Epigenetic and Transcriptional Shifts in Human Neural Stem Cells after Reprogramming into Induced Pluripotent Stem Cells and Subsequent Redifferentiation
Abstract
1. Introduction
2. Results
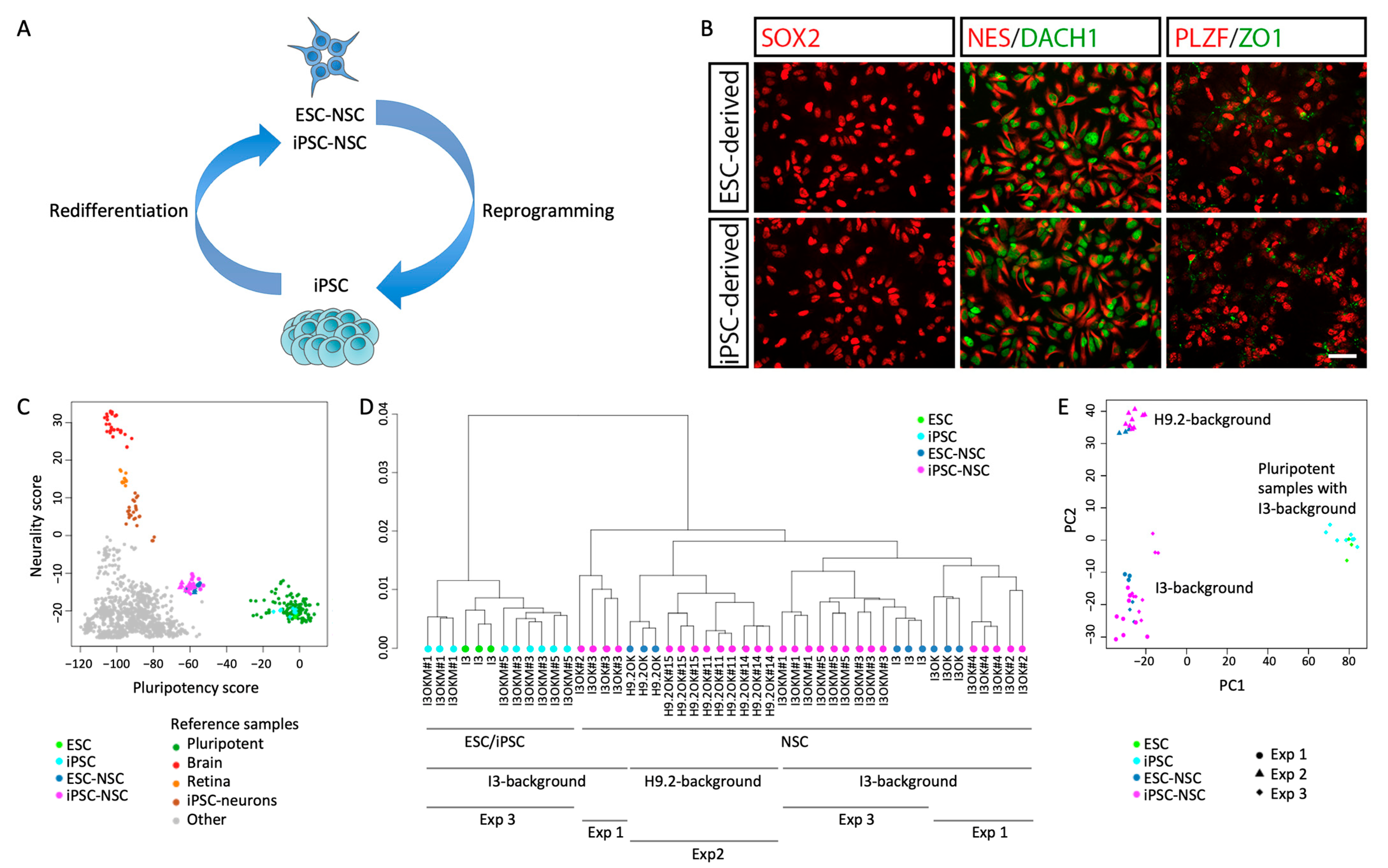
2.1. Reprogramming and Subsequent Re-Differentiation of Human NSCs
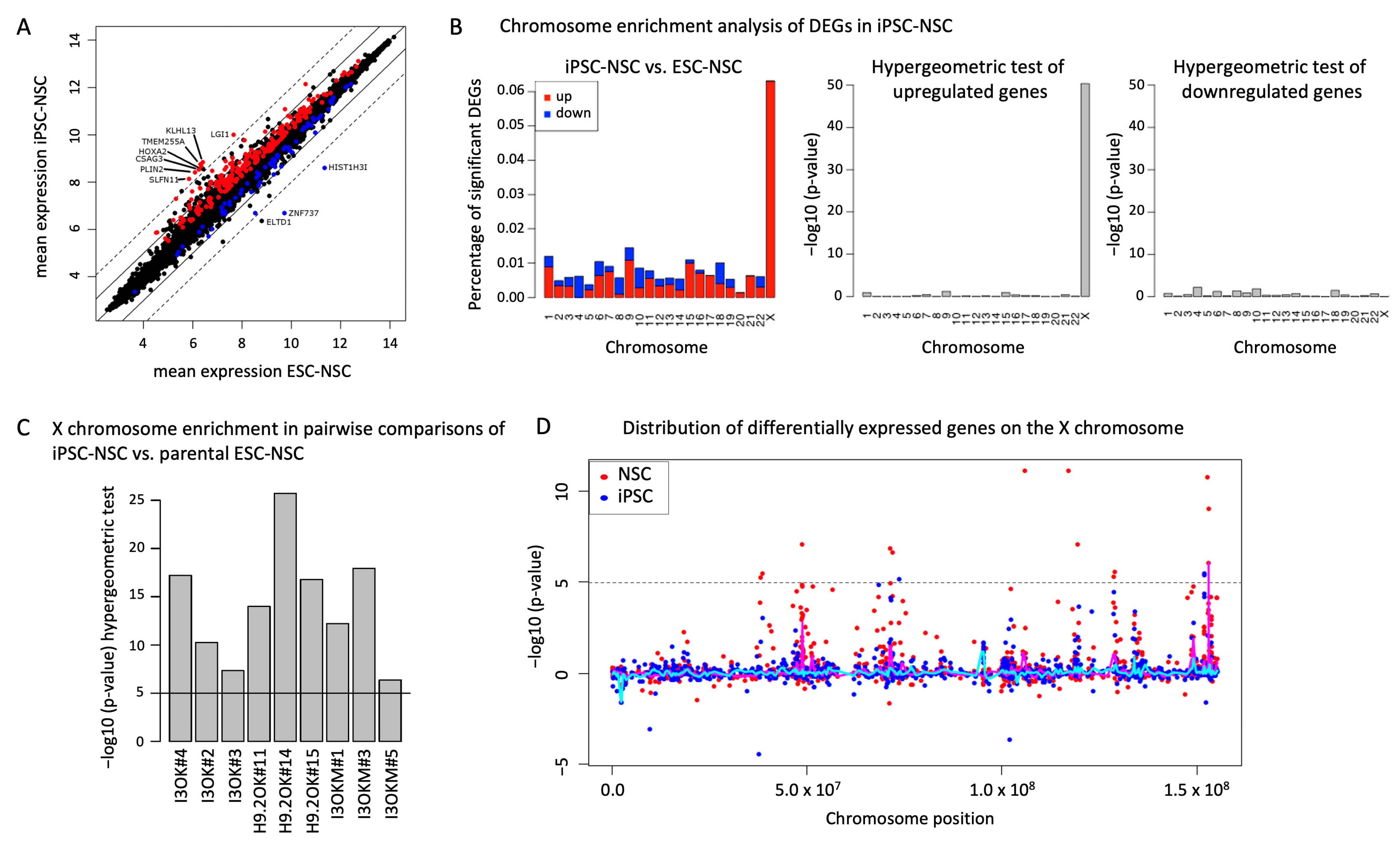
2.2. Maintenance of Autosomal and Alteration of X Chromosomal Gene Expression after Circular Reprogramming of Human NSCs
2.3. Alterations in X Chromosomal DNA Methylation during NSC Propagation through a Stable iPSC State
2.4. Biallelic X Chromosomal Gene Expression and X Chromosome Inactivation Status
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Reprogramming of NSCs to Pluripotency
4.3. Lentiviral Transduction of NSCs
4.4. In Vitro Differentiation of Pluripotent Cells into Three Germ Layers
4.5. Immunocytochemistry
4.6. Transcriptome Analysis
4.7. Whole Genome Methylation Analysis
4.8. SYBR Green-Based Quantitative Real-Time RT-PCR Analyses
4.9. Allele Specific Expression Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.N.; Yeo, G.W.; Kainohana, O.; Marsala, M.; Gage, F.H.; Muotri, A.R. Transcriptional Signature and Memory Retention of Human-Induced Pluripotent Stem Cells. PLoS ONE 2009, 4, e7076. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Lu, S.J.; Klimanskaya, I.; Gomes, I.; Kim, D.; Chung, Y.; Honig, G.R.; Kim, K.S.; Lanza, R. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 2010, 28, 704–712. [Google Scholar] [CrossRef] [PubMed]
- Guenther, M.G.; Frampton, G.M.; Soldner, F.; Hockemeyer, D.; Mitalipova, M.; Jaenisch, R.; Young, R.A. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 2010, 7, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.Y.; Weick, J.P.; Yu, J.; Ma, L.X.; Zhang, X.Q.; Thomson, J.A.; Zhang, S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef]
- Newman, A.M.; Cooper, J.B. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 2010, 7, 258–262. [Google Scholar] [CrossRef]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolo, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef]
- Bock, C.; Kiskinis, E.; Verstappen, G.; Gu, H.; Boulting, G.; Smith, Z.D.; Ziller, M.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011, 144, 439–452. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Kida, Y.S.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef]
- Mallon, B.S.; Hamilton, R.S.; Kozhich, O.A.; Johnson, K.R.; Fann, Y.C.; Rao, M.S.; Robey, P.G. Comparison of the molecular profiles of human embryonic and induced pluripotent stem cells of isogenic origin. Stem Cell Res. 2014, 12, 376–386. [Google Scholar] [CrossRef]
- Féraud, O.; Valogne, Y.; Melkus, M.W.; Zhang, Y.; Oudrhiri, N.; Haddad, R.; Daury, A.; Rocher, C.; Larbi, A.; Duquesnoy, P.; et al. Donor dependent variations in hematopoietic differentiation among embryonic and induced pluripotent stem cell lines. PLoS ONE 2016, 11, e0149291. [Google Scholar] [CrossRef][Green Version]
- Marei, H.E.; Althani, A.; Lashen, S.; Cenciarelli, C.; Hasan, A. Genetically unmatched human iPSC and ESC exhibit equivalent gene expression and neuronal differentiation potential. Sci. Rep. 2017, 7, 17504. [Google Scholar] [CrossRef]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A comparison of genetically matched cell lines reveals the equivalence of human iPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef]
- Buckberry, S.; Liu, X.; Poppe, D.; Tan, J.P.; Sun, G.; Chen, J.; Nguyen, T.V.; de Mendoza, A.; Pflueger, J.; Frazer, T.; et al. Transient naive reprogramming corrects hiPS cells functionally and epigenetically. Nature 2023, 620, 863–872. [Google Scholar] [CrossRef]
- Koch, P.; Opitz, T.; Steinbeck, J.A.; Ladewig, J.; Brüstle, O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. USA 2009, 106, 3225–3230. [Google Scholar] [CrossRef]
- Koch, P.; Breuer, P.; Peitz, M.; Jungverdorben, J.; Kesavan, J.; Poppe, D.; Doerr, J.; Ladewig, J.; Mertens, J.; Tüting, T.; et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado–Joseph disease. Nature 2011, 480, 543–546. [Google Scholar] [CrossRef]
- Maherali, N.; Ahfeldt, T.; Rigamonti, A.; Utikal, J.; Cowan, C.; Hochedlinger, K. A High-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell 2008, 3, 340–345. [Google Scholar] [CrossRef]
- Shao, K.; Koch, C.; Gupta, M.K.; Lin, Q.; Lenz, M.; Laufs, S.; Denecke, B.; Schmidt, M.; Linke, M.; Hennies, H.C.; et al. Induced pluripotent mesenchymal stromal cell clones retain donor-derived differences in DNA methylation profiles. Mol. Ther. 2013, 21, 240–250. [Google Scholar] [CrossRef]
- Leek, J.T.; Storey, J.D. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007, 3, 1724–1735. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Mekhoubad, S.; Bock, C.; de Boer, A.S.; Kiskinis, E.; Meissner, A.; Eggan, K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 2012, 10, 595–609. [Google Scholar] [CrossRef]
- Vallot, C.; Ouimette, J.-F.; Makhlouf, M.; Féraud, O.; Pontis, J.; Côme, J.; Martinat, C.; Bennaceur-Griscelli, A.; Lalande, M.; Rougeulle, C. Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell 2015, 16, 533–546. [Google Scholar] [CrossRef]
- Teichroeb, J.H.; Betts, D.H.; Vaziri, H. Suppression of the imprinted gene NNAT and X-chromosome gene activation in isogenic human iPS cells. PLoS ONE 2011, 6, e23436. [Google Scholar] [CrossRef]
- Nazor, K.L.; Altun, G.; Lynch, C.; Tran, H.; Harness, J.V.; Slavin, I.; Garitaonandia, I.; Müller, F.-J.; Wang, Y.-C.; Boscolo, F.S.; et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell 2012, 10, 620–634. [Google Scholar] [CrossRef]
- De Boni, L.; Gasparoni, G.; Haubenreich, C.; Tierling, S.; Schmitt, I.; Peitz, M.; Koch, P.; Walter, J.; Wüllner, U.; Brüstle, O. DNA methylation alterations in iPSC- and hESC-derived neurons: Potential implications for neurological disease modeling. Clin. Epigenet. 2018, 10, 13. [Google Scholar] [CrossRef]
- Genolet, O.; Monaco, A.A.; Dunkel, I.; Boettcher, M.; Schulz, E.G. Identification of X-chromosomal genes that drive sex differences in embryonic stem cells through a hierarchical CRISPR screening approach. Genome Biol. 2021, 22, 110. [Google Scholar] [CrossRef]
- Panda, A.; Zylicz, J.J.; Pasque, V. New insights into X-chromosome reactivation during reprogramming to pluripotency. Cells 2020, 9, 2706. [Google Scholar] [CrossRef]
- Naik, H.C.; Chandel, D.; Majumdar, S.; Arava, M.; Baro, R.; Bv, H.; Hari, K.; Parichitran; Avinchal; Jolly, M.K. Lineage-specific dynamics of erasure of X-upregulation during inactive-X reactivation. bioRxiv 2023. [Google Scholar] [CrossRef]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014, 15, 471–487. [Google Scholar] [CrossRef]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef]
- Sahakyan, A.; Kim, R.; Chronis, C.; Sabri, S.; Bonora, G.; Theunissen, T.W.; Kuoy, E.; Langerman, J.; Clark, A.T.; Jaenisch, R.; et al. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell 2016, 20, 87–101. [Google Scholar] [CrossRef]
- Pick, M.; Stelzer, Y.; Bar-Nur, O.; Mayshar, Y.; Eden, A.; Benvenisty, N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells 2009, 27, 2686–2690. [Google Scholar] [CrossRef]
- Stadtfeld, M.; Apostolou, E.; Akutsu, H.; Fukuda, A.; Follett, P.; Natesan, S.; Kono, T.; Shioda, T.; Hochedlinger, K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 2010, 465, 175–181. [Google Scholar] [CrossRef]
- Hiura, H.; Toyoda, M.; Okae, H.; Sakurai, M.; Miyauchi, N.; Sato, A.; Kiyokawa, N.; Okita, H.; Miyagawa, Y.; Akutsu, H.; et al. Stability of genomic imprinting in human induced pluripotent stem cells. BMC Genet. 2013, 14, 32. [Google Scholar] [CrossRef]
- Pastor, W.A.; Chen, D.; Liu, W.; Kim, R.; Sahakyan, A.; Lukianchikov, A.; Plath, K.; Jacobsen, S.E.; Clark, A.T. Naive human pluripotent cells feature a methylation landscape devoid of blastocyst or germline memory. Cell Stem Cell 2016, 18, 323–329. [Google Scholar] [CrossRef]
- Bar, S.; Schachter, M.; Eldar-Geva, T.; Benvenisty, N. Large-Scale Analysis of loss of imprinting in human pluripotent stem cells. Cell Rep. 2017, 19, 957–968. [Google Scholar] [CrossRef]
- Alders, M.; Ryan, A.; Hodges, M.D.J.; Bliek, J.; Feinberg, A.P.; Privitera, O.; Westerveld, A.; Little, P.F.R.; Mannens, M. Disruption of a novel imprinted zinc-finger gene, ZNF215, in Beckwith-Wiedemann Syndrome. Am. J. Hum. Genet. 2000, 66, 1473–1484. [Google Scholar] [CrossRef]
- Mertens, J.; Stüber, K.; Wunderlich, P.; Ladewig, J.; Kesavan, J.C.; Vandenberghe, R.; Vandenbulcke, M.; van Damme, P.; Walter, J.; Brüstle, O.; et al. APP processing in human pluripotent stem cell-derived neurons is resistant to NSAID-based γ-secretase modulation. Stem Cell Rep. 2013, 1, 491–498. [Google Scholar] [CrossRef]
- Müller, F.-J.; Schuldt, B.M.; Williams, R.; Mason, D.; Altun, G.; Papapetrou, E.P.; Danner, S.; E Goldmann, J.; Herbst, A.; O Schmidt, N.; et al. A bioinformatic assay for pluripotency in human cells. Nat. Methods 2011, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haubenreich, C.; Lenz, M.; Schuppert, A.; Peitz, M.; Koch, P.; Zenke, M.; Brüstle, O. Epigenetic and Transcriptional Shifts in Human Neural Stem Cells after Reprogramming into Induced Pluripotent Stem Cells and Subsequent Redifferentiation. Int. J. Mol. Sci. 2024, 25, 3214. https://doi.org/10.3390/ijms25063214
Haubenreich C, Lenz M, Schuppert A, Peitz M, Koch P, Zenke M, Brüstle O. Epigenetic and Transcriptional Shifts in Human Neural Stem Cells after Reprogramming into Induced Pluripotent Stem Cells and Subsequent Redifferentiation. International Journal of Molecular Sciences. 2024; 25(6):3214. https://doi.org/10.3390/ijms25063214
Chicago/Turabian StyleHaubenreich, Carolin, Michael Lenz, Andreas Schuppert, Michael Peitz, Philipp Koch, Martin Zenke, and Oliver Brüstle. 2024. "Epigenetic and Transcriptional Shifts in Human Neural Stem Cells after Reprogramming into Induced Pluripotent Stem Cells and Subsequent Redifferentiation" International Journal of Molecular Sciences 25, no. 6: 3214. https://doi.org/10.3390/ijms25063214
APA StyleHaubenreich, C., Lenz, M., Schuppert, A., Peitz, M., Koch, P., Zenke, M., & Brüstle, O. (2024). Epigenetic and Transcriptional Shifts in Human Neural Stem Cells after Reprogramming into Induced Pluripotent Stem Cells and Subsequent Redifferentiation. International Journal of Molecular Sciences, 25(6), 3214. https://doi.org/10.3390/ijms25063214






