Overexpression of REST Represses the Epithelial–Mesenchymal Transition Process and Decreases the Aggressiveness of Prostate Cancer Cells
Abstract
1. Introduction
1.1. Prostate Cancer
1.2. Castration-Resistant Prostate Cancer (CRPC)
1.3. The Neuroendocrine PCa Subtype (NEPC)
1.4. The Androgen Receptor (AR)
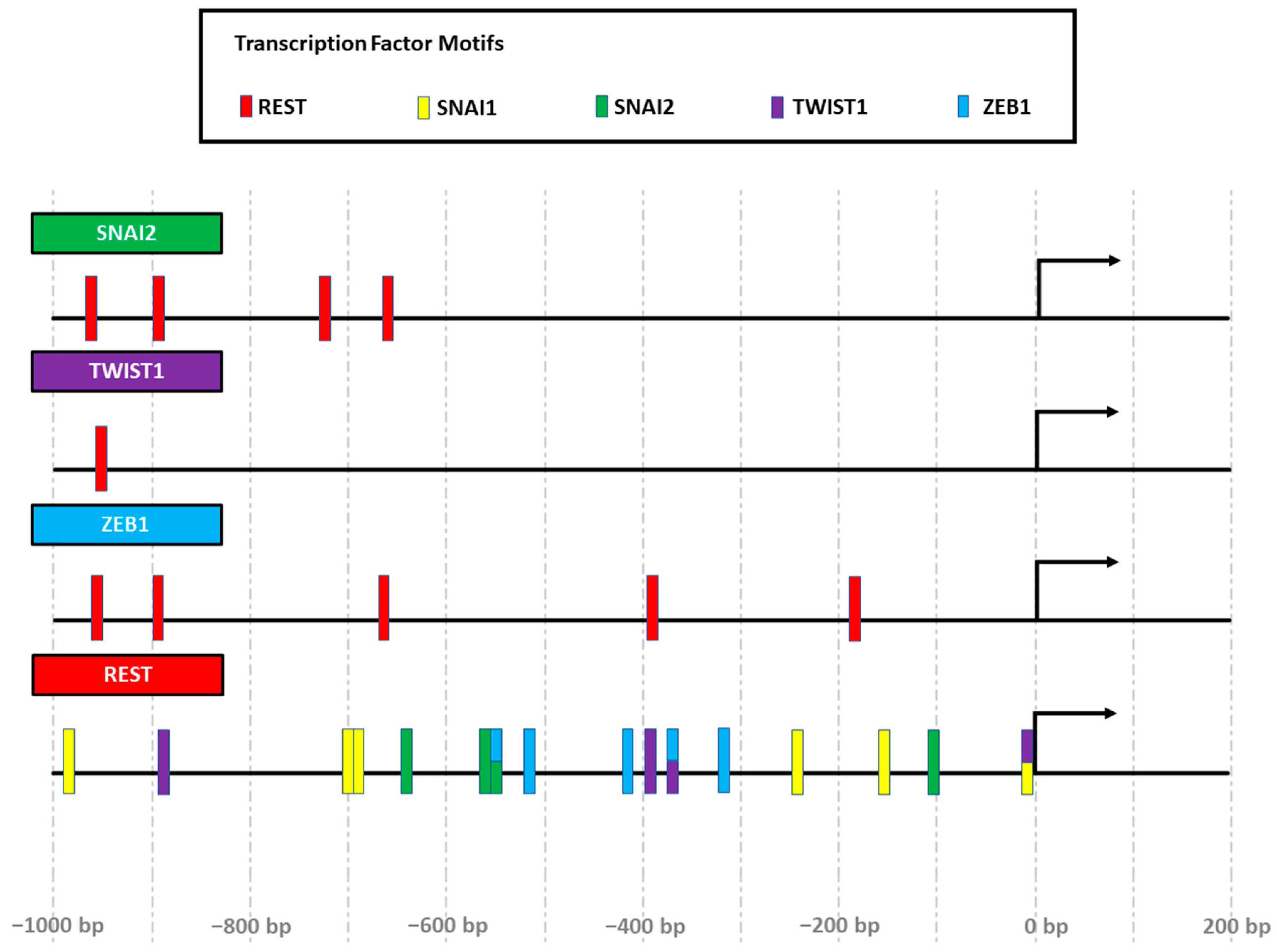
1.5. The RE-1 Silencing Transcription Factor (REST)
1.6. The Epithelial–Mesenchymal Transition (EMT)
2. Results
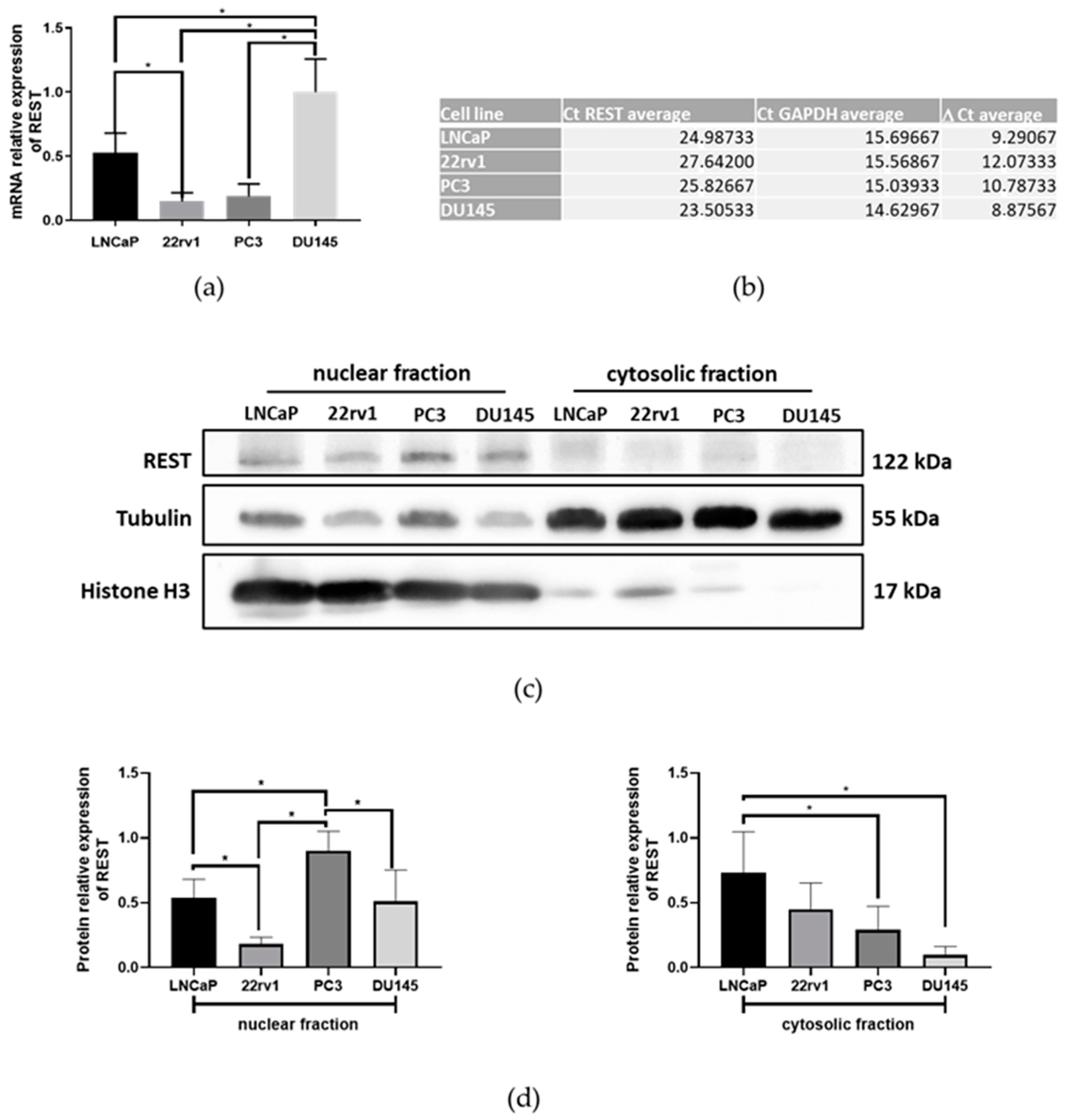
2.1. The Expression of REST in Prostate Cancer Cell Lines
2.2. The Overexpression of REST Produces Modifications in the Expression Levels of EMT Regulators and Epithelial–Mesenchymal Markers
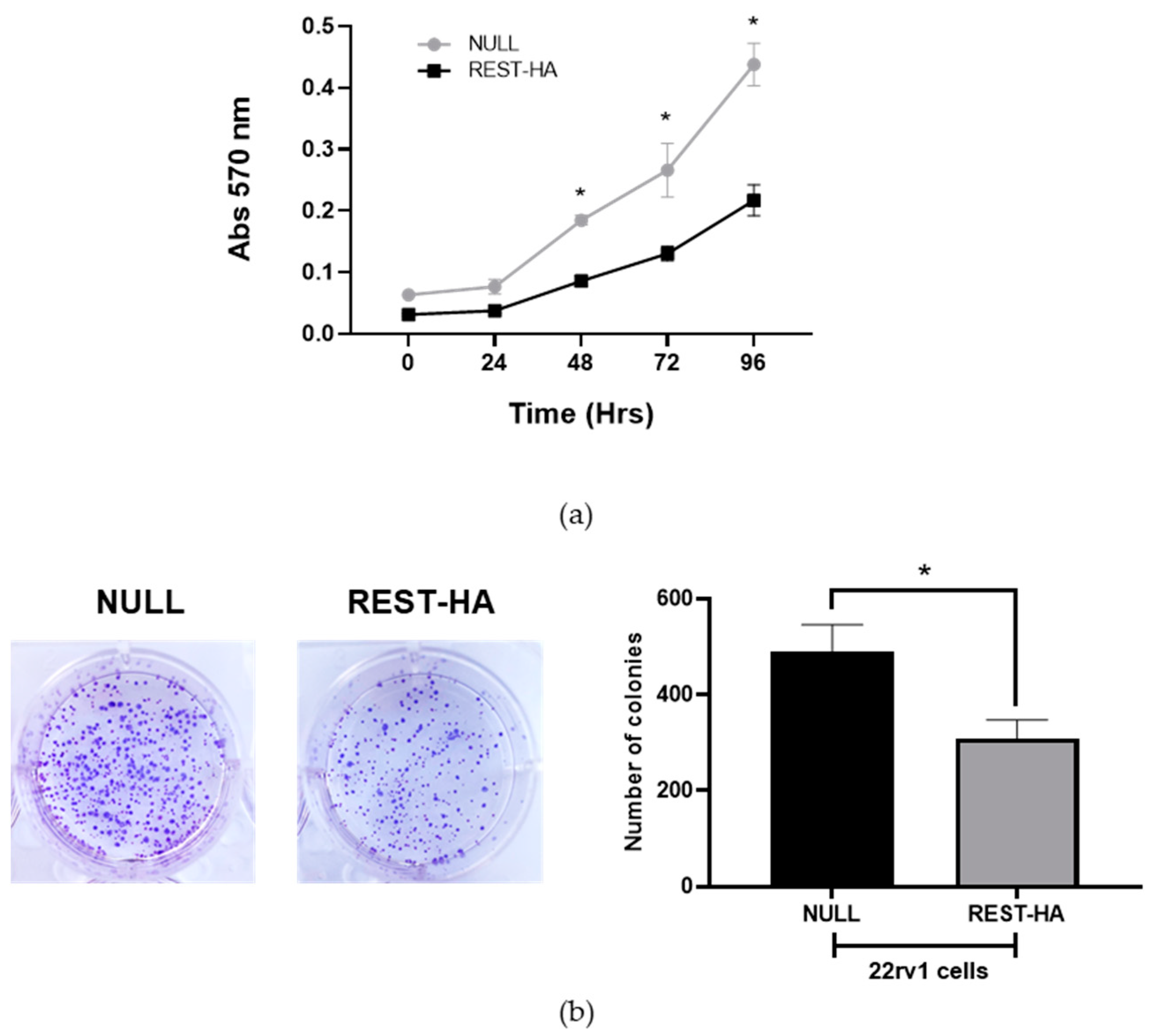
2.3. The Effect of REST Overexpression on the Proliferation and Clonogenic Capacity of 22rv1 Cells
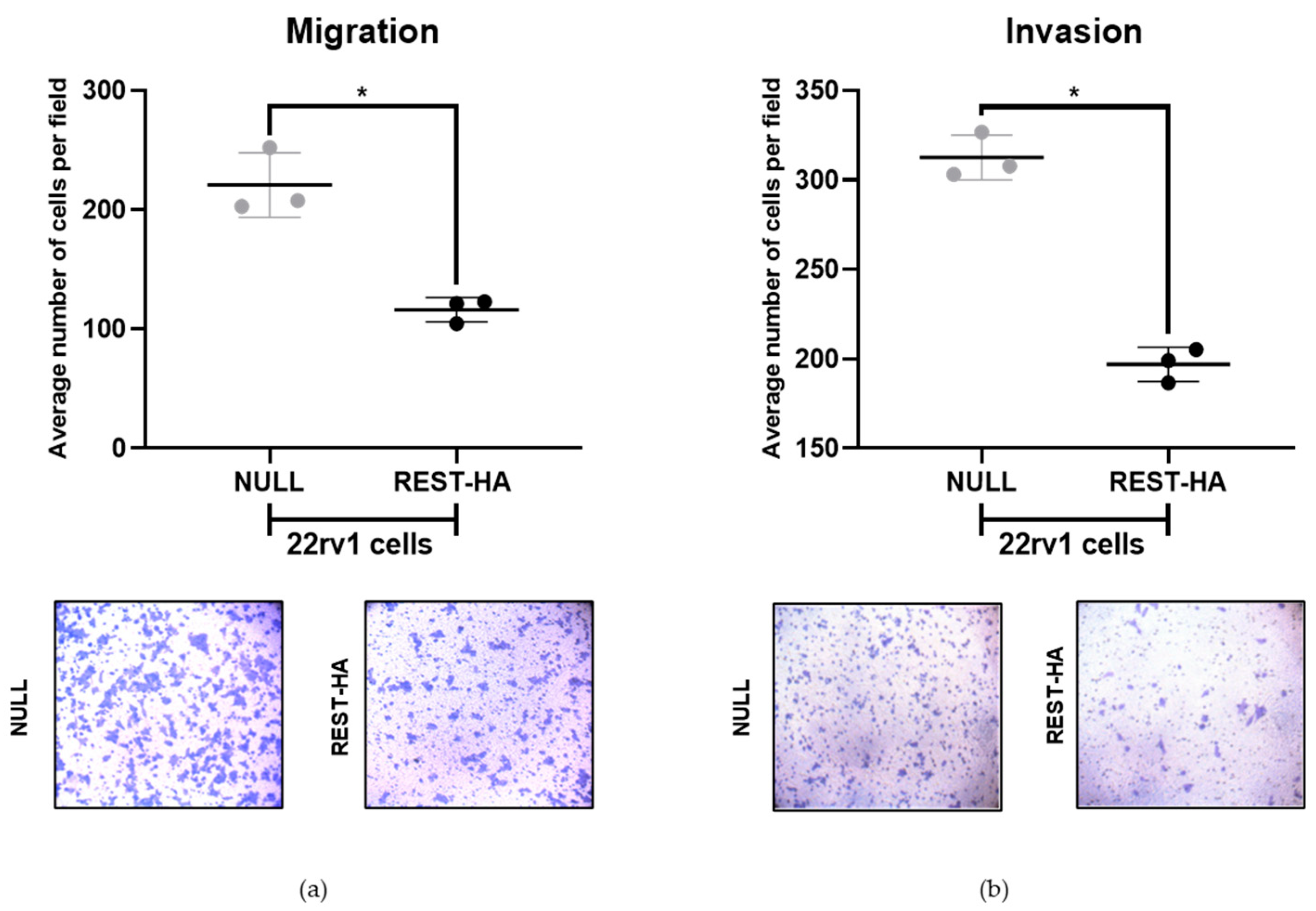
2.4. The Overexpression of REST in 22rv1 Cells Decreases Migration and Invasion
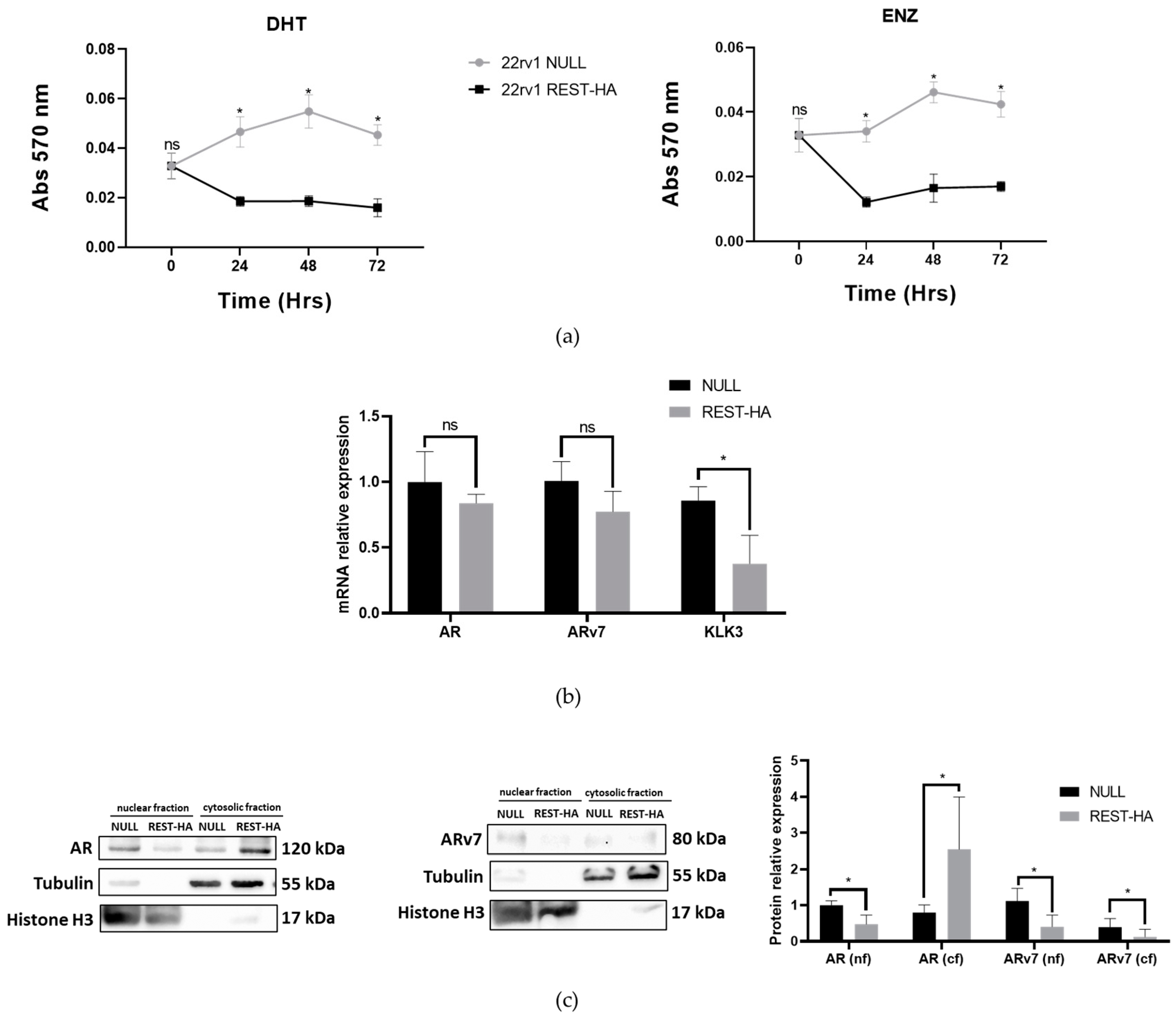
2.5. The Overexpression of REST in 22rv1 Cells Decreases Their Viability for Enzalutamide and Alters the Expression of AR and ARv7
2.6. Evaluation of EMT-Inducing Genes and Epithelial–Mesenchymal Markers
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Lentiviral Transduction
4.3. RNA Extraction and RT-qPCR
4.4. Protein Extraction and Western Blot
4.5. Clonogenicity Assays
4.6. Transmigration and Invasion Assays
4.7. Evaluation of the Viability of Enzalutamide
4.8. Statistical Analysis
5. Conclusions
6. Limitations of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Laversanne, M.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2024. Available online: https://gco.iarc.who.int/today (accessed on 21 February 2024).
- Barry, M.J.; Simmons, L.H. Prevention of Prostate Cancer Morbidity and Mortality: Primary Prevention and Early Detection. Med. Clin. N. Am. 2017, 101, 787–806. [Google Scholar] [CrossRef] [PubMed]
- Borkenhagen, J.F.; Eastwood, D.; Kilari, D.; See, W.A.; Van Wickle, J.D.; Lawton, C.A.; Hall, W.A. Digital Rectal Examination Remains a Key Prognostic Tool for Prostate Cancer: A National Cancer Database Review. J. Natl. Compr. Cancer Netw. 2019, 17, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Jiang, X.; Liang, X.; Jiang, G. Molecular and cellular mechanisms of castration resistant prostate cancer (Review). Oncol. Lett. 2018, 15, 6063–6076. [Google Scholar] [CrossRef] [PubMed]
- Copeland, B.T.; Pal, S.K.; Bolton, E.C.; Jones, J.O. The androgen receptor malignancy shift in prostate cancer. Prostate 2018, 78, 521–531. [Google Scholar] [CrossRef]
- Soundararajan, R.; Paranjape, A.N.; Maity, S.; Aparicio, A.; Mani, S.A. EMT, stemness and tumor plasticity in aggressive variant neuroendocrine prostate cancers. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 229–238. [Google Scholar] [CrossRef]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.S.K.; Varambally, S.; et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef]
- Yamada, Y.; Beltran, H. Clinical and Biological Features of Neuroendocrine Prostate Cancer. Curr. Oncol. Rep. 2021, 23, 15. [Google Scholar] [CrossRef]
- Bluemn, E.G.; Coleman, I.M.; Lucas, J.M.; Coleman, R.T.; Hernandez-Lopez, S.; Tharakan, R.; Bianchi-Frias, D.; Dumpit, R.F.; Kaipainen, A.; Corella, A.N.; et al. Androgen Receptor Pathway-Independent Prostate Cancer Is Sustained through FGF Signaling. Cancer Cell 2017, 32, 474–489.e6. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, R.; Huang, J.; Alumkal, J.J.; Zhang, L.; Feng, F.Y.; Thomas, G.V.; Weinstein, A.S.; Friedl, V.; Zhang, C.; Witte, O.N.; et al. Clinical and Genomic Characterization of Treatment-Emergent Small-Cell Neuroendocrine Prostate Cancer: A Multi-institutional Prospective Study. J. Clin. Oncol. 2018, 36, 2492–2503. [Google Scholar] [CrossRef] [PubMed]
- Vashchenko, N.; Abrahamsson, P.-A. Neuroendocrine differentiation in prostate cancer: Implications for new treatment modalities. Eur. Urol. 2005, 47, 147–155. [Google Scholar] [CrossRef]
- Hirano, D.; Okada, Y.; Minei, S.; Takimoto, Y.; Nemoto, N. Neuroendocrine differentiation in hormone refractory prostate cancer following androgen deprivation therapy. Eur. Urol. 2004, 45, 586–592. [Google Scholar] [CrossRef]
- Sasaki, T.; Komiya, A.; Suzuki, H.; Shimbo, M.; Ueda, T.; Akakura, K.; Ichikawa, T. Changes in chromogranin a serum levels during endocrine therapy in metastatic prostate cancer patients. Eur. Urol. 2005, 48, 224–230. [Google Scholar] [CrossRef]
- Tan, M.H.E.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E.-L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lu, J.; Ye, Z.; Hao, S.; Wang, L.; Kohli, M.; Tindall, D.J.; Li, B.; Zhu, R.; Wang, L.; et al. Androgen receptor splice variants bind to constitutively open chromatin and promote abiraterone-resistant growth of prostate cancer. Nucleic Acids Res. 2018, 46, 1895–1911. [Google Scholar] [CrossRef]
- Jernberg, E.; Bergh, A.; Wikström, P. Clinical relevance of androgen receptor alterations in prostate cancer. Endocr. Connect. 2017, 6, R146–R161. [Google Scholar] [CrossRef]
- Thiel, G.; Lietz, M.; Cramer, M. Biological activity and modular structure of RE-1-silencing transcription Factor (REST), a repressor of neuronal genes. J. Biol. Chem. 1998, 273, 26891–26899. [Google Scholar] [CrossRef]
- Thiel, G.; Ekici, M.; Rössler, O.G. RE-1 silencing transcription factor (REST): A regulator of neuronal development and neuronal/endocrine function. Cell Tissue Res. 2014, 359, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Bithell, A. REST: Transcriptional and epigenetic regulator. Epigenomics 2011, 3, 47–58. [Google Scholar] [CrossRef]
- Hermanson, O. Stem cells have different needs for REST. PLOS Biol. 2008, 6, e271. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Bao, S. Ubiquitination and deubiquitination of REST and its roles in cancers. FEBS Lett. 2012, 586, 1602–1605. [Google Scholar] [CrossRef] [PubMed]
- Lapuk, A.V.; Wu, C.; Wyatt, A.W.; McPherson, A.; McConeghy, B.J.; Brahmbhatt, S.; Mo, F.; Zoubeidi, A.; Anderson, S.; Bell, R.H.; et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J. Pathol. 2012, 227, 286–297. [Google Scholar] [CrossRef] [PubMed]
- Contreras, H.; Orellana-Serradell, O.; Herrera, D.; Castellon, E. The transcription factor ZEB1 promotes an aggressive phenotype in prostate cancer cell lines. Asian J. Androl. 2018, 20, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Contreras, H.; Orellana-Serradell, O.; Herrera, D.; Castellón, E. The transcription factor ZEB1 promotes chemoresistance in prostate cancer cell lines. Asian J. Androl. 2019, 21, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Odero-Marah, V.; Hawsawi, O.; Henderson, V.; Sweeney, J. Epithelial-Mesenchymal Transition (EMT) and Prostate Cancer. Adv. Exp. Med. Biol. 2018, 1095, 101–110. [Google Scholar] [PubMed]
- Dicken, H.; Hensley, P.J.; Kyprianou, N. Prostate tumor neuroendocrine differentiation via EMT: The road less traveled. Asian J. Urol. 2019, 6, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-T.; Lin, T.-P.; Campbell, M.; Pan, C.-C.; Lee, S.-H.; Lee, H.-C.; Yang, M.-H.; Kung, H.-J.; Chang, P.-C. REST is a crucial regulator for acquiring EMT-like and stemness phenotypes in hormone-refractory prostate cancer. Sci. Rep. 2017, 7, srep42795. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.S.; Li, J.; Stockert, J.A.; Herzog, B.; O’Connor, J.; Garzon-Manco, L.; Parsons, R.; Tewari, A.K.; Yadav, K.K. Induction of Neuroendocrine Differentiation in Prostate Cancer Cells by Dovitinib (TKI-258) and its Therapeutic Implications. Transl. Oncol. 2017, 10, 357–366. [Google Scholar] [CrossRef]
- Shimojo, M.; Hersh, L.B. REST/NRSF-interacting LIM domain protein, a putative nuclear translocation receptor. Mol. Cell. Biol. 2003, 23, 9025–9031. [Google Scholar] [CrossRef]
- Tai, S.; Sun, Y.; Squires, J.M.; Zhang, H.; Oh, W.K.; Liang, C.-Z.; Huang, J. PC3 is a cell line characteristic of prostatic small cell carcinoma. Prostate 2011, 71, 1668–1679. [Google Scholar] [CrossRef]
- Svensson, C.; Ceder, J.; Iglesias-Gato, D.; Chuan, Y.-C.; Pang, S.T.; Bjartell, A.; Martinez, R.M.; Bott, L.; Helczynski, L.; Ulmert, D.; et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 2014, 42, 999–1015. [Google Scholar] [CrossRef]
- Shi, X.-B.; Xue, L.; Ma, A.-H.; Tepper, C.G.; Gandour-Edwards, R.; Kung, H.-J.; White, R.W.D. Tumor suppressive miR-124 targets androgen receptor and inhibits proliferation of prostate cancer cells. Oncogene 2013, 32, 4130–4138. [Google Scholar] [CrossRef]
- Tivnan, A.; Zhao, J.; Johns, T.G.; Day, B.W.; Stringer, B.W.; Boyd, A.W.; Tiwari, S.; Giles, K.M.; Teo, C.; McDonald, K.L. The tumor suppressor microRNA, miR-124a, is regulated by epigenetic silencing and by the transcriptional factor, REST in glioblastoma. Tumor Biol. 2014, 35, 1459–1465. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.; Konno, Y.; Ihira, K.; Suzuki, F.; Yamada, T.; Kudo, M.; Yue, J.; et al. Suppression of iASPP-dependent aggressiveness in cervical cancer through reversal of methylation silencing of microRNA-124. Sci. Rep. 2016, 6, 35480. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-Y.; Zukin, R.S. REST, a master transcriptional regulator in neurodegenerative disease. Curr. Opin. Neurobiol. 2018, 48, 193–200. [Google Scholar] [CrossRef]
- Gloushankova, N.A.; Zhitnyak, I.Y.; Rubtsova, S.N. Role of Epithelial-Mesenchymal Transition in Tumor Progression. Biochemistry 2018, 83, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, V.; Montoya, C.A.; Donnenberg, V.S.; Sant, S. Interplay between tumor microenvironment and partial EMT as the driver of tumor progression. iScience 2021, 24, 102113. [Google Scholar] [CrossRef]
- Guo, Y.; Lu, X.; Chen, Y.; Clark, G.; Trent, J.; Cuatrecasas, M.; Emery, D.; Song, Z.-H.; Chariker, J.; Rouchka, E.; et al. Opposing roles of ZEB1 in the cytoplasm and nucleus control cytoskeletal assembly and YAP1 activity. Cell Rep. 2022, 41, 111452. [Google Scholar] [CrossRef] [PubMed]
- Mendez, M.G.; Kojima, S.; Goldman, R.D. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010, 24, 1838–1851. [Google Scholar] [CrossRef]
- Paulin, D.; Lilienbaum, A.; Kardjian, S.; Agbulut, O.; Li, Z. Vimentin: Regulation and pathogenesis. Biochimie 2022, 197, 96–112. [Google Scholar] [CrossRef]
- Paonessa, F.; Latifi, S.; Scarongella, H.; Cesca, F.; Benfenati, F. Specificity protein 1 (Sp1)-dependent activation of the synapsin I gene (SYN1) is modulated by RE1-silencing transcription factor (REST) and 5’-cytosine-phosphoguanine (CpG) methylation. J. Biol. Chem. 2013, 288, 3227–3239. [Google Scholar] [CrossRef] [PubMed]
- Tien, A.H.; Sadar, M.D. Keys to unlock androgen receptor translocation. J. Biol. Chem. 2019, 294, 8711–8712. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Hruszkewycz, A.; Scher, H.I.; Hildesheim, J.; Isaacs, J.; Yu, E.Y.; Kelly, K.; Lin, D.; Dicker, A.; Arnold, J.; et al. The Role of Lineage Plasticity in Prostate Cancer Therapy Resistance. Clin. Cancer Res. 2019, 25, 6916–6924. [Google Scholar] [CrossRef]
- Schaeffer, E.M.; Srinivas, S.; Adra, N.; An, Y.; Barocas, D.; Bitting, R.; Bryce, A.; Chapin, B.; Cheng, H.H.; D’Amico, A.V.; et al. Prostate Cancer, Version 4.2023, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2023, 21, 1067–1096. [Google Scholar] [CrossRef] [PubMed]
- Liao, R.S.; Ma, S.; Miao, L.; Li, R.; Yin, Y.; Raj, G.V. Androgen receptor-mediated non-genomic regulation of prostate cancer cell proliferation. Transl. Androl. Urol. 2013, 2, 187–196. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Fedor, H.L.; Lotan, T.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Westbrook, T.F.; Martin, E.S.; Schlabach, M.R.; Leng, Y.; Liang, A.C.; Feng, B.; Zhao, J.J.; Roberts, T.M.; Mandel, G.; Hannon, G.J.; et al. A genetic screen for candidate tumor suppressors identifies REST. Cell 2005, 121, 837–848. [Google Scholar] [CrossRef]
- Kreisler, A.; Strissel, P.L.; Strick, R.; Neumann, S.B.; Schumacher, U.; Becker, C.-M. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 2010, 29, 5828–5838. [Google Scholar] [CrossRef]
- Gunsalus, K.T.; Wagoner, M.P.; Meyer, K.; Potter, W.B.; Schoenike, B.; Kim, S.; Alexander, C.M.; Friedl, A.; Roopra, A. Induction of the RNA regulatory LIN28A is required for the growth and pathogenesis of RESTless breast tumors. Cancer Res. 2012, 72, 3207–3216. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Hu, H.-G.; Huang, J.; Zou, Q.; Wang, J.; Liu, M.-Q.; Zhao, Y.; Li, G.-Z.; Xue, S.; Wu, Z.-S. Expression and correlation of Twist and gelatinases in breast cancer. Exp. Ther. Med. 2013, 6, 97–100. [Google Scholar] [CrossRef]
- Lopez-Moncada, F.; Castellon, E.A.; Contreras, H.R. The Transcription Factors Zeb1 and Snail Induce Cell Malignancy and Cancer Stem Cell Phenotype in Prostate Cells, Increasing Androgen Synthesis Capacity and Therapy Resistance. Adv. Exp. Med. Biol. 2022, 1393, 51–64. [Google Scholar] [PubMed]
- Cloud, A.S.; Vargheese, A.M.; Gunewardena, S.; Shimak, R.M.; Ganeshkumar, S.; Kumaraswamy, E.; Jensen, R.A.; Chennathukuzhi, V.M. Loss of REST in breast cancer promotes tumor progression through estrogen sensitization, MMP24 and CEMIP overexpression. BMC Cancer 2022, 22, 180. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Indo, S.; Orellana-Serradell, O.; Torres, M.J.; Castellón, E.A.; Contreras, H.R. Overexpression of REST Represses the Epithelial–Mesenchymal Transition Process and Decreases the Aggressiveness of Prostate Cancer Cells. Int. J. Mol. Sci. 2024, 25, 3332. https://doi.org/10.3390/ijms25063332
Indo S, Orellana-Serradell O, Torres MJ, Castellón EA, Contreras HR. Overexpression of REST Represses the Epithelial–Mesenchymal Transition Process and Decreases the Aggressiveness of Prostate Cancer Cells. International Journal of Molecular Sciences. 2024; 25(6):3332. https://doi.org/10.3390/ijms25063332
Chicago/Turabian StyleIndo, Sebastián, Octavio Orellana-Serradell, María José Torres, Enrique A. Castellón, and Héctor R. Contreras. 2024. "Overexpression of REST Represses the Epithelial–Mesenchymal Transition Process and Decreases the Aggressiveness of Prostate Cancer Cells" International Journal of Molecular Sciences 25, no. 6: 3332. https://doi.org/10.3390/ijms25063332
APA StyleIndo, S., Orellana-Serradell, O., Torres, M. J., Castellón, E. A., & Contreras, H. R. (2024). Overexpression of REST Represses the Epithelial–Mesenchymal Transition Process and Decreases the Aggressiveness of Prostate Cancer Cells. International Journal of Molecular Sciences, 25(6), 3332. https://doi.org/10.3390/ijms25063332







