Differential Gene Regulatory Network Analysis between Azacitidine-Sensitive and -Resistant Cell Lines
Abstract
1. Introduction
2. Results
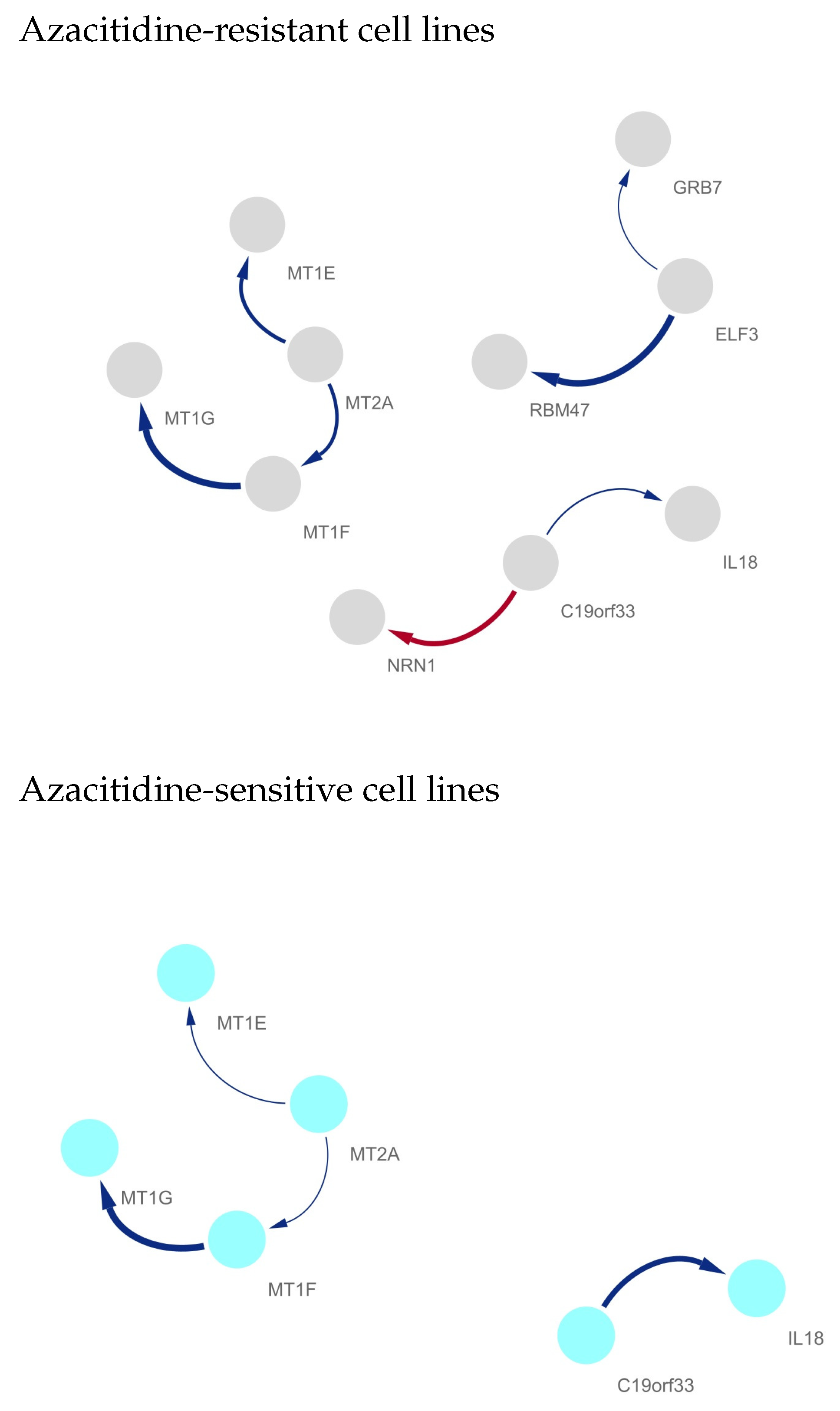
Differentially Regulated Gene Network Identification between Azacitidine-Resistant and -Sensitive Cell Lines
- ELF3
- IL18Saadi et al. [19] evaluated the expression levels of IL18 in AML patients according to their response to treatment and showed that expression levels of IL18 were increased in AML patients who did not respond to therapy compared to those patients who respond to therapy. Furthermore, it was observed that the expression levels of IL18 were significantly increased in high-risk groups of AML patients [19]. A correlation was observed between the levels of IL18 and the prognosis of AML, i.e., higher levels of IL18 were correlated with worse prognosis of AML [20]. The expression profiles of IL-6 and IL18 were considered as prognostic markers for AML [19]. Furthermore, the higher expression levels of IL18 and its receptor induced drug resistance in AML [19]. From the results, Saadi et al. [19] suggested that IL18 is an important prognostic marker in AML and control of the expression and regulation of IL18 may play key roles in the management of AML. Chen et al. [21] showed that overexpression of IL18 might reflect the convergence of several important unfavorable prognostic factors in AML. Song et al. [22] also suggested that high circulating levels of IL18 are a potential predictor for a decreased risk of AML. It was also demonstrated that the variant GT genotype of IL18 rs1946518 led to poorer survival rates in AML [23].
- Metallothionein (MT) gene familyExposure of cells in culture to 5-azacytidine stimulates the expression of the metallothionein gene [26]. Stallings et al. [28] showed that 5-azacytidine-induced conversion to cadmium resistance is correlated with early S-phase replication of inactive metallothionein genes in synchronized cadmium-sensitive cells. Increased MT-I expression is a poor prognostic marker for AML [24]. MT was identified as a drug-resistance-related protein and was expressed in leukemic cells in more than half of cases of newly diagnosed AML [24]. Patricia et al. [25] showed that MT1 is critical for the growth and survival of DNMT3A;NPM1-mutant AML cells and nominated MT1 as a key marker for the treatment and prevention of DNMT3A;NPM1-mutant AML.
- RBM475-azacytidine-resistant metabolic adaptable cells have several other alterations in RBM47 expression [31]. RBM47 was identified as an important AML-specific RBP gene, and Saha et al. [30] revealed that RBM47 is a potential candidate for therapeutic intervention toward effective eradication of leukemic stem cells in AML.
3. Discussion
4. Method for Differential Gene Network Analysis
4.1. Azacitidine Sensitivity-Specific Gene Regulatory Network Estimation
4.2. Differentially Regulated Gene Network Identification in Azacitidine-Resistant Cell Lines
4.2.1. Existing Method for Identifying Differentially Coexpressed Gene Set: DiffCoEx
- Step 1.
- For the drug-sensitive (-resistant) cell lines, the correlation matrix is computed, where the entry of is given as a correlation between and genes as follows,where and ( and ) are expression levels of the and genes in drug-sensitive (-resistant) cell lines.
- Step 2.
- Compute adjacency difference matrixwhere is a parameter that can emphasize large correlation differences in the matrix .
- Step 3.
- Compute dissimilarity matrix based on the topological overlap [41],The small value of indicates that the and genes both show considerable correlation changes within the same group of genes. This implies that the groups of genes corresponding to small values of were differentially coexpressed between phenotypes.
4.2.2. Differentially Regulated Gene Network Identification from the Azacitidine Sensitivity-Specific Gene Networks
- Step 1: Paired cell linesConstruct paired cell lines based on randomly selected drug-sensitive and -resistant cell lines,where and are randomly selected azacitidine sensitivity values from the drug-sensitive and -resistant cell lines, respectively.
- Step 2: Edge weightFrom the estimated and -specific gene networks, the edge weight and are computedwhere and are the varying coefficients of the regulator gene on target gene in the cell line.
- Step 3: Adjacency difference matrixCompute the adjacency difference matrix for the paired cell lines (i.e., ),where is a tuning emphasizing large correlation differences in the matrix . In practice, it is advisable to select optimal value of because the result heavily depends on the value of .
- Step 4: Dissimilarity matrix for cell lineCompute the dissimilarity matrix for the paired cell lines,Compute the average of the entries in the dissimilarity matrix
- Step 5: Dissimilarity measure of all cell linesCompute the dissimilarity measure of the cell line-specific gene network analysis as follows:
- Step 6. Statistical significanceShuffle cell lines into drug-sensitive and -resistant groups and construct permutation paired cell lines,For the permutation paired cell lines , Steps 2–5 are conducted, and then the dissimilarity measure is computed for . The permutation p value is computed as follows,where is the indicate function.
- Step 7. Identifying differentially regulated gene networksFor the significance level , we identify differentially regulated gene networks that satisfy
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moreno, V.Y.; Badar, T. Clinical Utility of azacitidine in the Management of Acute Myeloid Leukemia: Update on Patient Selection and Reported Outcomes. Cancer Manag. Res. 2022, 14, 3527–3538. [Google Scholar] [CrossRef]
- Schuh, A.C.; Döhner, H.; Pleyer, L.; Seymour, J.F.; Fenaux, P.; Dombret, H. Azacitidine in adult patients with acute myeloid leukemia. Crit. Rev. Oncol. Hematol. 2017, 116, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Sripayap, P.; Nagai, T.; Uesawa, M.; Kobayashi, H.; Tsukahara, T.; Ohmine, K.; Muroi, K.; Ozawa, K. Mechanisms of resistance to azacitidine in human leukemia cell lines. Exp. Hematol. 2014, 42, 294–306.e2. [Google Scholar] [CrossRef] [PubMed]
- Gruber, E.; Franich, R.L.; Shortt, J.; Johnstone, R.W.; Kats, L.M. Distinct and overlapping mechanisms of resistance to azacytidine and guadecitabine in acute myeloid leukemia. Leukemia 2020, 34, 3388–3392. [Google Scholar] [CrossRef] [PubMed]
- Šimoničová, K.; Janotka, L.; Kavcova, H.; Sulova, Z.; Messingerova, L.; Breier, A. Resistance of Leukemia Cells to 5-Azacytidine: Different Responses to the Same Induction Protocol. Cancers 2023, 15, 3063. [Google Scholar] [CrossRef]
- Minařík, L.; Pimková, K.; Kokavec, J.; Schaffartziková, A.; Vellieux, F.; Kulvait, V.; Daumová, L.; Dusilková, N.; Jonášová, A.; Vargová, K.S.; et al. Analysis of 5-Azacytidine Resistance Models Reveals a Set of Targetable Pathways. Cells 2022, 11, 223. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Park, S.; Choi, H.; Kim, H.J.; Kwon, Y.R.; Ryu, D.; Kim, M.; Kim, T.M.; Kim, Y.J. Gene expression signatures associated with sensitivity to azacitidine in myelodysplastic syndromes. Sci. Rep. 2020, 10, 19555. [Google Scholar] [CrossRef]
- Aloraini, A.; ElSawy, K.M. Potential breast anticancer drug targets revealed by differential gene regulatory network analysis and molecular docking: Neoadjuvant docetaxel drug as a case study. Cancer Inform. 2018, 17, 1176935118755354. [Google Scholar] [CrossRef]
- Basso, K.; Margolin, A.A.; Stolovitzky, G.; Klein, U.; Dalla-Favera, R.; Califano, A. Reverse engineering of regulatory networks in human b cells. Nat. Genet. 2005, 37, 382–390. [Google Scholar] [CrossRef]
- Imoto, S.; Tamada, Y.; Savoie, C.J.; Miyano, S. Analysis of gene networks for drug target discovery and validation. Methods Mol. Biol. 2007, 360, 33–56. [Google Scholar]
- Madhamshettiwar, P.; Maetschke, S.; Davis, M.; Reverter, A.; Ragan, M.A. Gene regulatory network inference: Evaluation and application to ovarian cancer allows the prioritization of drug targets. Genome Med. 2012, 4, 41. [Google Scholar] [CrossRef]
- Grimes, T.; Potter, S.S.; Datta, S. Integrating gene regulatory pathways into differential network analysis of gene expression data. discovery and validation. Sci. Rep. 2019, 9, 5479. [Google Scholar] [CrossRef]
- Tu, J.J.; Ou-Yang, L.; Zhu, Y.; Yan, H.; Qin, H.; Zhang, X.F. Differential network analysis by simultaneously considering changes in gene interactions and gene expression. discovery and validation. Bioinformatics 2021, 37, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Yang, F.; Wang, J.; Karikomi, M.; Yin, Y.; Sun, J.; Wen, T.; Nie, Q. DNF: A differential network flow method to identify rewiring drivers for gene regulatory networks. discovery and validation. Neurocomputing 2020, 410, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Mall, R.; Cerulo, L.; Bensmail, H.; Iavarone, A.; Ceccarelli, M. Detection of statistically significant network changes in complex biological networks. discovery and validation. BMC Syst. Biol. 2017, 11, 32. [Google Scholar] [CrossRef]
- Park, H.; Imoto, S.; Miyano, S. Comprehensive information-based differential gene regulatory networks analysis (CIdrgn): Application to gastric cancer and chemotherapy-responsive gene network identification. PLoS ONE 2023, 18, e0286044. [Google Scholar] [CrossRef]
- Guo, S.; Li, B.; Xu, X.; Wang, W.; Wang, S.; Lv, T.; Wang, H. Construction of a 14-lncRNA risk score system predicting survival of children with acute myelocytic leukemia. Exp. Ther. Med. 2020, 20, 1521–1531. [Google Scholar] [CrossRef]
- Li, H.; Chiappinelli, K.B.; Guzzetta, A.A.; Easwaran, H.; Yen, R.W.; Vatapalli, R.; Topper, M.J.; Luo, J.; Connolly, R.M.; Azad, N.S.; et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget 2014, 5, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Saadi, M.I.; Ramzi, M.; Hosseinzadeh, M.; Ebrahimi, N.; Owjfard, M.; Abdolyousefi, E.N.; Hesami, Z.; Valibeigi, B.; Zareei, N.; Tavasolian, F.; et al. Expression Levels of Il-6 and IL18 in Acute Myeloid Leukemia and Its Relation with Response to Therapy and Acute GvHD After Bone Marrow Transplantation. Indian J. Surg. Oncol. 2021, 12, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Y.; Zheng, G.G.; Ma, X.T.; Li, G.; Zhang, F.K.; Wu, K.F. Clinical significance of IL18 gene over-expression in AML. Leuk Res. 2002, 26, 887–892. [Google Scholar] [CrossRef]
- Chen, C.C.; Tzeng, H.E.; Kuo, C.C.; Lim, S.N.S.; Hsu, P.C.; Hsu, Y.N.; Chin, Y.T.; Chang, W.S.; Wang, C.H.; Tsai, C.W.; et al. Significant Contribution of Interleukin-18 Genotypes to Childhood Acute Lymphocytic Leukemia Risk in Taiwanese. Anticancer Res. 2022, 42, 5283–5290. [Google Scholar] [CrossRef]
- Song, J.; Li, A.; Qian, Y.; Liu, B.; Lv, L.; Ye, D.; Sun, X.; Mao, Y. Genetically predicted circulating levels of cytokines and the risk of cancer. Front. Immunol. 2022, 13, 886144. [Google Scholar] [CrossRef]
- Wang, H.; Hua, M.; Wang, S.; Yu, J.; Chen, C.; Zhao, X.; Zhang, C.; Zhong, C.; Wang, R.; He, N.; et al. Genetic polymorphisms of IL18 rs1946518 and IL-1β rs16944 are associated with prognosis and survival of acute myeloid leukemia. Inflamm. Res. 2017, 66, 249–258. [Google Scholar] [CrossRef]
- Takahashi, S. Molecular functions of metallothionein and its role in hematological malignancies. J. Hematol. Oncol. 2012, 5, 41. [Google Scholar] [CrossRef]
- Patricia, A.; Colom, D.; Chih-Hsing, C.; Jayna, J.M.; Nathan, S.; Mohammad Daud Khan, H.; Leighton, G.; Jennifer, J.T. Metallothionein 1 (MT1) Is Selectively Required for DNMT3A;NPM1-Mutant Acute Myeloid Leukemia Proliferation. Blood 2022, 140 (Suppl. S1), 9103–9104. [Google Scholar]
- Waalkes, M.P.; Poirier, L.A. Induction of hepatic metallothionein following 5-azacytidine administration. Toxicol. Appl. Pharmacol. 1985, 79, 47–53. [Google Scholar] [CrossRef]
- Thomas, D.J.; Angle, C.R.; Swanson, S.A. Effect of 5-azacytidine on metallothionein inducibility and sensitivity to lethality of cadmium in rat osteosarcoma (ROS 17/2.8) cells. Toxicology 1990, 65, 169–178. [Google Scholar] [CrossRef]
- Stallings, R.L.; Crawford, B.D.; Tobey, R.A.; Tesmer, J.; Hildebrand, C.E. 5-Azacytidine-induced conversion to cadmium resistance correlates with early S phase replication of inactive metallothionein genes in synchronized CHO cells. Somat. Cell Mol. Genet. 1986, 12, 423–432. [Google Scholar] [CrossRef]
- Schmidt, C.J.; Hamer, D.H. Cell specificity and an effect of ras on human metallothionein gene expression. Proc. Natl. Acad. Sci. USA 1986, 83, 3346–3350. [Google Scholar] [CrossRef]
- Saha, S.; Murmu, K.C.; Biswas, M.; Chakraborty, S.; Basu, J.; Madhulika, S.; Kolapalli, S.P.; Chauhan, S.; Sengupta, A.; Prasad, P. Transcriptomic Analysis Identifies RNA Binding Proteins as Putative Regulators of Myelopoiesis and Leukemia. Front. Oncol. 2019, 9, 692. [Google Scholar] [CrossRef]
- Singh, B.; Sarli, V.N.; Lucci, A. Inhibition of resistant triple-negative breast cancer cells with low-dose 6-mercaptopurine and 5-azacitidine. Oncotarget 2021, 12, 626–637. [Google Scholar] [CrossRef]
- Nehme, A.; Dakik, H.; Picou, F.; Cheok, M.; Preudhomme, C.; Dombret, H.; Lambert, J.; Gyan, E.; Pigneux, A.; Récher, C.; et al. Horizontal meta-analysis identifies common deregulated genes across AML subgroups providing a robust prognostic signature. Blood Adv. 2020, 4, 5322–5335. [Google Scholar] [CrossRef]
- Cheng, Q.; Shi, H.; Wang, H.; Wang, J.; Liu, Y. Asplatin enhances drug efficacy by altering the cellular response. Metallomics 2016, 8, 672–678. [Google Scholar] [CrossRef]
- Mellor, H.R.; Callaghan, R. Resistance to chemotherapy in cancer: A complex and integrated cellular response. Pharmacology 2008, 81, 275–300. [Google Scholar] [CrossRef]
- Zheng, X.; Chou, P.M.; Mirkin, B.L.; Rebbaa, A. Senescence-initiated reversal of drug resistance: Specific role of cathepsin L. Cancer Res. 2004, 64, 1773–1780. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R. Varying-Coefficient Models. J. R. Stat. Soc. Ser. B 1993, 4, 757–796. [Google Scholar] [CrossRef]
- Shimamura, T.; Imoto, S.; Shimada, Y.; Hosono, Y.; Niida, A.; Nagasaki, M.; Yamaguchi, R.; Takahashi, T.; Miyano, S. A novel network profiling analysis reveals system changes in epithelial-mesenchymal transition. PLoS ONE 2011, 6, e20804. [Google Scholar] [CrossRef]
- Tesson, B.M.; Breitling, R.; Jansen, R.C. DiffCoEx: A simple and sensitive method to find differentially coexpressed gene modules. BMC Bioinform. 2010, 6, 497. [Google Scholar] [CrossRef]
- Ravasz, E.; Somera, A.L.; Mongru, D.A.; Oltvai, Z.N.; Barabasi, A.L. Hierarchical Organization of Modularity in Metabolic Networks. Science 2002, 297, 1551–1555. [Google Scholar] [CrossRef]


| Genes | AML | Azacitidine | |||
|---|---|---|---|---|---|
| Yes/No | Evidence | Yes/No | Evidence | ||
| C19orf33 | No | - | No | - | |
| ELF3 | Yes | [17] | Yes | [18] | |
| GRB7 | No | - | No | - | |
| IL18 | Yes | [19,20,21,22,23] | No | - | |
| Metallothionein family | MT1E | Yes | [24,25] | Yes | [26,27,28,29] |
| MT1F | |||||
| MT1G | |||||
| MT2A | |||||
| NRN1 | No | - | No | - | |
| RBM47 | Yes | [30] | Yes | [31] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, H.; Miyano, S. Differential Gene Regulatory Network Analysis between Azacitidine-Sensitive and -Resistant Cell Lines. Int. J. Mol. Sci. 2024, 25, 3302. https://doi.org/10.3390/ijms25063302
Park H, Miyano S. Differential Gene Regulatory Network Analysis between Azacitidine-Sensitive and -Resistant Cell Lines. International Journal of Molecular Sciences. 2024; 25(6):3302. https://doi.org/10.3390/ijms25063302
Chicago/Turabian StylePark, Heewon, and Satoru Miyano. 2024. "Differential Gene Regulatory Network Analysis between Azacitidine-Sensitive and -Resistant Cell Lines" International Journal of Molecular Sciences 25, no. 6: 3302. https://doi.org/10.3390/ijms25063302
APA StylePark, H., & Miyano, S. (2024). Differential Gene Regulatory Network Analysis between Azacitidine-Sensitive and -Resistant Cell Lines. International Journal of Molecular Sciences, 25(6), 3302. https://doi.org/10.3390/ijms25063302





