [CrossRef]
- Garcia-Leon, J.A.; Caceres-Palomo, L.; Sanchez-Mejias, E.; Mejias-Ortega, M.; Nuñez-Diaz, C.; Fernandez-Valenzuela, J.J.; Sanchez-Varo, R.; Davila, J.C.; Vitorica, J.; Gutierrez, A. Human Pluripotent Stem Cell-Derived Neural Cells as a Relevant Platform for Drug Screening in Alzheimer’s Disease. Int. J. Mol. Sci. 2020, 21, 6867. [Google Scholar] [CrossRef]
- Hoffmann, A.; Ziller, M.; Spengler, D. Progress in iPSC-Based Modeling of Psychiatric Disorders. Int. J. Mol. Sci. 2019, 20, 4896. [Google Scholar] [CrossRef]
- Li, L.; Shi, Y. When glia meet induced pluripotent stem cells (iPSCs). Mol. Cell. Neurosci. 2020, 109, 103565. [Google Scholar] [CrossRef]
- Wu, Y.-C.; Sonninen, T.-M.; Peltonen, S.; Koistinaho, J.; Lehtonen, Š. Blood-Brain Barrier and Neurodegenerative Diseases-Modeling with iPSC-Derived Brain Cells. Int. J. Mol. Sci. 2021, 22, 7710. [Google Scholar] [CrossRef]
- Pong, S.; Karmacharya, R.; Sofman, M.; Bishop, J.R.; Lizano, P. The Role of Brain Microvascular Endothelial Cell and Blood-Brain Barrier Dysfunction in Schizophrenia. Complex Psychiatry 2020, 6, 30–46. [Google Scholar] [CrossRef]
- Jung, M.; Hartmann, C.; Ehrhardt, T.; Peter, L.-M.; Abid, C.L.; Harwardt, B.; Hirschfeld, J.; Claus, C.; Haferkamp, U.; Pless, O.; et al. Generation of a set of induced pluripotent stem cell lines from two Alzheimer disease patients carrying APOE4 (MLUi007-J; MLUi008-A) and healthy old donors carrying APOE3 (MLUi009-A; MLUi010-B) to study APOE in aging and disease. Stem Cell Res. 2023, 69, 103072. [Google Scholar] [CrossRef]
- Jung, M.; Jung, J.-S.; Pfeifer, J.; Hartmann, C.; Ehrhardt, T.; Abid, C.L.; Kintzel, J.; Puls, A.; Navarrete Santos, A.; Hollemann, T.; et al. Neuronal Stem Cells from Late-Onset Alzheimer Patients Show Altered Regulation of Sirtuin 1 Depending on Apolipoprotein E Indicating Disturbed Stem Cell Plasticity. Mol. Neurobiol. 2023, 61, 1562–1579. [Google Scholar] [CrossRef]
- Haferkamp, U.; Hartmann, C.; Abid, C.L.; Brachner, A.; Höchner, A.; Gerhartl, A.; Harwardt, B.; Leckzik, S.; Leu, J.; Metzger, M.; et al. Human isogenic cells of the neurovascular unit exert transcriptomic cell type-specific effects on a blood-brain barrier in vitro model of late-onset Alzheimer disease. Fluids Barriers CNS 2023, 20, 78. [Google Scholar] [CrossRef]
- Bergau, N.; Maul, S.; Rujescu, D.; Simm, A.; Navarrete Santos, A. Reduction of Glycolysis Intermediate Concentrations in the Cerebrospinal Fluid of Alzheimer’s Disease Patients. Front. Neurosci. 2019, 13, 871. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shah, W.; Holland, P.; Carbonetto, S. Integrins and dystroglycan regulate astrocyte wound healing: The integrin beta1 subunit is necessary for process extension and orienting the microtubular network. Dev. Neurobiol. 2008, 68, 559–574. [Google Scholar] [CrossRef] [PubMed]
- Dzwonek, J.; Wilczynski, G.M. CD44: Molecular interactions, signaling and functions in the nervous system. Front. Cell. Neurosci. 2015, 9, 175. [Google Scholar] [CrossRef]
- Barraud, P.; Stott, S.; Møllgård, K.; Parmar, M.; Björklund, A. In vitro characterization of a human neural progenitor cell coexpressing SSEA4 and CD133. J. Neurosci. Res. 2007, 85, 250–259. [Google Scholar] [CrossRef]
- Ampofo, E.; Schmitt, B.M.; Menger, M.D.; Laschke, M.W. The regulatory mechanisms of NG2/CSPG4 expression. Cell. Mol. Biol. Lett. 2017, 22, 4. [Google Scholar] [CrossRef]
- Chen, J.; Luo, Y.; Hui, H.; Cai, T.; Huang, H.; Yang, F.; Feng, J.; Zhang, J.; Yan, X. CD146 coordinates brain endothelial cell-pericyte communication for blood-brain barrier development. Proc. Natl. Acad. Sci. USA 2017, 114, E7622–E7631. [Google Scholar] [CrossRef]
- Sharygin, D.; Koniaris, L.G.; Wells, C.; Zimmers, T.A.; Hamidi, T. Role of CD14 in human disease. Immunology 2023, 169, 260–270. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.; Deng, X.; Ma, Y.; Huang, S.; Wang, Y. Association of CD14-260 (-159) C/T and Alzheimer’s disease: Systematic review and trial sequential analyses. J. Neural Transm. 2018, 125, 1313–1318. [Google Scholar] [CrossRef]
- Tanaka, T.; Matsuda, T.; Hayes, L.N.; Yang, S.; Rodriguez, K.; Severance, E.G.; Yolken, R.H.; Sawa, A.; Eaton, W.W. Infection and inflammation in schizophrenia and bipolar disorder. Neurosci. Res. 2017, 115, 59–63. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, D.; Zhang, D.; Zuo, X.; Yao, L.; Liu, T.; Ge, X.; He, C.; Zhou, Y.; Shen, Z. Causal role of immune cells in schizophrenia: Mendelian randomization (MR) study. BMC Psychiatry 2023, 23, 590. [Google Scholar] [CrossRef] [PubMed]
- Baur, K.; Abdullah, Y.; Mandl, C.; Hölzl-Wenig, G.; Shi, Y.; Edelkraut, U.; Khatri, P.; Hagenston, A.M.; Irmler, M.; Beckers, J.; et al. A novel stem cell type at the basal side of the subventricular zone maintains adult neurogenesis. EMBO Rep. 2022, 23, e54078. [Google Scholar] [CrossRef] [PubMed]
- Schrode, N.; Ho, S.-M.; Yamamuro, K.; Dobbyn, A.; Huckins, L.; Matos, M.R.; Cheng, E.; Deans, P.J.M.; Flaherty, E.; Barretto, N.; et al. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 2019, 51, 1475–1485. [Google Scholar] [CrossRef]
- Vacchi, E.; Burrello, J.; Di Silvestre, D.; Burrello, A.; Bolis, S.; Mauri, P.; Vassalli, G.; Cereda, C.W.; Farina, C.; Barile, L.; et al. Immune profiling of plasma-derived extracellular vesicles identifies Parkinson disease. Neurol. Neuroimmunol. Neuroinflamm. 2020, 7, e866. [Google Scholar] [CrossRef]
- Kamphuis, W.; Kooijman, L.; Schetters, S.; Orre, M.; Hol, E.M. Transcriptional profiling of CD11c-positive microglia accumulating around amyloid plaques in a mouse model for Alzheimer’s disease. Biochim. Biophys. Acta 2016, 1862, 1847–1860. [Google Scholar] [CrossRef]
- Davies, T.A.; Long, H.J.; Sgro, K.; Rathbun, W.H.; McMenamin, M.E.; Seetoo, K.; Tibbles, H.; Billingslea, A.M.; Fine, R.E.; Fishman, J.B.; et al. Activated Alzheimer disease platelets retain more beta amyloid precursor protein. Neurobiol. Aging 1997, 18, 147–153. [Google Scholar] [CrossRef]
- Jaudon, F.; Thalhammer, A.; Cingolani, L.A. Integrin adhesion in brain assembly: From molecular structure to neuropsychiatric disorders. Eur. J. Neurosci. 2021, 53, 3831–3850. [Google Scholar] [CrossRef]
- Kuwano, Y.; Kamio, Y.; Kawai, T.; Katsuura, S.; Inada, N.; Takaki, A.; Rokutan, K. Autism-associated gene expression in peripheral leucocytes commonly observed between subjects with autism and healthy women having autistic children. PLoS ONE 2011, 6, e24723. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; Keefe, R.S.E.; Lange, L.A.; Lange, E.M.; Stroup, T.S.; Lieberman, J.; Maness, P.F. NCAM1 and neurocognition in schizophrenia. Biol. Psychiatry 2007, 61, 902–910. [Google Scholar] [CrossRef]
- Giegling, I.; Chiesa, A.; Mandelli, L.; Gibiino, S.; Hartmann, A.M.; Möller, H.-J.; Schneider, B.; Schnabel, A.; Maurer, K.; de Ronchi, D.; et al. Influence of neuronal cell adhesion molecule (NCAM1) variants on suicidal behaviour and correlated traits. Psychiatry Res. 2010, 179, 222–225. [Google Scholar] [CrossRef]
- Hidese, S.; Hattori, K.; Sasayama, D.; Tsumagari, T.; Miyakawa, T.; Matsumura, R.; Yokota, Y.; Ishida, I.; Matsuo, J.; Yoshida, S.; et al. Cerebrospinal fluid neuroplasticity-associated protein levels in patients with psychiatric disorders: A multiplex immunoassay study. Transl. Psychiatry 2020, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Kucharska-Mazur, J.; Tarnowski, M.; Dołęgowska, B.; Budkowska, M.; Pędziwiatr, D.; Jabłoński, M.; Pełka-Wysiecka, J.; Kazimierczak, A.; Ratajczak, M.Z.; Samochowiec, J. Novel evidence for enhanced stem cell trafficking in antipsychotic-naïve subjects during their first psychotic episode. J. Psychiatr. Res. 2014, 49, 18–24. [Google Scholar] [CrossRef]
- Giovanazzi, A.; van Herwijnen, M.J.C.; Kleinjan, M.; van der Meulen, G.N.; Wauben, M.H.M. Surface protein profiling of milk and serum extracellular vesicles unveils body fluid-specific signatures. Sci. Rep. 2023, 13, 8758. [Google Scholar] [CrossRef]
- Hernandez, L.; Ward, L.J.; Arefin, S.; Barany, P.; Wennberg, L.; Söderberg, M.; Bruno, S.; Cantaluppi, V.; Stenvinkel, P.; Kublickiene, K. Blood-Brain Barrier Biomarkers before and after Kidney Transplantation. Int. J. Mol. Sci. 2023, 24, 6628. [Google Scholar] [CrossRef]
- Collino, F.; Lopes, J.A.; Tapparo, M.; Tortelote, G.G.; Kasai-Brunswick, T.H.; Lopes, G.M.C.; Almeida, D.B.; Skovronova, R.; Wendt, C.H.C.; Miranda, K.R.d.; et al. Extracellular Vesicles Derived from Induced Pluripotent Stem Cells Promote Renoprotection in Acute Kidney Injury Model. Cells 2020, 9, 453. [Google Scholar] [CrossRef]
- Rujescu, D.; Hartmann, A.M.; Giegling, I.; Konte, B.; Herrling, M.; Himmelein, S.; Strupp, M. Genome-Wide Association Study in Vestibular Neuritis: Involvement of the Host Factor for HSV-1 Replication. Front. Neurol. 2018, 9, 591. [Google Scholar] [CrossRef]
- First, M.B.; Spitzer, R.L.; Gibbon, M.; Williams, J.B.W. Structured Clinical Interview for DSM-IV Axis I Disorders—Patient Edition (SCID-I/P, Version 2.0); Biometrics Research Department: New York, NY, USA, 1998. [Google Scholar]
- Marshall, C.R.; Howrigan, D.P.; Merico, D.; Thiruvahindrapuram, B.; Wu, W.; Greer, D.S.; Antaki, D.; Shetty, A.; Holmans, P.A.; Pinto, D.; et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar] [CrossRef] [PubMed]
- McKhann, G.; Drachman, D.; Folstein, M.; Katzman, R.; Price, D.; Stadlan, E.M. Clinical diagnosis of Alzheimer’s disease: Report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology 1984, 34, 939–944. [Google Scholar] [CrossRef]
- First, M.B.; Gibbon, M.; Spitzer, R.L.; Williams, J.B.W.; Benjamin, L.S. Structured Clinical Interview for DSM-IV Axis II Personality Disorders (SCID-II); American Psychiatric Press: Washington, DC, USA, 1997. [Google Scholar]
- Rice, J.P.; Reich, T.; Bucholz, K.K.; Neuman, R.J.; Fishman, R.; Rochberg, N.; Hesselbrock, V.M.; Nurnberger, J.I.; Schuckit, M.A.; Begleiter, H. Comparison of direct interview and family history diagnoses of alcohol dependence. Alcohol. Clin. Exp. Res. 1995, 19, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, P.; Glatza, M.; Hemmer, K.; Tsytsyura, Y.; Thiel, C.S.; Höing, S.; Moritz, S.; Parga, J.A.; Wagner, L.; Bruder, J.M.; et al. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS ONE 2013, 8, e59252. [Google Scholar] [CrossRef]
- Douvaras, P.; Sun, B.; Wang, M.; Kruglikov, I.; Lallos, G.; Zimmer, M.; Terrenoire, C.; Zhang, B.; Gandy, S.; Schadt, E.; et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Rep. 2017, 8, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293.e9. [Google Scholar] [CrossRef] [PubMed]
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.-H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016, 22, 1358–1367. [Google Scholar] [CrossRef]
- Lippmann, E.S.; Al-Ahmad, A.; Azarin, S.M.; Palecek, S.P.; Shusta, E.V. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 2014, 4, 4160. [Google Scholar] [CrossRef]
- Appelt-Menzel, A.; Cubukova, A.; Günther, K.; Edenhofer, F.; Piontek, J.; Krause, G.; Stüber, T.; Walles, H.; Neuhaus, W.; Metzger, M. Establishment of a Human Blood-Brain Barrier Co-culture Model Mimicking the Neurovascular Unit Using Induced Pluri- and Multipotent Stem Cells. Stem Cell Rep. 2017, 8, 894–906. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio PBC: Boston, MA, USA, 2024; Available online: http://www.rstudio.com/ (accessed on 20 March 2024).
| Group | HC | LOAD | SCZ | |
|---|---|---|---|---|
| Cell lines | MLUi009-A WISCi004-B WAi001-A | MLUi007-J MLUi007-H | MLUi001-M MLUi002-G | |
| Cell type | Astrocytes | 2 | 2 | - |
| BCECs | 4 | 6 | - | |
| MGCs | 2 | 1 | 2 | |
| NSCs | 2 | - | 2 | |
| NPCs | 10 | - | 4 | |
| Neurons | 3 | - | 5 | |
| Total | 23 | 9 | 15 | |
| Cell Type | Single Cell Type vs. Any Other in HC | LOAD vs. HC, SCZ |
|---|---|---|
| Astrocytes | ↑CD29 *, ↑CD44 *, ↑CD49e *, ↑MCSP 1 | ↑CD1c * ↑CD2 * ↑CD11c ↓CD41b 2 ↓CD56 2 ↑CD62p |
| BCECs | n.s. | |
| Microglia | ↑CD44 *, ↑CD146 | |
| Neurons | n.s. | |
| NPCs | n.s. | |
| NSCs | ↑SSEA4 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xavier, G.; Navarrete Santos, A.; Hartmann, C.; Santoro, M.L.; Flegel, N.; Reinsch, J.; Majer, A.; Ehrhardt, T.; Pfeifer, J.; Simm, A.; et al. Comparison of Extracellular Vesicles from Induced Pluripotent Stem Cell-Derived Brain Cells. Int. J. Mol. Sci. 2024, 25, 3575. https://doi.org/10.3390/ijms25073575
Xavier G, Navarrete Santos A, Hartmann C, Santoro ML, Flegel N, Reinsch J, Majer A, Ehrhardt T, Pfeifer J, Simm A, et al. Comparison of Extracellular Vesicles from Induced Pluripotent Stem Cell-Derived Brain Cells. International Journal of Molecular Sciences. 2024; 25(7):3575. https://doi.org/10.3390/ijms25073575
Chicago/Turabian StyleXavier, Gabriela, Alexander Navarrete Santos, Carla Hartmann, Marcos L. Santoro, Nicole Flegel, Jessica Reinsch, Annika Majer, Toni Ehrhardt, Jenny Pfeifer, Andreas Simm, and et al. 2024. "Comparison of Extracellular Vesicles from Induced Pluripotent Stem Cell-Derived Brain Cells" International Journal of Molecular Sciences 25, no. 7: 3575. https://doi.org/10.3390/ijms25073575
APA StyleXavier, G., Navarrete Santos, A., Hartmann, C., Santoro, M. L., Flegel, N., Reinsch, J., Majer, A., Ehrhardt, T., Pfeifer, J., Simm, A., Hollemann, T., Belangero, S. I., Rujescu, D., & Jung, M. (2024). Comparison of Extracellular Vesicles from Induced Pluripotent Stem Cell-Derived Brain Cells. International Journal of Molecular Sciences, 25(7), 3575. https://doi.org/10.3390/ijms25073575