Investigating Vα7.2+/CD161− T Cell and MAIT Cell Profiles Using Flow Cytometry in Healthy Subjects and Subjects with Atopic Dermatitis
Abstract
1. Introduction
2. Results
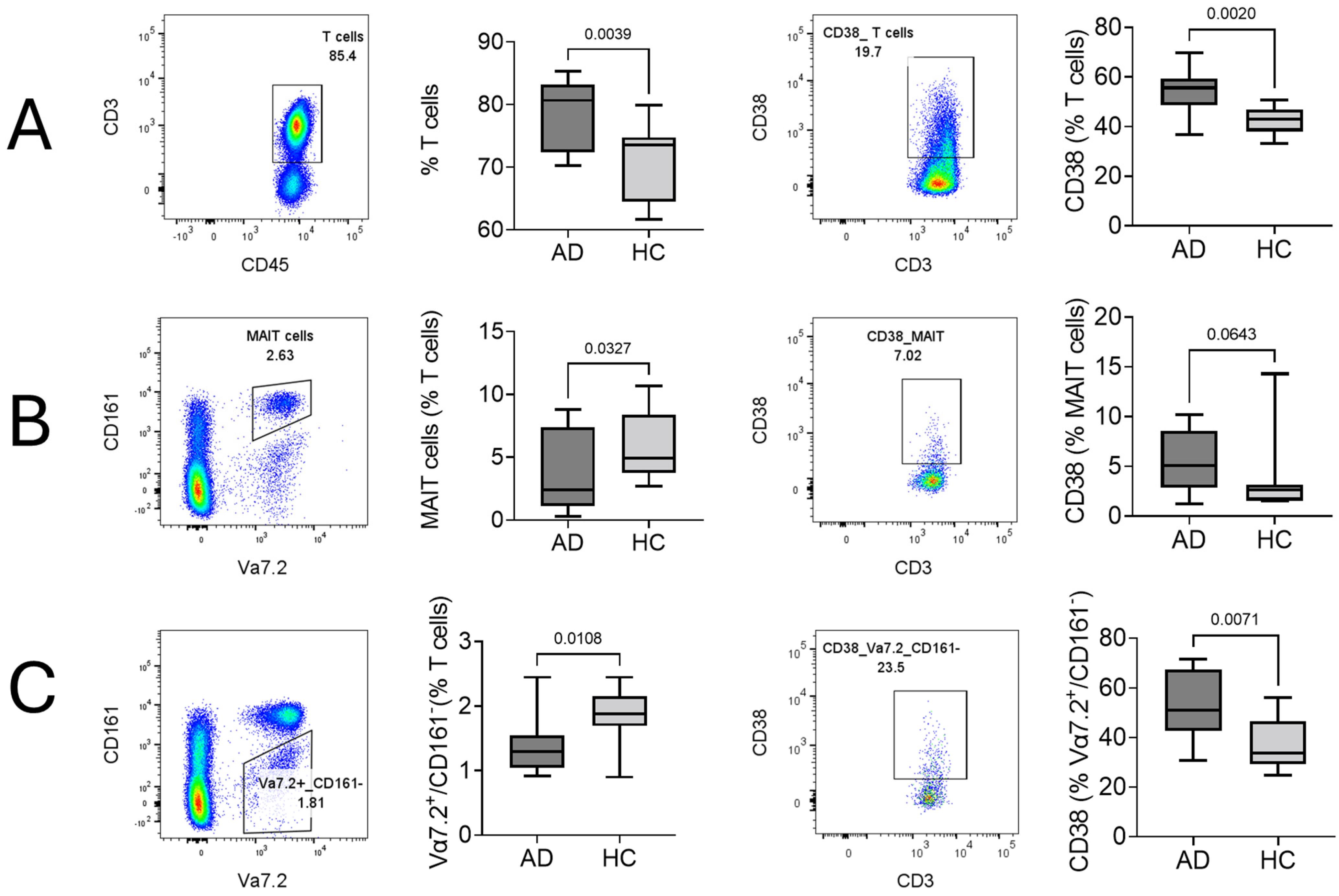
2.1. Cellular Surface Marker Characterization of T Cells, MAIT Cells, and Vα7.2+/CD161− T Cells in AD
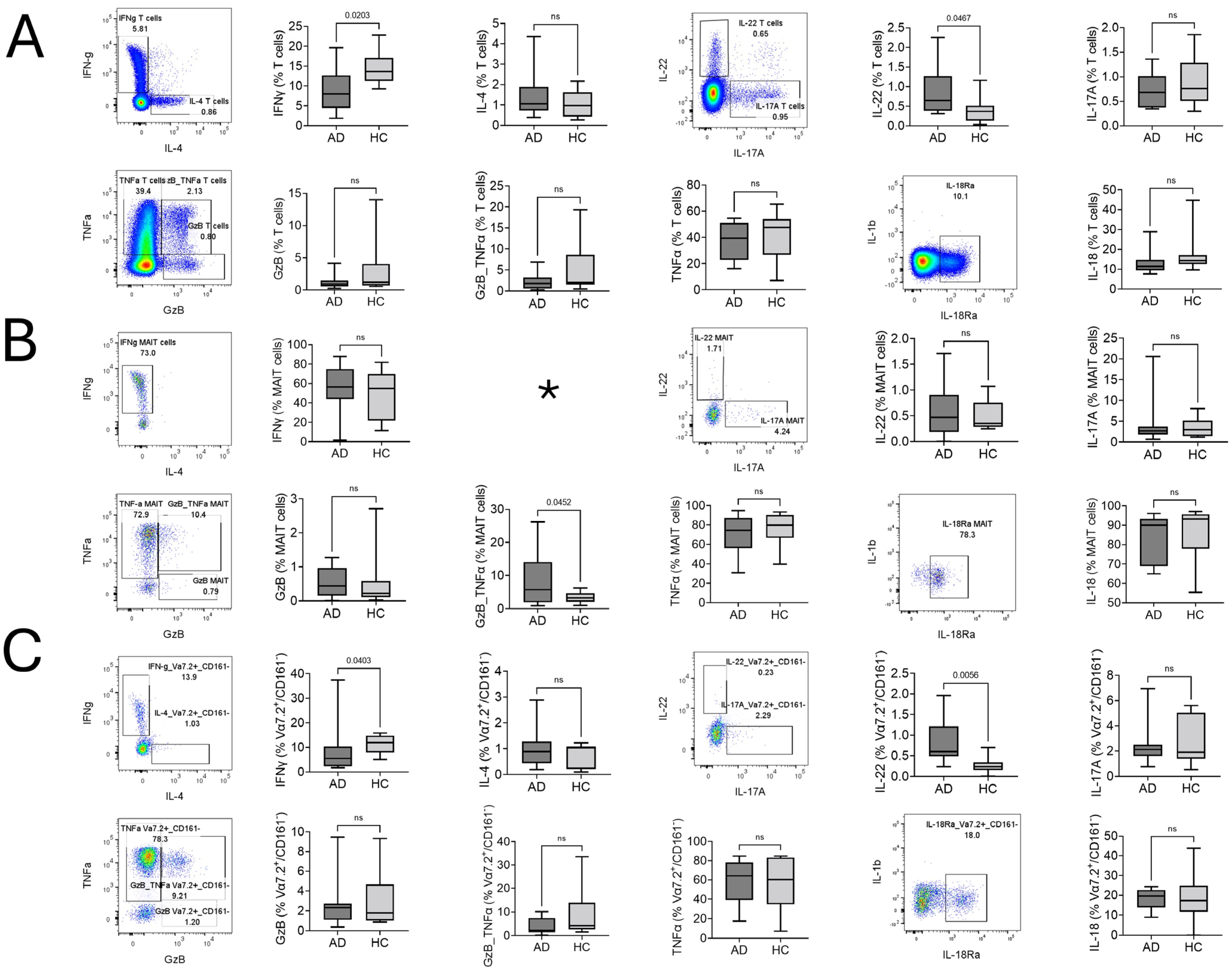
2.2. Type of T Cell, MAIT Cell, and Vα7.2+/CD161− T Cell Response in AD
2.3. Comparison of MAIT and Vα7.2+/CD161− T Cells
2.4. Multidimensional T Cell Analysis
3. Discussion
4. Materials and Methods
4.1. Study Ethics
4.2. Study Participants
4.3. Sample Collection and Complete Blood Counts
4.4. Peripheral Blood Mononuclear Cell (PBMC) Isolation and In Vitro Stimulation
4.5. Flow Cytometry
4.5.1. Surface Staining
4.5.2. Intracellular Staining
4.6. Data Analysis
4.6.1. Sequential Gating of T Cells, MAIT Cells, and Vα7.2+/CD161− T Cells
4.6.2. Multidimensional Data Analysis
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfrey, D.I.; Uldrich, A.P.; Mccluskey, J.; Rossjohn, J.; Moody, D.B. The burgeoning family of unconventional T cells. Nat. Immunol. 2015, 16, 1114–1124. [Google Scholar] [CrossRef]
- Tilloy, F.; Treiner, E.; Park, S.H.; Garcia, C.; Lemonnier, F.; de la Salle, H.; Bendelac, A.; Bonneville, M.; Lantz, O. An invariant T cell receptor alpha chain defines a novel TAP-independent major histocompatibility complex class Ib-restricted alpha/beta T cell subpopulation in mammals. J. Exp. Med. 1999, 189, 1907–1921. [Google Scholar] [CrossRef]
- Reantragoon, R.; Corbett, A.J.; Sakala, I.G.; Gherardin, N.A.; Furness, J.B.; Chen, Z.; Eckle, S.B.G.; Uldrich, A.P.; Birkinshaw, R.W.; Patel, O.; et al. Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells. J. Exp. Med. 2013, 210, 2305–2320. [Google Scholar] [CrossRef]
- Lanier, L.L.; Chang, C.; Phillips, J.H. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J. Immunol. 1994, 153, 2417–2428. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Dejbakhsh-Jones, S.; Strober, S. Expression of CD161 (NKR-P1A) Defines Subsets of Human CD4 and CD8 T Cells with Different Functional Activities1. J. Immunol. 2006, 176, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Wyrożemski, Ł.; Qiao, S.-W. Immunobiology and conflicting roles of the human CD161 receptor in T cells. Scand. J. Immunol. 2021, 94, e13090. [Google Scholar] [CrossRef] [PubMed]
- Park, D.; Kim, H.G.; Kim, M.; Park, T.; Ha, H.-H.; Lee, D.H.; Park, K.-S.; Park, S.J.; Lim, H.J.; Lee, C.H. Differences in the molecular signatures of mucosal-associated invariant T cells and conventional T cells. Sci. Rep. 2019, 9, 7094. [Google Scholar] [CrossRef]
- López-Sagaseta, J.; Dulberger, C.L.; Crooks, J.E.; Parks, C.D.; Luoma, A.M.; McFedries, A.; Van Rhijn, I.; Saghatelian, A.; Adams, E.J. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc. Natl. Acad. Sci. USA 2013, 110, E1771–E1778. [Google Scholar] [CrossRef] [PubMed]
- Dusseaux, M.; Martin, E.; Serriari, N.; Péguillet, I.; Premel, V.; Louis, D.; Milder, M.; Le Bourhis, L.; Soudais, C.; Treiner, E.; et al. Human MAIT cells are xenobiotic-resistant, tissue-targeted, CD161 hi IL-17-secreting T cells. Blood 2011, 117, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Szaraz-Szeles, M.; Mezei, Z.; Barath, S.; Hevessy, Z. Age-dependent frequency of unconventional T cells in a healthy adult Caucasian population: A combinational study of invariant natural killer T cells, γδ T cells, and mucosa-associated invariant T cells. GeroScience 2022, 44, 2047–2060. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Szaraz-Szeles, M.; Mezei, Z.; Barath, S.; Hevessy, Z. Gender-dependent frequency of unconventional T cells in a healthy adult Caucasian population: A combinational study of invariant NKT cells, γδ T cells, and mucosa-associated invariant T cells. J. Leukoc. Biol. 2022, 112, 1155–1165. [Google Scholar] [CrossRef]
- Barathan, M.; Mohamed, R.; Vadivelu, J.; Chang, L.Y.; Saeidi, A.; Yong, Y.K.; Ravishankar Ram, M.; Gopal, K.; Velu, V.; Larsson, M.; et al. Peripheral loss of CD8(+) CD161(++) TCRVα7·2(+) mucosal-associated invariant T cells in chronic hepatitis C virus-infected patients. Eur. J. Clin. Investig. 2016, 46, 170–180. [Google Scholar] [CrossRef]
- Leeansyah, E.; Ganesh, A.; Quigley, M.F.; Sönnerborg, A.; Andersson, J.; Hunt, P.W.; Somsouk, M.; Deeks, S.G.; Martin, J.N.; Moll, M.; et al. Activation, exhaustion, and persistent decline of the antimicrobial MR1-restricted MAIT-cell population in chronic HIV-1 infection. Blood 2013, 121, 1124–1135. [Google Scholar] [CrossRef]
- Gallagher, C.; Mahon, J.M.; O’Neill, C.; Cassidy, F.C.; Dunbar, H.; De Barra, C.; Cadden, C.; Pisarska, M.M.; Wood, N.A.W.; Masterson, J.C.; et al. Mucosal-Associated Invariant T Cells Are Altered in Patients with Hidradenitis Suppurativa and Contribute to the Inflammatory Milieu. J. Investig. Dermatol. 2023, 143, 1094–1097.e2. [Google Scholar] [CrossRef]
- Teunissen, M.B.M.; Yeremenko, N.G.; Baeten, D.L.P.; Chielie, S.; Spuls, P.I.; de Rie, M.A.; Lantz, O.; Res, P.C.M. The IL-17A-Producing CD8+ T-Cell Population in Psoriatic Lesional Skin Comprises Mucosa-Associated Invariant T Cells and Conventional T Cells. J. Investig. Dermatol. 2014, 134, 2898–2907. [Google Scholar] [CrossRef]
- Boothe, D.; Patel, N.; Feldman, S.R. Management of atopic dermatitis. Adherence in atopic Dermatitis. Introduction. Adv. Exp. Med. Biol. 2017, 1027, 139–159. [Google Scholar] [CrossRef]
- Schuler, C.F.; Billi, A.C.; Maverakis, E.; Tsoi, L.C.; Gudjonsson, J.E. Novel insights into atopic dermatitis. J. Allergy Clin. Immunol. 2023, 151, 1145–1154. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, S.; Beck, L.A.; Bieber, T.; Kabashima, K.; Irvine, A.D. Atopic dermatitis. Nat. Rev. Dis. Prim. 2018, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Humeau, M.; Boniface, K.; Bodet, C. Cytokine-Mediated Crosstalk Between Keratinocytes and T Cells in Atopic Dermatitis. Front. Immunol. 2022, 13, 801579. [Google Scholar] [CrossRef] [PubMed]
- Bantz, S.K.; Zhu, Z.; Zheng, T. The Atopic March: Progression from Atopic Dermatitis to Allergic Rhinitis and Asthma. J. Clin. Cell. Immunol. 2014, 5, 202. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, K.; Woods, K.; Pellefigues, C.; Cait, A.; O’Sullivan, D.; Gell, K.; Marshall, A.J.; Anderson, R.J.; Li, Y.; Schmidt, A.; et al. MR1-dependent immune surveillance of the skin contributes to pathogenesis and is a photobiological target of UV light therapy in a mouse model of atopic dermatitis. Allergy Eur. J. Allergy Clin. Immunol. 2021, 76, 3155–3170. [Google Scholar] [CrossRef] [PubMed]
- Kjer-Nielsen, L.; Patel, O.; Corbett, A.J.; Le Nours, J.; Meehan, B.; Liu, L.; Bhati, M.; Chen, Z.; Kostenko, L.; Reantragoon, R.; et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012, 491, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Patrycja, O.; Klaudia, S.; Joanna, E.; Małgorzata, U.; Agata, W.-P.; Wioletta, B.-R.; Dariusz, Ś.; Joanna, N. Staphylococcus aureus from Atopic Dermatitis Patients: Its Genetic Structure and Susceptibility to Phototreatment. Microbiol. Spectr. 2023, 11, e04598-22. [Google Scholar] [CrossRef]
- Le Bourhis, L.; Martin, E.; Péguillet, I.; Guihot, A.; Froux, N.; Coré, M.; Lévy, E.; Dusseaux, M.; Meyssonnier, V.; Premel, V.; et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 2010, 11, 701–708. [Google Scholar] [CrossRef]
- Sandberg, J.K.; Norrby-Teglund, A.; Leeansyah, E. Bacterial deception of MAIT cells in a cloud of superantigen and cytokines. PLoS Biol. 2017, 15, e2003167. [Google Scholar] [CrossRef] [PubMed]
- du Halgouet, A.; Darbois, A.; Alkobtawi, M.; Mestdagh, M.; Alphonse, A.; Premel, V.; Yvorra, T.; Colombeau, L.; Rodriguez, R.; Zaiss, D.; et al. Role of MR1-driven signals and amphiregulin on the recruitment and repair function of MAIT cells during skin wound healing. Immunity 2023, 56, 78–92.e6. [Google Scholar] [CrossRef] [PubMed]
- Malavasi, F.; Deaglio, S.; Funaro, A.; Ferrero, E.; Horenstein, A.L.; Ortolan, E.; Vaisitti, T.; Aydin, S. Evolution and Function of the ADP Ribosyl Cyclase/CD38 Gene Family in Physiology and Pathology. Physiol. Rev. 2008, 88, 841–886. [Google Scholar] [CrossRef] [PubMed]
- Seneviratne, S.L.; Black, A.P.; Jones, L.; Bailey, A.S.; Ogg, G.S. The role of skin-homing T cells in extrinsic atopic dermatitis. QJM An Int. J. Med. 2007, 100, 19–27. [Google Scholar] [CrossRef][Green Version]
- Orfali, R.L.; Yoshikawa, F.S.Y.; Oliveira, L.M.D.S.; Pereira, N.Z.; de Lima, J.F.; Ramos, Y.Á.L.; Duarte, A.J.D.S.; Sato, M.N.; Aoki, V. Staphylococcal enterotoxins modulate the effector CD4+ T cell response by reshaping the gene expression profile in adults with atopic dermatitis. Sci. Rep. 2019, 9, 13082. [Google Scholar] [CrossRef]
- Nograles, K.E.; Zaba, L.C.; Shemer, A.; Fuentes-Duculan, J.; Cardinale, I.; Kikuchi, T.; Ramon, M.; Bergman, R.; Krueger, J.G.; Guttman-Yassky, E. IL-22-producing “T22” T cells account for upregulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J. Allergy Clin. Immunol. 2009, 123, 1244–1252.e2. [Google Scholar] [CrossRef]
- Pan, Y.; Du, D.; Wang, L.; Wang, X.; He, G.; Jiang, X. The Role of T Helper 22 Cells in Dermatological Disorders. Front. Immunol. 2022, 13, 911546. [Google Scholar] [CrossRef]
- Gittler, J.K.; Shemer, A.; Suárez-Fariñas, M.; Fuentes-Duculan, J.; Gulewicz, K.J.; Wang, C.Q.F.; Mitsui, H.; Cardinale, I.; de Guzman Strong, C.; Krueger, J.G.; et al. Progressive activation of T(H)2/T(H)22 cytokines and selective epidermal proteins characterizes acute and chronic atopic dermatitis. J. Allergy Clin. Immunol. 2012, 130, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, A.; Leeansyah, E.; Introini, A.; Paquin-Proulx, D.; Hasselrot, K.; Andersson, E.; Broliden, K.; Sandberg, J.K.; Tjernlund, A. MAIT cells reside in the female genital mucosa and are biased towards IL-17 and IL-22 production in response to bacterial stimulation. Mucosal Immunol. 2017, 10, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, Q.; Zhang, L.-X. Granzyme B: A novel therapeutic target for treatment of atopic dermatitis. Indian J. Dermatol. Venereol. Leprol. 2023, 89, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.T.; Zeglinski, M.R.; Richardson, K.C.; Santacruz, S.; Hiroyasu, S.; Wang, C.; Zhao, H.; Shen, Y.; Sehmi, R.; Lima, H.; et al. Granzyme B Contributes to Barrier Dysfunction in Oxazolone-Induced Skin Inflammation through E-Cadherin and FLG Cleavage. J. Investig. Dermatol. 2021, 141, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Lopatnikova, J.A.; Alshevskaya, A.A.; Krugleeva, O.L.; Nepomnyschih, V.M.; Gladkikh, V.S.; Lukinov, V.L.; Karaulov, A.V.; Sennikov, S.V. Expression of TNFα Receptors on Immunocompetent Cells Is Increased in Atopic Dermatitis. Int. Arch. Allergy Immunol. 2017, 174, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Held, K.; Bhonsle-Deeng, L.; Siewert, K.; Sato, W.; Beltrán, E.; Schmidt, S.; Rühl, G.; Ng, J.K.M.; Engerer, P.; Moser, M.; et al. αβ T-cell receptors from multiple sclerosis brain lesions show MAIT cell–related features. Neurol.—Neuroimmunol. Neuroinflamm. 2015, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Font, L.; Mayer, J.U.; Old, S.; Hermans, I.F.; Irish, J.; Price, K.M. High-Dimensional Data Analysis Algorithms Yield Comparable Results for Mass Cytometry and Spectral Flow Cytometry Data. Cytom. Part A J. Int. Soc. Anal. Cytol. 2020, 97, 824–831. [Google Scholar] [CrossRef]
- Rahim, A.; Meskas, J.; Drissler, S.; Yue, A.; Lorenc, A.; Laing, A.; Saran, N.; White, J.; Abeler-Dörner, L.; Hayday, A.; et al. High throughput automated analysis of big flow cytometry data. Methods 2018, 134–135, 164–176. [Google Scholar] [CrossRef]
- Liu, P.; Liu, S.; Fang, Y.; Xue, X.; Zou, J.; Tseng, G.; Konnikova, L. Recent Advances in Computer-Assisted Algorithms for Cell Subtype Identification of Cytometry Data. Front. Cell Dev. Biol. 2020, 8, 234. [Google Scholar] [CrossRef]
- Baumgaertner, P.; Sankar, M.; Herrera, F.; Benedetti, F.; Barras, D.; Thierry, A.-C.; Dangaj, D.; Kandalaft, L.E.; Coukos, G.; Xenarios, I.; et al. Unsupervised Analysis of Flow Cytometry Data in a Clinical Setting Captures Cell Diversity and Allows Population Discovery. Front. Immunol. 2021, 12, 633910. [Google Scholar] [CrossRef]
- Fergusson, J.R.; Fleming, V.M.; Klenerman, P. CD161-expressing human T cells. Front. Immunol. 2011, 2, 36. [Google Scholar] [CrossRef]
- Roesner, L.M.; Floess, S.; Witte, T.; Olek, S.; Huehn, J.; Werfel, T. Foxp3(+) regulatory T cells are expanded in severe atopic dermatitis patients. Allergy 2015, 70, 1656–1660. [Google Scholar] [CrossRef] [PubMed]
- Samochocki, Z.; Alifier, M.; Bodera, P.; Jeziorkowska, R.; Rosiak, E.; Jurkiewicz, B.; Glińska, O.; Gliński, W.; Stankiewicz, W. T-regulatory cells in severe atopic dermatitis: Alterations related to cytokines and other lymphocyte subpopulations. Arch. Dermatol. Res. 2012, 304, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Cassius, C.; Branchtein, M.; Battistella, M.; Amode, R.; Lepelletier, C.; Jachiet, M.; de Masson, A.; Frumholtz, L.; Chasset, F.; Amoura, Z.; et al. Persistent deficiency of mucosal-associated invariant T cells during dermatomyositis. Rheumatology 2020, 59, 2282–2286. [Google Scholar] [CrossRef] [PubMed]
- Hijnen, D.J. Shifting paradigms in the immunology of atopic dermatitis. J. Allergy Clin. Immunol. 2020, 145, 1360–1362. [Google Scholar] [CrossRef] [PubMed]
- McInnes, L.; Healy, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Van Gassen, S.; Callebaut, B.; Van Helden, M.J.; Lambrecht, B.N.; Demeester, P.; Dhaene, T.; Saeys, Y. FlowSOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytom. Part A J. Int. Soc. Anal. Cytol. 2015, 87, 636–645. [Google Scholar] [CrossRef]


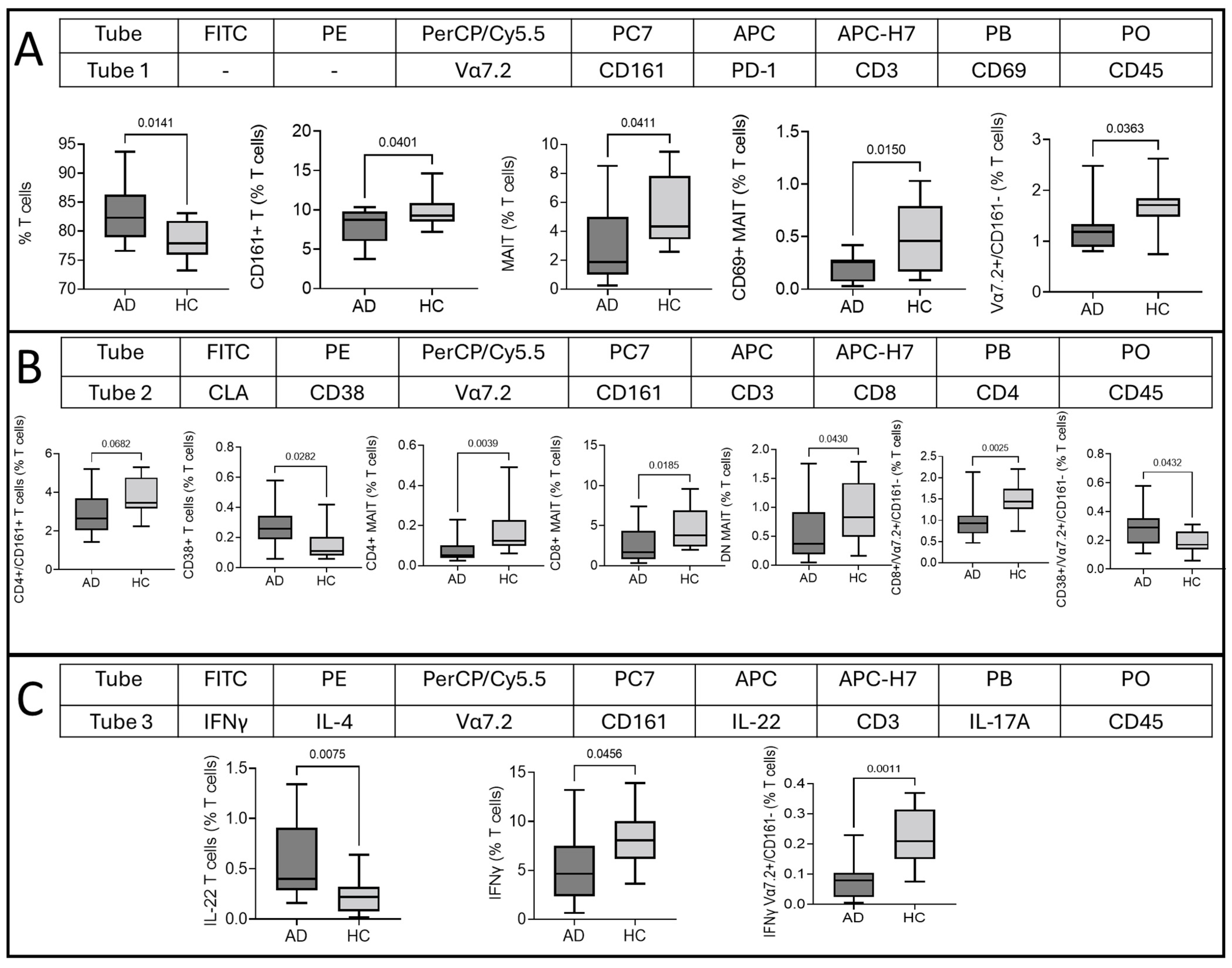
| Tube | FITC | PE | PerCP/Cy5.5 | PC7 | APC | APC-H7 | PB | PO |
|---|---|---|---|---|---|---|---|---|
| Tube 1 | - | - | Vα7.2 | CD161 | PD-1 | CD3 | CD69 | CD45 |
| Tube 2 | CLA | CD38 | Vα7.2 | CD161 | CD3 | CD8 | CD4 | CD45 |
| Tube 3 | IFN-γ | IL-4 | Vα7.2 | CD161 | IL-22 | CD3 | IL-17A | CD45 |
| Tube 4 | IL-1β | TNF-α | Vα7.2 | CD161 | IL-18Rα | CD3 | GzB | CD45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.; Gaspar, K.; Szegedi, A.; Sajtos, L.; Barath, S.; Hevessy, Z. Investigating Vα7.2+/CD161− T Cell and MAIT Cell Profiles Using Flow Cytometry in Healthy Subjects and Subjects with Atopic Dermatitis. Int. J. Mol. Sci. 2024, 25, 3486. https://doi.org/10.3390/ijms25063486
Singh P, Gaspar K, Szegedi A, Sajtos L, Barath S, Hevessy Z. Investigating Vα7.2+/CD161− T Cell and MAIT Cell Profiles Using Flow Cytometry in Healthy Subjects and Subjects with Atopic Dermatitis. International Journal of Molecular Sciences. 2024; 25(6):3486. https://doi.org/10.3390/ijms25063486
Chicago/Turabian StyleSingh, Parvind, Krisztian Gaspar, Andrea Szegedi, Laszlo Sajtos, Sandor Barath, and Zsuzsanna Hevessy. 2024. "Investigating Vα7.2+/CD161− T Cell and MAIT Cell Profiles Using Flow Cytometry in Healthy Subjects and Subjects with Atopic Dermatitis" International Journal of Molecular Sciences 25, no. 6: 3486. https://doi.org/10.3390/ijms25063486
APA StyleSingh, P., Gaspar, K., Szegedi, A., Sajtos, L., Barath, S., & Hevessy, Z. (2024). Investigating Vα7.2+/CD161− T Cell and MAIT Cell Profiles Using Flow Cytometry in Healthy Subjects and Subjects with Atopic Dermatitis. International Journal of Molecular Sciences, 25(6), 3486. https://doi.org/10.3390/ijms25063486






