Development of Genome-Wide Intron Length Polymorphism (ILP) Markers in Tea Plant (Camellia sinensis) and Related Applications for Genetics Research
Abstract
1. Introduction
2. Results
2.1. Tea ILP Marker Development
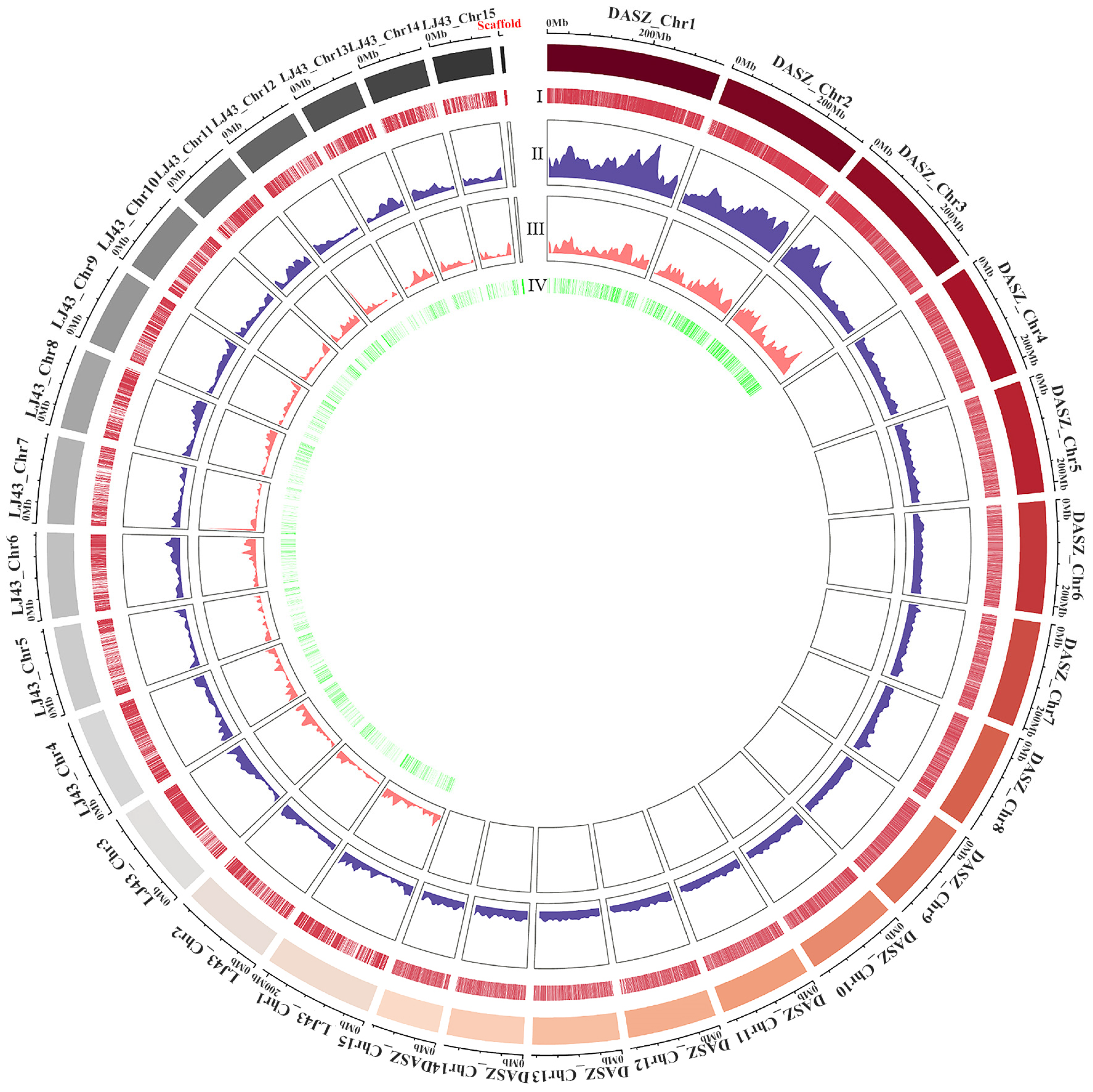
2.1.1. Number, Distribution, and Density of ILP Markers in Tea Trees
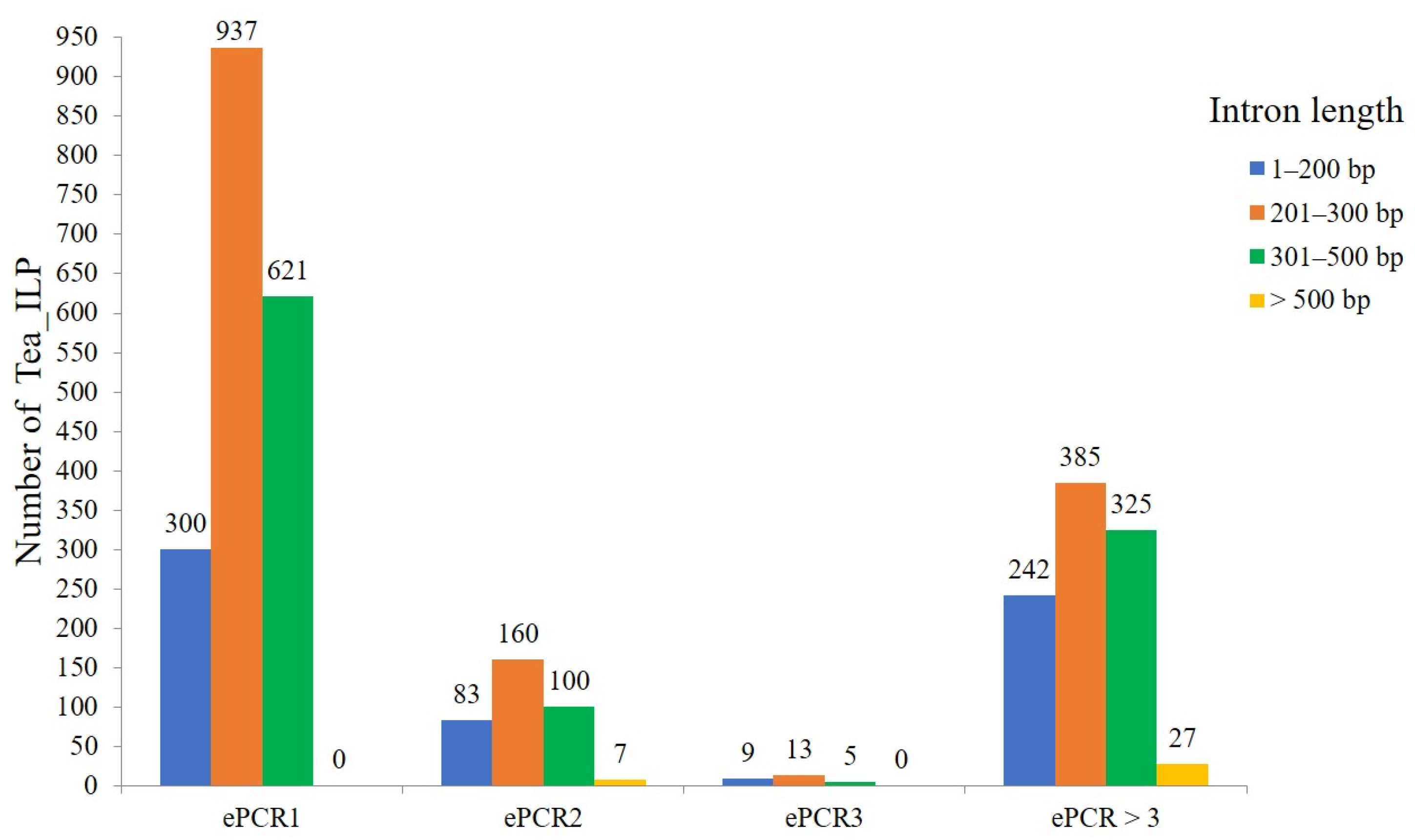
2.1.2. Differential Distribution of Intron Length of Tea ILP
2.1.3. Experimental Validation of the Tea ILP Marker
2.2. Cross-Transferability of Tea ILP Markers among Different Plant Species
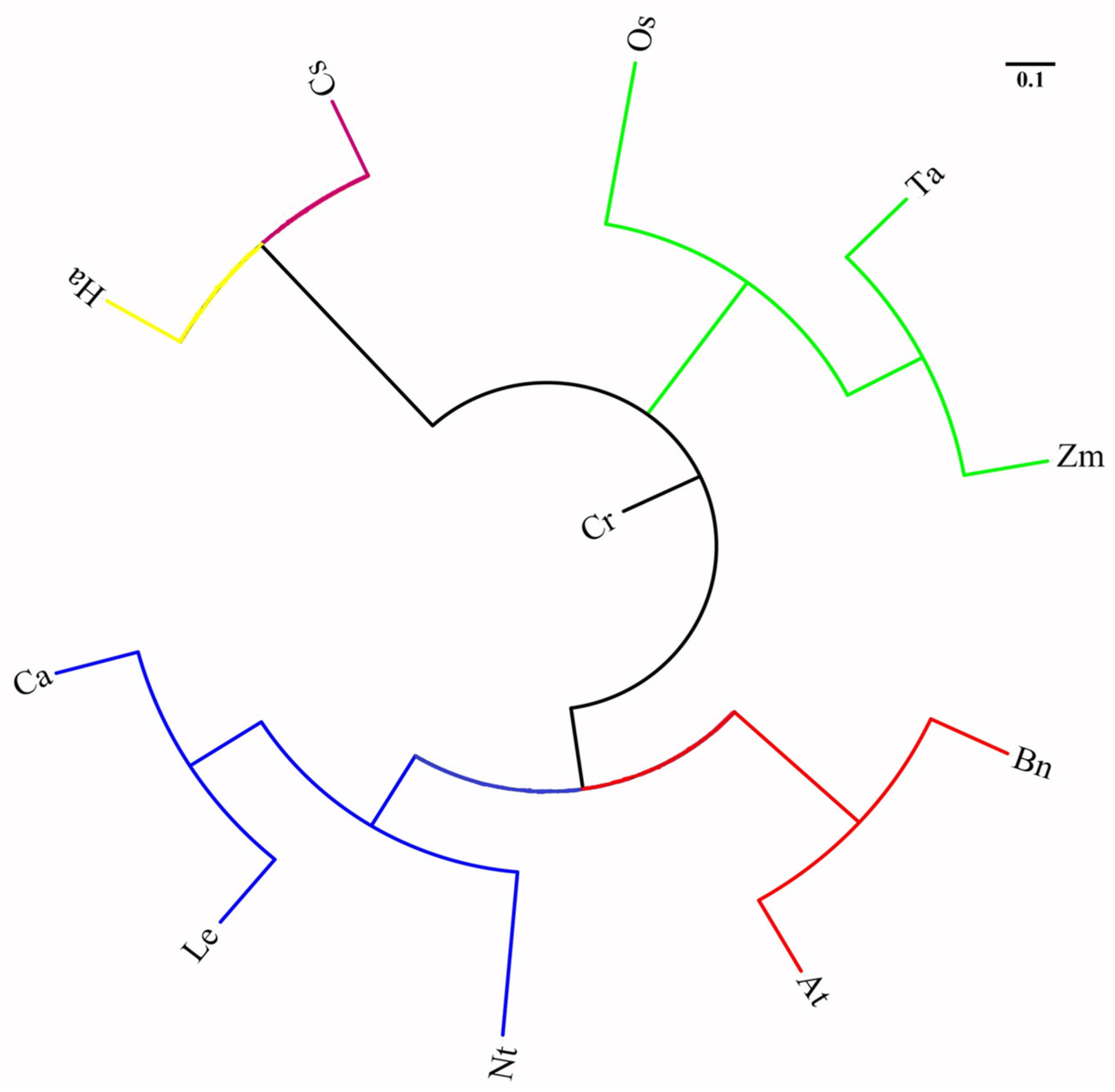
2.2.1. Analysis of the Cross-Transferability of Tea ILP Markers among 11 Plant Species
2.2.2. Analysis of Genetic Relationships in 11 Plant Species by Tea ILP Markers
2.3. Genetic Diversity and Population Structure of C. tetracocca by 40 ILP Molecular Markers
2.3.1. Genetic Diversity and Genetic Differentiation Analysis of Cultivated C. tetracoccain Puan
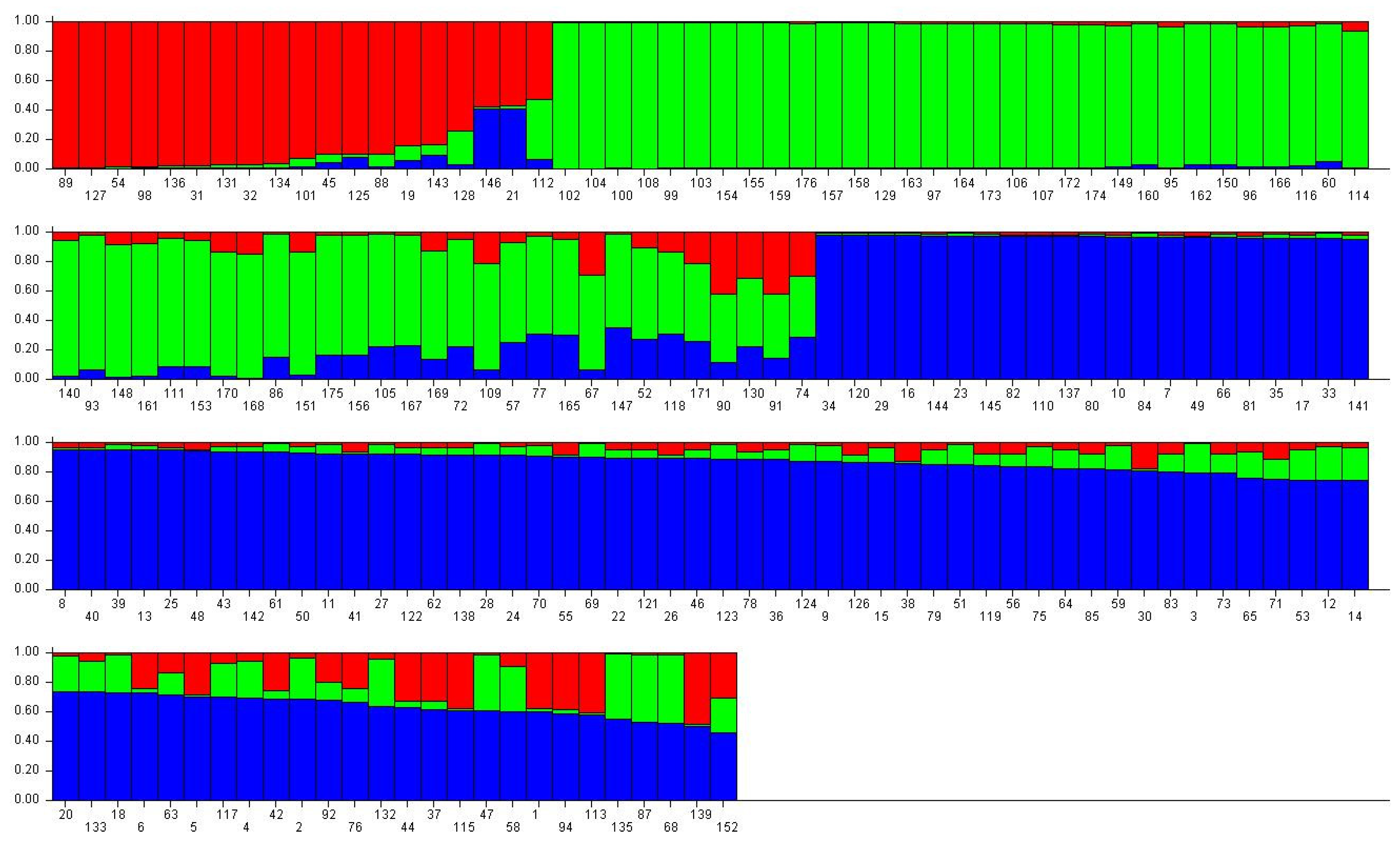
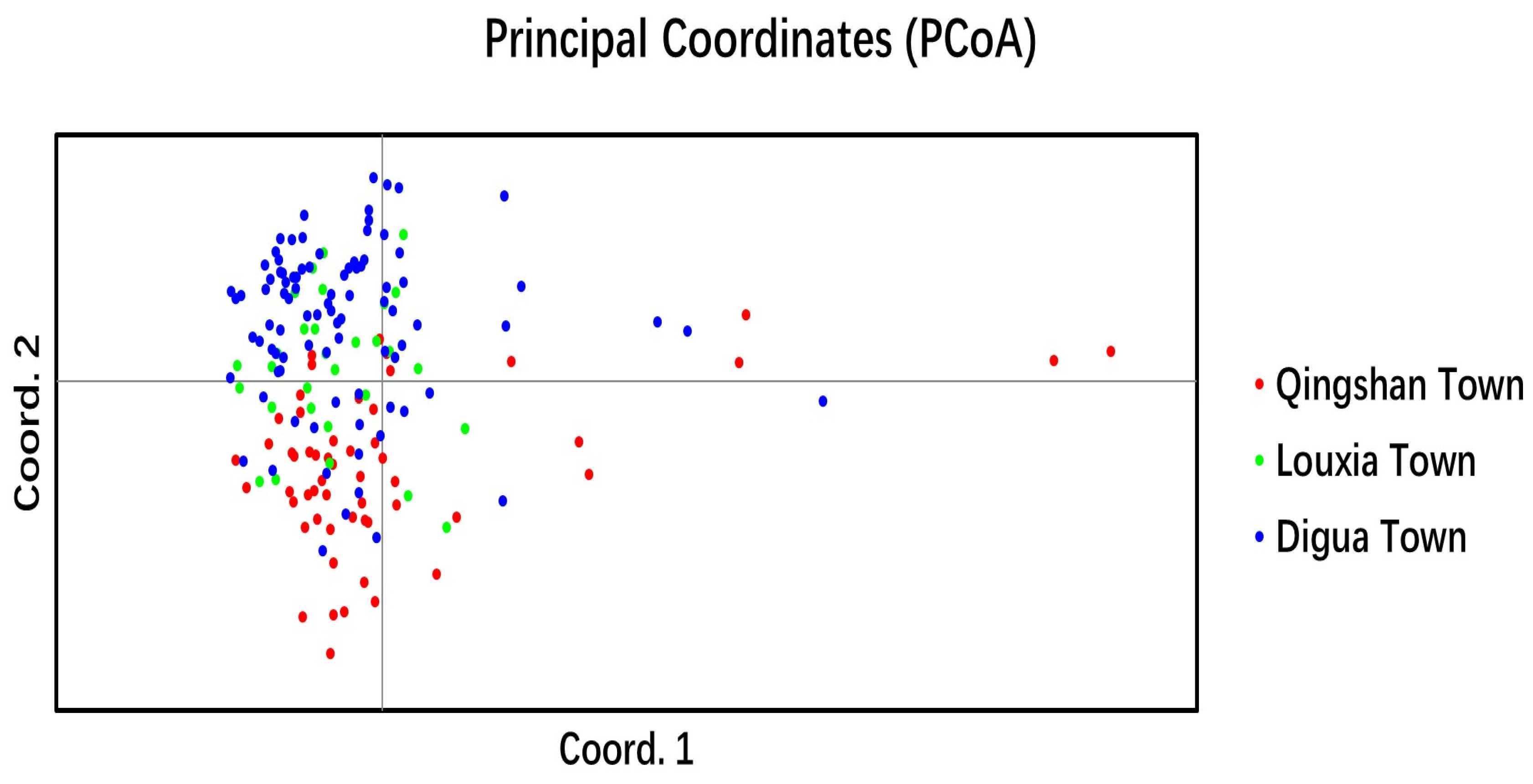
2.3.2. Genetic Relationship and Genetic Structure of Cultivated C. tetracocca Population in Puan
3. Discussion
4. Materials and Methods
4.1. Plant Materials and DNA Extraction
4.2. Source of Sequences
4.3. ILP Marker Development and ePCR Analysis
4.4. ILP Molecular Marker Detection
4.5. Cross-Transferability of Tea ILP
4.6. Genetic Diversity and Population Structure Analysis of C. tetracocca
4.7. Data Statistics and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kanwar, J.; Taskeen, M.; Mohammad, I.; Huo, C.; Chan, T.H.; Dou, Q.P. Recent advances on tea polyphenols. Front. Biosci. 2012, E4, 111–131. [Google Scholar] [CrossRef]
- Pervin, M.; Unno, K.; Ohishi, T.; Tanabe, H.; Miyoshi, N.; Nakamura, Y. Benefificial effects of green tea catechins on neurodegenerative diseases. Molecules 2018, 23, 1297. [Google Scholar] [CrossRef]
- Mancini, E.; Beglinger, C.; Drewe, J.; Zanchi, D.; Lang, U.E.; Borgwardt, S. Green tea effects on cognition, mood and human brain function: A systematic review. Phytomedicine 2017, 34, 26–37. [Google Scholar] [CrossRef]
- Chen, L.; Yao, M.Z.; Wang, X.C.; Yang, Y.J. Tea genetic resources in China. Int. J. Tea Sci. 2012, 8, 55–64. [Google Scholar]
- Yao, M.-Z.; Ma, C.-L.; Qiao, T.-T.; Jin, J.-Q.; Chen, L. Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet. Genomes 2012, 8, 205–220. [Google Scholar] [CrossRef]
- Wang, L.; Xun, H.; Aktar, S.; Zhang, R.; Wu, L.; Ni, D.; Wei, K.; Wang, L. Development of SNP markers for original analysis and germplasm identification in Camellia sinensis. Plants 2022, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Shehasen, M.Z. Tea plant (Camellia Sinensis) breeding mechanisms role in genetic improvement and production of major producing countries. Int. J. Res. Stud. Sci. Eng. Technol. 2019, 6, 10–20. [Google Scholar]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Li, J.-W.; Li, H.; Liu, Z.-W.; Wang, Y.-X.; Chen, Y.; Yang, N.; Hu, Z.-H.; Li, T.; Zhuang, J. Molecular markers in tea plant (Camellia sinensis): Applications to evolution, genetic identification, and molecular breeding. Plant Physiol. Biochem. 2023, 198, 107704. [Google Scholar] [CrossRef]
- Gunasekare, M.T.K. Applications of molecular markers to the genetic improvement of Camellia sinensis L. (tea)—A review. J. Hortic. Sci. Biotechnol. 2007, 82, 161–169. [Google Scholar] [CrossRef]
- Wight, W. Tea classification revised. Curr. Sci. 1962, 31, 298–299. [Google Scholar]
- Chang, H. Thea—A section of beveragial tea trees of the genus Camellia. Acta. Sci. Nat. Univ. Sunyats 1981, 1, 87–99. [Google Scholar]
- Zhang, Z.; Min, X.; Wang, Z.; Wang, Y.; Liu, Z.; Liu, W. Genome-wide development and utilization of novel intron-length polymorphic (ILP) markers in Medicago sativa. Mol. Breed. 2017, 37, 87. [Google Scholar] [CrossRef]
- Xia, E.; Li, F.; Tong, W.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Tai, Y.; She, G.; et al. The tea plant reference genome and improved gene annotation using long-read and paired-end sequencing data. Sci. Data 2019, 6, 122. [Google Scholar] [CrossRef]
- Zhao, D.-W.; Yang, J.-B.; Yang, S.-X.; Kato, K.; Luo, J.-P. Genetic diversity and domestication origin of tea plant Camellia taliensis (Theaceae) as revealed by microsatellite markers. BMC Plant Biol. 2014, 14, 14. [Google Scholar] [CrossRef]
- Kaundun, S.S.; Zhyvoloup, A.; Park, Y.-G. Evaluation of the genetic diversity among elite tea (Camellia sinensis var. sinensis) accessions using RAPD markers. Euphytica 2000, 115, 7–16. [Google Scholar] [CrossRef]
- Zhang, X.-W.; Wang, Z.-D.; Liu, Y.-H.; Mou, J.-M. Intronic polymorphism markers and their use in molecular breeding of tobacco species. Anhui Agric. Sci. 2008, 36, 3147–3148, 3159. [Google Scholar]
- Yang, L.; Jin, G.; Zhao, X.; Zheng, Y.; Xu, Z.; Wu, W. PIP: A database of potential intron polymorphism markers. Bioinformatics 2007, 23, 2174–2177. [Google Scholar] [CrossRef]
- Srivastava, R.; Bajaj, D.; Sayal, Y.-K.; Meher, P.-K.; Upadhyaya, H.-D.; Kumar, R.; Tripathi, S.; Bharadwaj, C.; Rao, A.-R.; Parida, S.-K. Genome-wide development and deployment of informative intron-spanning and intron-length polymorphism markers for genomics-assisted breeding applications in chickpea. Plant Sci. 2016, 252, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Xie, F.-M.; Chen, L.-Y.; Zhao, X.-Q.; Jojee, L.; Madonna, D. Comparative analysis of genetic diversity and structure in rice using ILP and SSR markers. Rice Sci. 2010, 17, 257–268. [Google Scholar] [CrossRef]
- Muthamilarasan, M.; Suresh, B.V.; Pandey, G.; Kumari, K.; Parida, S.K.; Prasad, M. Development of 5123 intron-length polymorphic markers for large-scale genotyping applications in Foxtail millet. DNA Res. 2013, 21, 41–52. [Google Scholar] [CrossRef]
- Jayaswall, K.; Sharma, H.; Bhandawat, A.; Sagar, R.; Yadav, V.-K.; Sharma, V.; Mahajan, V.; Roy, J.; Singh, M. Development of intron length polymorphic (ILP) markers in onion (Allium cepa L.), and their cross-species transferability in garlic (A. sativum L.) and wild relatives. Genet. Resour. Crop Evol. 2019, 66, 1379–1388. [Google Scholar] [CrossRef]
- Stelmach, K.; Macko-Podgórni, M.; Machaj, G.; Grzebelus, D. Miniature inverted repeat transposable element insertions provide a source of intron length polymorphism markers in the Carrot (Daucus carota L.). Front. Plant Sci. 2017, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, M.; Mondal, T.-K.; Chand, P.-K. Biotechnological advances in tea (Camellia sinensis [L.] O. Kuntze): A review. Plant Cell Rep. 2015, 35, 255–287. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, Y.; Qiu, H.; Guo, Y.; Wan, H.; Zhang, X.; Scossa, F.; Alseekh, S.; Zhang, Q.; Wang, P.; et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 2020, 11, 3719. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.-H.; Zhang, H.-B.; Sheng, J.; Li, K.; Zhang, Q.-J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The Tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.D.; Zheng, C.; Ma, J.-Q.; Jiang, C.-K.; Ercisli, S.; Yao, M.-Z.; Chen, L. The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic. Res. 2020, 7, 63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-J.; Li, W.; Li, K.; Nan, H.; Shi, C.; Zhang, Y.; Dai, Z.-Y.; Lin, Y.-L.; Yang, X.-L.; Tong, Y.; et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol. Plant 2020, 13, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Yu, J.; Jin, S.; Chen, S.; Yue, C.; Wang, W.; Gao, S.; Cao, H.; Zheng, Y.; Gu, M.; et al. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic. Res. 2021, 8, 107. [Google Scholar] [CrossRef]
- Matsumoto, S.; Takeuchi, A.; Hayatsu, M.; Kondo, S. Molecular cloning of phenylalanine ammonia-lyase cDNA and classification of varieties and cultivars of tea plants (Camellia sinensis) using the tea PAL cDNA probe. Theor. Appl. Genet. 1994, 89, 671–675. [Google Scholar] [CrossRef]
- Matsumoto, S.; Kiriiwa, Y.; Yamaguchi, S. The Korean tea plant (Camellia sinensis): RFLP analysis of genetic diversity and relationship to Japanese tea. Breed. Sci. 2004, 54, 231–237. [Google Scholar] [CrossRef]
- Kaundun, S.-S.; Matsumoto, S. Identification of Processed Japanese Green Tea Based on Polymorphisms Generated by STS−RFLP Analysis. J. Agric. Food Chem. 2003, 51, 1765–1770. [Google Scholar] [CrossRef]
- Chen, L.J.; Zhang, S.Q.; Yin, J.; Song, Q.F.; Niu, S.Z.; Zhao, J.Y.; Chen, D.P.; Wang, S.W.; Geng, G.D. SNP analysis of the genetic evolution of ancient tea trees in Huaxi, Guiyang. J. Southwest Univ. (Nat. Sci. Dep. Acad. Ed.) 2019, 41, 33–40. [Google Scholar]
- Taniguchi, F.; Kimura, K.; Saba, T.; Ogino, A.; Yamaguchi, S.; Tanaka, J. Worldwide core collections of tea (Camellia sinensis) based on SSR markers. Tree Genet. Genomes 2014, 10, 1555–1565. [Google Scholar] [CrossRef]
- Jo, B.-S.; Choi, S.-S. Introns: The functional benefits of introns in genomes. Genom. Inform. 2015, 13, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.; Bhandawat, A.; Rahim, M.S.; Kumar, P.; Choudhoury, M.P.; Roy, J. Novel intron length polymorphic (ILP) markers from starch biosynthesis genes reveal genetic relationships in Indian wheat varieties and related species. Mol. Biol. Rep. 2020, 47, 3485–3500. [Google Scholar] [CrossRef] [PubMed]
- Feltus, F.-A.; Singh, H.-P.; Lohithaswa, H.-C.; Schulze, S.-R.; Silva, T.-D.; Paterson, A.-H. A comparative genomics strategy for targeted discovery of single-nucleotide polymorphisms and conserved-noncoding sequences in orphan crops. Plant Phys. 2006, 140, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Gowd, T.-Y.-M.; Deo, C.; Manjunathagowda, D.-C.; Mahajan, V.; Dutta, R.; Bhutia, N.-D.; Singh, B.; Mounika, V. Deployment of Intron Length Polymorphic (ILP) markers in dissipating diversity of Allium species. S. Afr. J. Bot. 2023, 160, 157–165. [Google Scholar] [CrossRef]
- Li, D.; Wang, L.; Liu, X.; Cui, Y.; Wang, X.; Wang, K.; Li, J. Development and characterization of 159 polymorphic EST-SSR markers for the Chinese endemic genus Metasequoia (Cupressaceae). Am. J. Bot. 2013, 100, e386–e390. [Google Scholar]
- Wang, X.-S.; Zhao, X.-Q.; Zhu, J.; Wu, W.-R. Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Res. 2005, 12, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. Genome-Wide Identification and Utilization of Intron-Length Polymorphic (ILP) Markers in Cleistogenes Songorica; Lanzhou University: Lanzhou, China, 2019. [Google Scholar]
- Cai, C.; Wu, S.; Niu, E.; Cheng, C.; Guo, W. Identification of genes related to salt stress tolerance using intro-length polymorphic markers, association mapping and virus-induced gene silencing in cotton. Sci. Rep. 2017, 7, 528. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Ma, Y.; Wang, Q.; Li, Y.; Liu, W. Transcriptome-wide development and utilisation of novel intron-length polymorphic markers in common vetch (Vicia sativa subsp. sativa). Crop Pasture Sci. 2021, 72, 1048–1057. [Google Scholar] [CrossRef]
- Wang, F.; Cheng, X.; Cheng, X.; Li, W.; Huang, X. Genetic diversity of the wild ancient tea tree (Camellia taliensis) populations at different altitudes in Qianjiazhai. PLoS ONE 2023, 18, e0283189. [Google Scholar] [CrossRef]
- Hu, C.-Y.; Tsai, Y.-Z.; Lin, S.-F. Development of STS and CAPS markers for variety identification and genetic diversity analysis of tea germplasm in Taiwan. Bot. Stud. 2014, 55, 12. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Wu, A.; Hou, Y.; An, Y.; Wei, C. Construction of fingerprinting for tea plant (Camellia sinensis) accessions using new genomic SSR markers. Mol. Breeding 2017, 37, 93. [Google Scholar] [CrossRef]
- Lessa, E.P. Rapid surveying of DNA sequence variation in natural populations. Mol. Biol. Evol. 1992, 9, 323–330. [Google Scholar]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Shen, C.-W.; Ning, Z.-X.; Huang, J.-A.; Chen, D.; Li, J.-X. Genetic diversity of Camellia sinensis germplasm in Guangdong Province based on morphological parameters and SRAP markers. Chin. J. Appl. Ecol. 2009, 20, 1551–1558. [Google Scholar]
- Chen, X.; Zhang, Y.; Li, J.; Xi, Y.; Zhang, Y. Genetic diversity analysis of tea germplasm in Shaanxi province based on SCoT marker. J. Tea Sci. 2016, 36, 131–138. [Google Scholar]
- Tang, Y.H.; Guo, C.F.; Zhang, M.Q. A method for extracting genomic DNA of tea tree—A modified CTAB method. J. Fujian Inst. Educ. 2007, 1, 99–101. [Google Scholar]
- Zhang, X.; Chen, S.; Shi, L.; Gong, D.; Zhang, S.; Zhao, Q.; Zhan, D.; Vasseur, L.; Wang, Y.; Yu, J.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef]
- Schuler, G.D. Sequence Mapping by Electronic PCR. Genome Res. 2016, 7, 541–550. [Google Scholar] [CrossRef]
- Fan, W.; Gai, H.; Sun, X.; Yang, A.; Zhang, J.; Ren, M. SSR data format conversion software DataFormater. Mol. Plant Breed. 2016, 14, 265–270. [Google Scholar]
- Rohlf, F. NTSYS-pc: Numerical Naxonomy and Nultivariate Nnalysis Nystem: Version 2.1; Exceter Software; Applied Biostatistics Inc.: New York, NY, USA, 2000. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C. Microsoft window based freeware for population genetic analysis. Popgene Ver. 1999, 1, 31. [Google Scholar]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
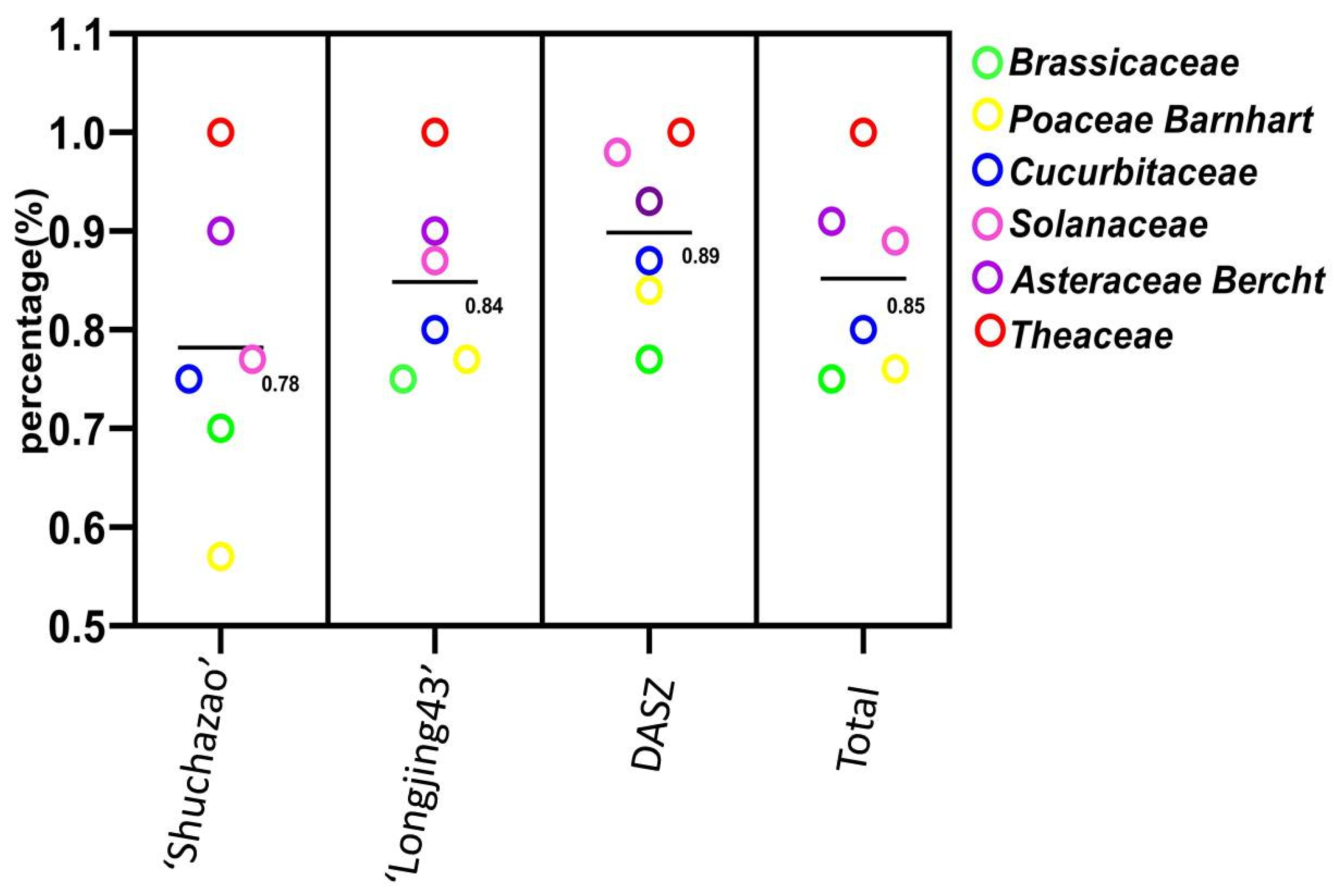

| Origin of Primers | Primers Synthesis | Amplified Primers (%) | Polymorphic Primers (%) |
|---|---|---|---|
| ‘Shuchazao’ [15] | 39 | 35 (89.74) | 9 (23.80) |
| ‘Longjing 43’ [9] | 91 | 87 (95.60) | 38 (41.76) |
| DASZ [26] | 100 | 91 (91.00) | 65 (65.00) |
| Total | 230 | 213 (92.61) | 112 (48.70) |
| Marker ID | Na | Ne | I | Obs-Ho | Obs-He | Exp-Ho | Exp-He | H | PIC% |
|---|---|---|---|---|---|---|---|---|---|
| Tea_ILP1116 | 6.00 | 3.89 | 1.53 | 1.00 | 0.00 | 0.26 | 0.74 | 0.74 | 70.71 |
| Tea_ILP1418 | 4.00 | 2.13 | 0.83 | 0.01 | 0.99 | 0.47 | 0.53 | 0.53 | 41.91 |
| Tea_ILP1396 | 4.00 | 2.16 | 0.92 | 0.81 | 0.19 | 0.46 | 0.54 | 0.54 | 46.58 |
| Tea_ILP1000 | 5.00 | 3.29 | 1.31 | 1.00 | 0.00 | 0.30 | 0.70 | 0.70 | 63.95 |
| Tea_ILP1589 | 3.00 | 1.87 | 0.71 | 0.34 | 0.66 | 0.53 | 0.47 | 0.46 | 37.12 |
| Tea_ILP900 | 2.00 | 1.32 | 0.41 | 0.72 | 0.28 | 0.76 | 0.24 | 0.24 | 21.40 |
| Tea_ILP1097 | 2.00 | 1.97 | 0.69 | 1.00 | 0.00 | 0.51 | 0.49 | 0.49 | 37.11 |
| Tea_ILP1023 | 6.00 | 3.54 | 1.42 | 0.95 | 0.05 | 0.28 | 0.72 | 0.72 | 66.92 |
| Tea_ILP1222 | 3.00 | 1.76 | 0.77 | 0.76 | 0.24 | 0.57 | 0.43 | 0.43 | 39.22 |
| Tea_ILP1192 | 4.00 | 1.59 | 0.73 | 0.80 | 0.20 | 0.63 | 0.37 | 0.37 | 34.69 |
| Tea_ILP1073 | 3.00 | 1.26 | 0.39 | 0.80 | 0.20 | 0.80 | 0.20 | 0.20 | 18.71 |
| Tea_ILP591 | 7.00 | 5.00 | 1.72 | 1.00 | 0.00 | 0.20 | 0.80 | 0.80 | 77.16 |
| Tea_ILP1158 | 4.00 | 3.86 | 1.37 | 0.47 | 0.53 | 0.26 | 0.74 | 0.74 | 69.26 |
| Tea_ILP072 | 2.00 | 1.62 | 0.57 | 0.48 | 0.52 | 0.62 | 0.38 | 0.38 | 30.99 |
| Tea_ILP015 | 4.00 | 3.07 | 1.16 | 1.00 | 0.00 | 0.32 | 0.68 | 0.67 | 60.45 |
| Tea_ILP290 | 2.00 | 1.78 | 0.63 | 1.00 | 0.00 | 0.56 | 0.44 | 0.44 | 34.21 |
| Tea_ILP380 | 5.00 | 3.74 | 1.40 | 0.53 | 0.47 | 0.27 | 0.74 | 0.73 | 68.66 |
| Tea_ILP450 | 3.00 | 1.52 | 0.57 | 0.59 | 0.41 | 0.66 | 0.34 | 0.34 | 29.16 |
| Tea_ILP202 | 3.00 | 2.48 | 0.98 | 1.00 | 0.00 | 0.40 | 0.60 | 0.60 | 51.77 |
| Tea_ILP284 | 4.00 | 2.68 | 1.07 | 1.00 | 0.00 | 0.37 | 0.63 | 0.63 | 55.01 |
| Tea_ILP1875 | 8.00 | 6.07 | 1.92 | 1.00 | 0.00 | 0.16 | 0.84 | 0.84 | 81.51 |
| Tea_ILP1946 | 4.00 | 3.14 | 1.20 | 1.00 | 0.00 | 0.32 | 0.68 | 0.68 | 61.60 |
| Tea_ILP1986 | 10.00 | 7.63 | 2.11 | 1.00 | 0.00 | 0.13 | 0.87 | 0.87 | 85.48 |
| Tea_ILP2114 | 7.00 | 3.01 | 1.32 | 1.00 | 0.00 | 0.33 | 0.67 | 0.67 | 61.18 |
| Tea_ILP2142 | 4.00 | 1.31 | 0.49 | 0.97 | 0.03 | 0.77 | 0.24 | 0.23 | 21.94 |
| Tea_ILP2171 | 5.00 | 2.67 | 1.24 | 1.00 | 0.00 | 0.37 | 0.63 | 0.63 | 58.70 |
| Tea_ILP1923 | 3.00 | 2.08 | 0.78 | 0.10 | 0.90 | 0.48 | 0.52 | 0.52 | 40.57 |
| Tea_ILP1924 | 5.00 | 1.35 | 0.60 | 1.00 | 0.00 | 0.74 | 0.26 | 0.26 | 25.02 |
| Tea_ILP1945 | 4.00 | 2.11 | 0.86 | 0.26 | 0.74 | 0.47 | 0.53 | 0.53 | 43.27 |
| Tea_ILP1951 | 3.00 | 1.78 | 0.76 | 0.56 | 0.44 | 0.56 | 0.44 | 0.44 | 38.52 |
| Tea_ILP1967 | 4.00 | 3.11 | 1.18 | 1.00 | 0.00 | 0.32 | 0.68 | 0.68 | 61.01 |
| Tea_ILP1982 | 5.00 | 1.46 | 0.66 | 1.00 | 0.00 | 0.68 | 0.32 | 0.32 | 29.83 |
| Tea_ILP1991 | 4.00 | 1.85 | 0.75 | 0.40 | 0.60 | 0.54 | 0.46 | 0.46 | 37.74 |
| Tea_ILP2017 | 6.00 | 2.83 | 1.34 | 1.00 | 0.00 | 0.35 | 0.65 | 0.65 | 61.13 |
| Tea_ILP2551 | 2.00 | 1.54 | 0.54 | 0.55 | 0.45 | 0.65 | 0.35 | 0.35 | 28.96 |
| Tea_ILP3195 | 5.00 | 2.71 | 1.15 | 1.00 | 0.00 | 0.37 | 0.63 | 0.63 | 55.95 |
| Tea_ILP1959 | 3.00 | 2.00 | 0.74 | 0.15 | 0.85 | 0.50 | 0.50 | 0.50 | 38.65 |
| Tea_ILP3087 | 3.00 | 1.15 | 0.27 | 0.88 | 0.13 | 0.87 | 0.13 | 0.13 | 12.10 |
| Tea_ILP1953 | 4.00 | 2.97 | 1.13 | 1.00 | 0.00 | 0.34 | 0.66 | 0.66 | 59.18 |
| Tea_ILP2343 | 4.00 | 2.02 | 0.92 | 1.00 | 0.00 | 0.49 | 0.51 | 0.50 | 45.10 |
| Mean | 4.23 | 2.58 | 0.98 | 0.78 | 0.22 | 0.47 | 0.53 | 0.53 | 47.56 |
| Population | Sample Size | Longitude | Latitude | Elevation (m) | Ground Diameter (cm) | Na | Ne | I | H | PPB% |
|---|---|---|---|---|---|---|---|---|---|---|
| Qingshan Town | 55 | 104.96–104.97 | 25.42–25.43 | 1697–1721 | 18–39 | 4.1 | 2.57 | 1.01 | 0.55 | 100 |
| Louxia Town | 30 | 104.98–104.99 | 25.40–25.42 | 1574–1588 | 23–44 | 3.13 | 2.25 | 0.84 | 0.48 | 100 |
| Digua Town | 91 | 104.98–104.99 | 25.75–25.76 | 1675–1875 | 16–28 | 3.68 | 2.43 | 0.9 | 0.5 | 100 |
| Popgene | AMOVA | |||||||
|---|---|---|---|---|---|---|---|---|
| Fis | Fit | Fst | Nm | Source of Variation | df | SS | Var. Components | PMV (%) |
| 0.56 | 0.57 | 0.04 | 5.47 | Among populations | 2 | 183.68 | 1.35 | 6.40% |
| Within populations | 173 | 3419.25 | 19.76 | 93.60% | ||||
| Population | Qingshan Town | Louxia Town | Digua Town |
|---|---|---|---|
| Qingshan Town | 1.00 | 0.92 | 0.93 |
| Louxia Town | 0.09 | 1.00 | 0.94 |
| Digua Town | 0.07 | 0.06 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, Y.; He, X.; Zu, F.; Huang, X.; Yin, S.; Wang, L.; Geng, F.; Cheng, X. Development of Genome-Wide Intron Length Polymorphism (ILP) Markers in Tea Plant (Camellia sinensis) and Related Applications for Genetics Research. Int. J. Mol. Sci. 2024, 25, 3241. https://doi.org/10.3390/ijms25063241
Shen Y, He X, Zu F, Huang X, Yin S, Wang L, Geng F, Cheng X. Development of Genome-Wide Intron Length Polymorphism (ILP) Markers in Tea Plant (Camellia sinensis) and Related Applications for Genetics Research. International Journal of Molecular Sciences. 2024; 25(6):3241. https://doi.org/10.3390/ijms25063241
Chicago/Turabian StyleShen, Yuan, Xiaoying He, Feng Zu, Xiaoxia Huang, Shihua Yin, Lifei Wang, Fang Geng, and Xiaomao Cheng. 2024. "Development of Genome-Wide Intron Length Polymorphism (ILP) Markers in Tea Plant (Camellia sinensis) and Related Applications for Genetics Research" International Journal of Molecular Sciences 25, no. 6: 3241. https://doi.org/10.3390/ijms25063241
APA StyleShen, Y., He, X., Zu, F., Huang, X., Yin, S., Wang, L., Geng, F., & Cheng, X. (2024). Development of Genome-Wide Intron Length Polymorphism (ILP) Markers in Tea Plant (Camellia sinensis) and Related Applications for Genetics Research. International Journal of Molecular Sciences, 25(6), 3241. https://doi.org/10.3390/ijms25063241





