METACASPASE8 (MC8) Is a Crucial Protein in the LSD1-Dependent Cell Death Pathway in Response to Ultraviolet Stress
Abstract
1. Introduction
2. Results
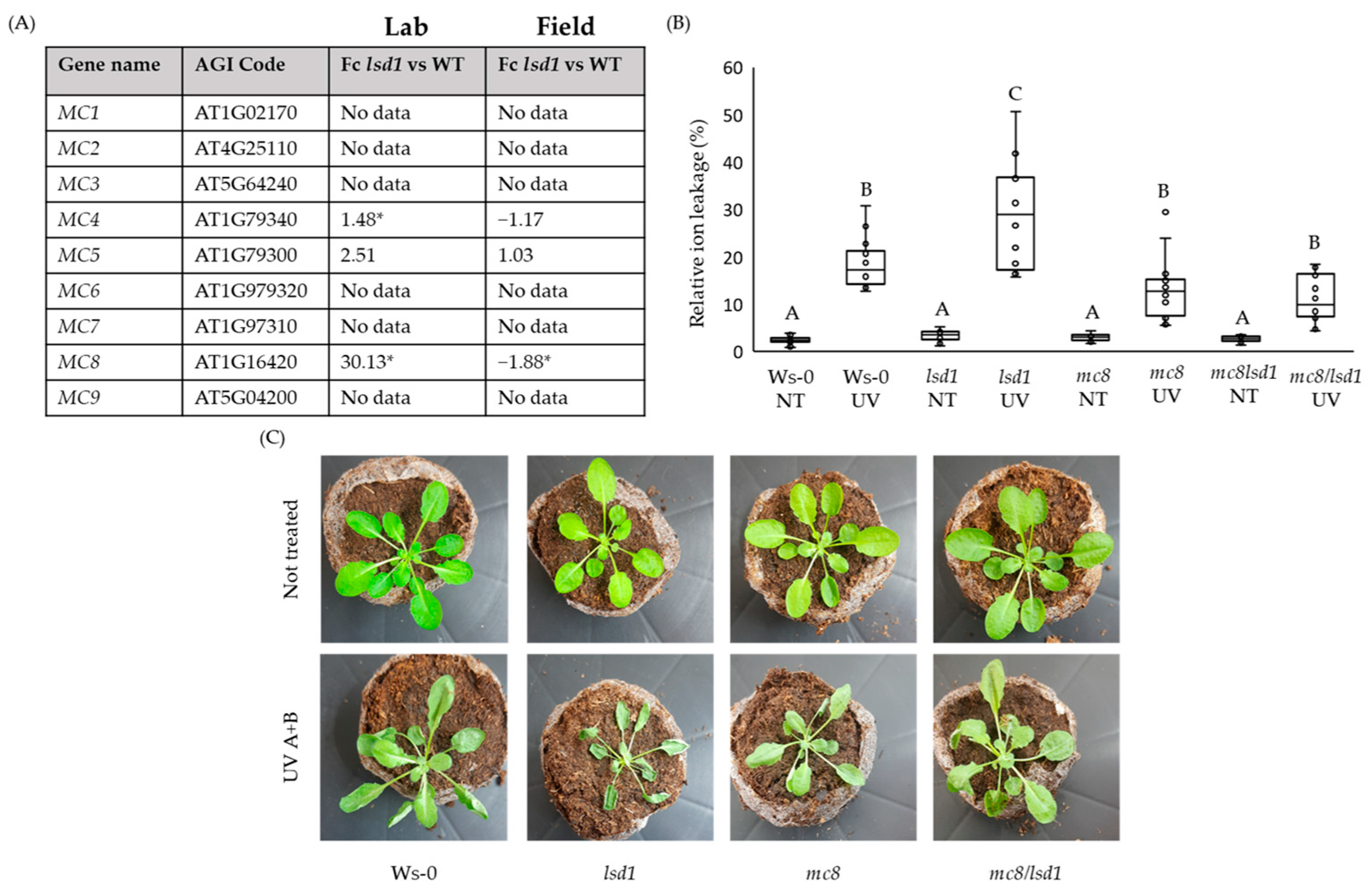
2.1. MC8 Is Important in the LSD1-Dependent Cell Death Pathway
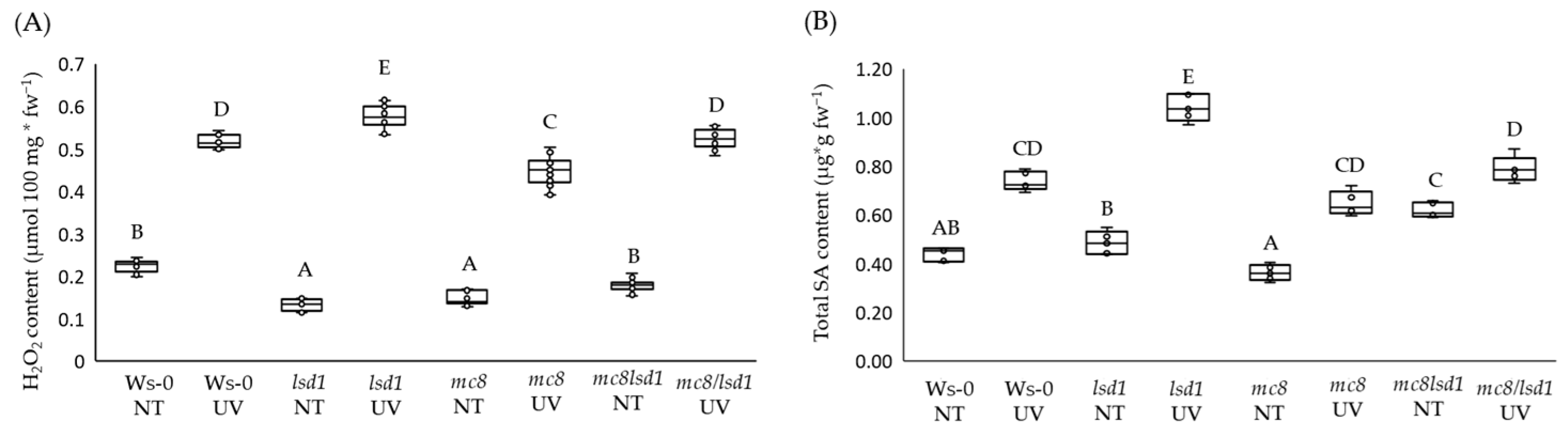
2.2. MC8 Affects LSD1-Dependent Foliar ROS and SA Levels
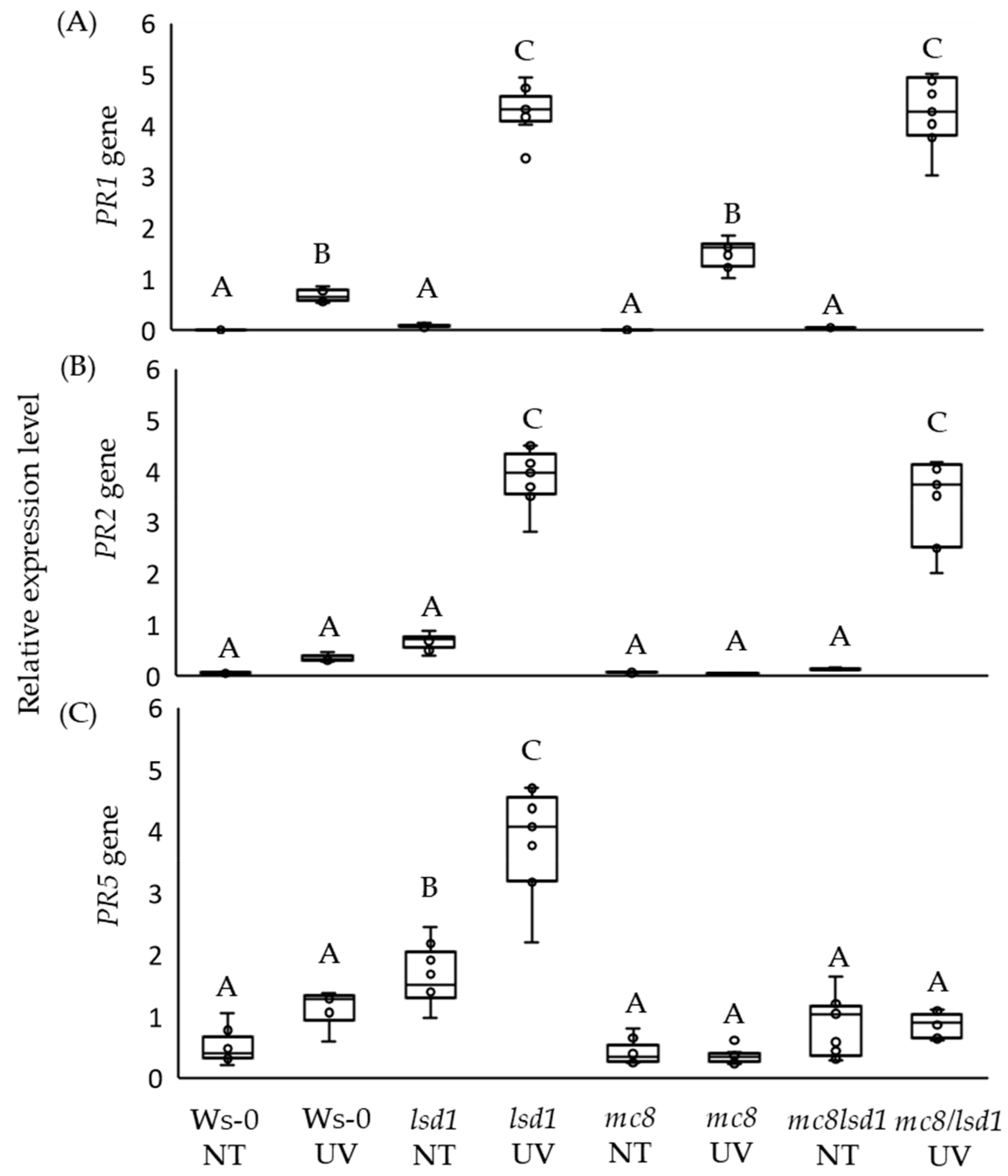
2.3. High PR5 Gene Expression Level in lsd1 Is Reverted by the Mutation in MC8
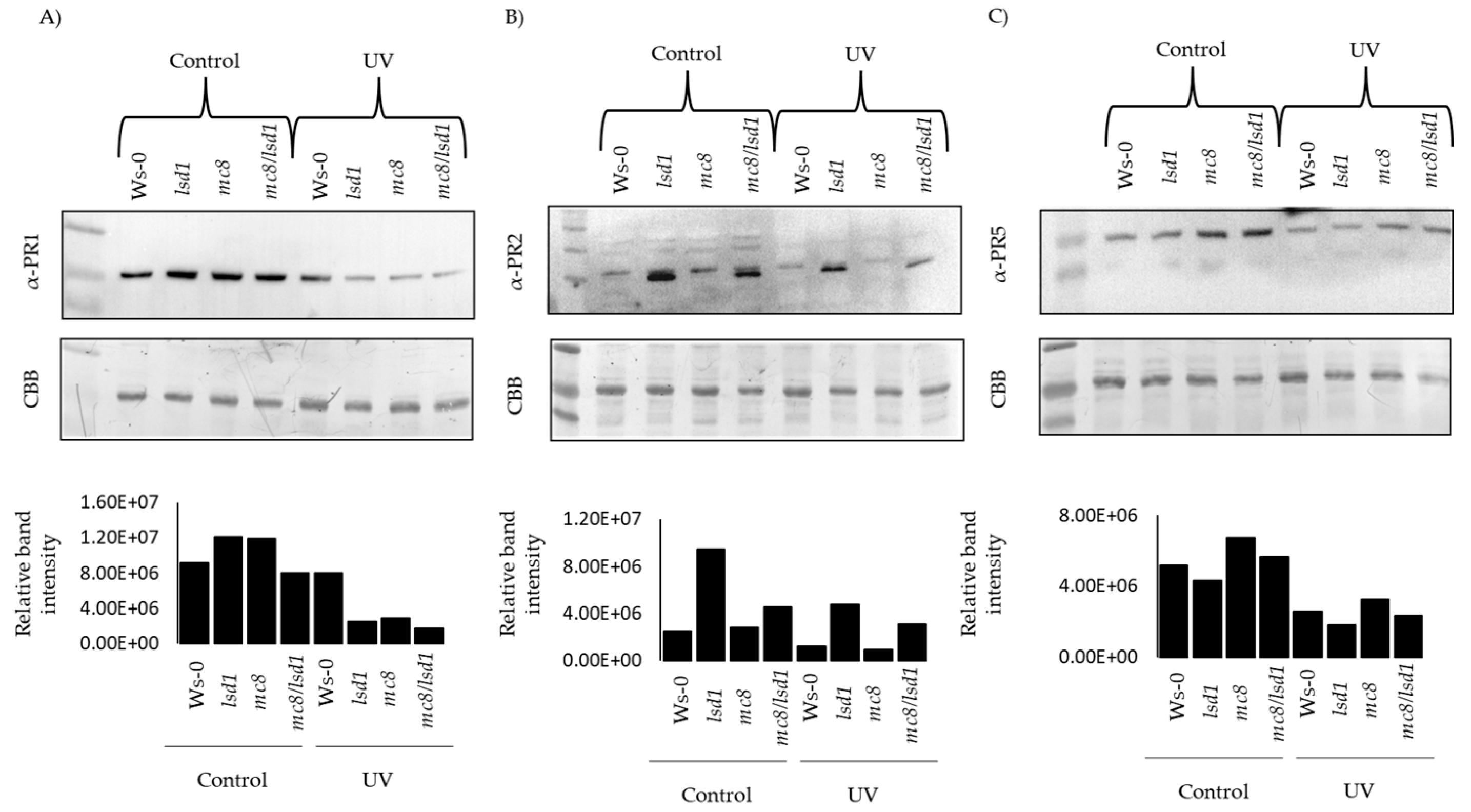
2.4. PR Proteins Are Degraded in Response to UV Stress
2.5. Prediction of MC8 Interaction
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Growing Conditions
4.3. Stress Induction
4.4. Ion Leakage Measurement
4.5. Determination of H2O2 and SA Contents
4.6. RNA Isolation, cDNA Synthesis and RT-PCR Analysis
4.7. Protein Extraction and Western Blot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mühlenbock, P.; Plaszczyca, M.; Plaszczyca, M.; Mellerowicz, E.; Karpinski, S. Lysigenous Aerenchyma Formation in Arabidopsis Is Controlled by LESION SIMULATING DISEASE1. Plant Cell 2007, 19, 3819–3830. [Google Scholar] [CrossRef] [PubMed]
- Mühlenbock, P.; Szechynska-Hebda, M.; Plaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpinska, B.; Karpinski, S. Chloroplast Signaling and LESION SIMULATING DISEASE1 Regulate Crosstalk between Light Acclimation and Immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef] [PubMed]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light Acclimation, Retrograde Signalling, Cell Death and Immune Defences in Plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Kruk, J.; Górecka, M.; Karpińska, B.; Karpiński, S. Evidence for Light Wavelength-Specific Photoelectrophysiological Signaling and Memory of Excess Light Episodes in Arabidopsis. Plant Cell 2010, 22, 2201–2218. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Dong, X. How Do Plants Achieve Immunity? Defence without Specialized Immune Cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef]
- Fukuda, H. Programmed Cell Death of Tracheary Elements as a Paradigm in Plants. Plant Mol. Biol. 2000, 44, 245–253. [Google Scholar] [CrossRef]
- Domínguez, F.; Cejudo, F.J. Programmed Cell Death (PCD): An Essential Process of Cereal Seed Development and Germination. Front Plant Sci 2014, 5, 366. [Google Scholar] [CrossRef]
- Jabs, T.; Dietrich, R.A.; Dangl, J.L. Initiation of Runaway Cell Death in an Arabidopsis Mutant by Extracellular Superoxide. Science 1996, 273, 1853–1856. [Google Scholar] [CrossRef]
- Lorrain, S.; Vailleau, F.; Balagué, C.; Roby, D. Lesion Mimic Mutants: Keys for Deciphering Cell Death and Defense Pathways in Plants? Trends Plant Sci. 2003, 8, 263–271. [Google Scholar] [CrossRef]
- Vandenabeele, S.; Vanderauwera, S.; Vuylsteke, M.; Rombauts, S.; Langebartels, C.; Seidlitz, H.K.; Zabeau, M.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Catalase Deficiency Drastically Affects Gene Expression Induced by High Light in Arabidopsis Thaliana. Plant J. 2004, 39, 45–58. [Google Scholar] [CrossRef]
- Dietrich, R.A.; Delaney, T.P.; Uknes, S.J.; Ward, E.R.; Ryals, J.A.; Dangl, J.L. Arabidopsis Mutants Simulating Disease Resistance Response. Cell 1994, 77, 565–577. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Karpiński, S. Programmed Cell Death as a Response to High Light, UV and Drought Stress in Plants. In Abiotic Stress—Plant Responses and Applications in Agriculture; InTech: Rijeka, Croatia; Shanghai, China, 2013. [Google Scholar] [CrossRef]
- Mateo, A.; Funck, D.; Mühlenbock, P.; Kular, B.; Mullineaux, P.M.; Karpinski, S. Controlled Levels of Salicylic Acid Are Required for Optimal Photosynthesis and Redox Homeostasis. J. Exp. Bot. 2006, 57, 1795–1807. [Google Scholar] [CrossRef]
- Chai, T.; Zhou, J.; Liu, J.; Xing, D. LSD1 and HY5 Antagonistically Regulate Red Light Induced-Programmed Cell Death in Arabidopsis. Front. Plant Sci. 2015, 6, 292. [Google Scholar] [CrossRef] [PubMed]
- Bernacki, M.J.; Mielecki, J.; Antczak, A.; Drożdżek, M.; Witoń, D.; Dąbrowska-Bronk, J.; Gawroński, P.; Burdiak, P.; Marchwicka, M.; Rusaczonek, A.; et al. Biotechnological Potential of the Stress Response and Plant Cell Death Regulators Proteins in the Biofuel Industry. Cells 2023, 12, 2018. [Google Scholar] [CrossRef] [PubMed]
- Wituszyńska, W.; Ślesak, I.; Vanderauwera, S.; Szechyńska-Hebda, M.; Kornaś, A.; Kelen, K.V.D.; Mühlenbock, P.; Karpińska, B.; Maćkowski, S.; Breusegem, F.V.; et al. LESION SIMULATING DISEASE1, ENHANCED DISEASE SUSCEPTIBILITY1, and PHYTOALEXIN DEFICIENT4 Conditionally Regulate Cellular Signaling Homeostasis, Photosynthesis, Water Use Efficiency, and Seed Yield in Arabidopsis. Plant Physiol. 2013, 161, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Szechyńska-Hebda, M.; Czarnocka, W.; Hebda, M.; Karpiński, S. PAD4, LSD1 and EDS1 Regulate Drought Tolerance, Plant Biomass Production, and Cell Wall Properties. Plant Cell Rep. 2016, 35, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Li, Y.; Zhang, X.; Zuo, J.; Yang, S. The Arabidopsis LSD1 Gene Plays an Important Role in the Regulation of Low Temperature-Dependent Cell Death. New Phytol. 2010, 187, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Bernacki, M.J.; Rusaczonek, A.; Czarnocka, W.; Karpiński, S. Salicylic Acid Accumulation Controlled by LSD1 Is Essential in Triggering Cell Death in Response to Abiotic Stress. Cells 2021, 10, 962. [Google Scholar] [CrossRef] [PubMed]
- Aviv, D.H.; Rustérucci, C.; Iii, B.F.H.; Dietrich, R.A.; Parker, J.E.; Dangl, J.L. Runaway Cell Death, but Not Basal Disease Resistance, in Lsd1 Is SA- and NIM1/NPR1-Dependent. Plant J. 2002, 29, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Rustérucci, C.; Aviv, D.H.; Holt, B.F.; Dangl, J.L.; Parker, J.E. The Disease Resistance Signaling Components EDS1 and PAD4 Are Essential Regulators of the Cell Death Pathway Controlled by LSD1 in Arabidopsis. Plant Cell 2001, 13, 2211–2224. [Google Scholar] [CrossRef]
- Bernacki, M.J.; Czarnocka, W.; Rusaczonek, A.; Witoń, D.; Kęska, S.; Czyż, J.; Szechyńska-Hebda, M.; Karpiński, S. LSD1, EDS1 and PAD4-dependent conditional correlation among salicylic acid, hydrogen peroxide, water use efficiency, and seed yield in Arabidopsis thaliana. Physiol. Plant. 2018, 165, 369–382. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Szechyńska-Hebda, M.; Sobczak, M.; Rusaczonek, A.; Kozłowska-Makulska, A.; Witoń, D.; Karpiński, S. Lesion Simulating Disease 1 and Enhanced Disease Susceptibility 1 Differentially Regulate UV-C-Induced Photooxidative Stress Signalling and Programmed Cell Death in Arabidopsis Thaliana. Plant Cell Environ. 2015, 38, 315–330. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Mu, J.; Zuo, J. LESION SIMULATING DISEASE1 Interacts with Catalases to Regulate Hypersensitive Cell Death in Arabidopsis1[C][W]. Plant Physiol. 2013, 163, 1059–1070. [Google Scholar] [CrossRef]
- Kliebenstein, D.J.; Dietrich, R.A.; Martin, A.C.; Last, R.L.; Dangl, J.L. LSD1 Regulates Salicylic Acid Induction of Copper Zinc Superoxide Dismutase in Arabidopsis Thaliana. Mol. Plant. Microbe. Interact. 1999, 12, 1022–1026. [Google Scholar] [CrossRef]
- Feys, B.J.; Moisan, L.J.; Newman, M.-A.; Parker, J.E. Direct Interaction between the Arabidopsis Disease Resistance Signaling Proteins, EDS1 and PAD4. EMBO J. 2001, 20, 5400–5411. [Google Scholar] [CrossRef]
- Czarnocka, W.; Van Der Kelen, K.; Willems, P.; Szechyńska-Hebda, M.; Shahnejat-Bushehri, S.; Balazadeh, S.; Rusaczonek, A.; Mueller-Roeber, B.; Van Breusegem, F.; Karpiński, S. The Dual Role of LESION SIMULATING DISEASE 1 as a Condition-Dependent Scaffold Protein and Transcription Regulator. Plant Cell Environ. 2017, 40, 2644–2662. [Google Scholar] [CrossRef]
- Parker, J.E.; Holub, E.B.; Frost, L.N.; Falk, A.; Gunn, N.D.; Daniels, M.J. Characterization of Eds1, a Mutation in Arabidopsis Suppressing Resistance to Peronospora Parasitica Specified by Several Different RPP Genes. Plant Cell 1996, 8, 2033–2046. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J.; Rogers, E.E.; Ausubel, F.M. Isolation of Arabidopsis Mutants with Enhanced Disease Susceptibility by Direct Screening. Genetics 1996, 143, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Falk, A.; Feys, B.J.; Frost, L.N.; Jones, J.D.G.; Daniels, M.J.; Parker, J.E. EDS1, an Essential Component of R Gene-Mediated Disease Resistance in Arabidopsis Has Homology to Eukaryotic Lipases. Proc. Natl. Acad. Sci. USA 1999, 96, 3292–3297. [Google Scholar] [CrossRef] [PubMed]
- Jirage, D.; Tootle, T.L.; Reuber, T.L.; Frost, L.N.; Feys, B.J.; Parker, J.E.; Ausubel, F.M.; Glazebrook, J. Arabidopsis Thaliana PAD4 Encodes a Lipase-like Gene That Is Important for Salicylic Acid Signaling. Proc. Natl. Acad. Sci. USA 1999, 96, 13583–13588. [Google Scholar] [CrossRef] [PubMed]
- Mateo, A.; Mühlenbock, P.; Rustérucci, C.; Chang, C.C.-C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. LESION SIMULATING DISEASE 1 Is Required for Acclimation to Conditions That Promote Excess Excitation Energy. Plant Physiol. 2004, 136, 2818–2830. [Google Scholar] [CrossRef]
- Dietrich, R.A.; Richberg, M.H.; Schmidt, R.; Dean, C.; Dangl, J.L. A Novel Zinc Finger Protein Is Encoded by the Arabidopsis LSD1 Gene and Functions as a Negative Regulator of Plant Cell Death. Cell 1997, 88, 685–694. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-Finger Transcription Factors in Plants. CMLS Cell. Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis Type I Metacaspases Control Cell Death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef]
- Fagundes, D.; Bohn, B.; Cabreira, C.; Leipelt, F.; Dias, N.; Bodanese-Zanettini, M.H.; Cagliari, A. Caspases in Plants: Metacaspase Gene Family in Plant Stress Responses. Funct. Integr. Genom. 2015, 15, 639–649. [Google Scholar] [CrossRef]
- Vercammen, D.; van de Cotte, B.; De Jaeger, G.; Eeckhout, D.; Casteels, P.; Vandepoele, K.; Vandenberghe, I.; Van Beeumen, J.; Inzé, D.; Van Breusegem, F. Type II Metacaspases Atmc4 and Atmc9 of Arabidopsis Thaliana Cleave Substrates after Arginine and Lysine. J. Biol. Chem. 2004, 279, 45329–45336. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Drury, G.E.; Rotari, V.I.; Gordon, A.; Willer, M.; Farzaneh, T.; Woltering, E.J.; Gallois, P. Metacaspase-8 Modulates Programmed Cell Death Induced by Ultraviolet Light and H2O2 in Arabidopsis. J. Biol. Chem. 2008, 283, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.I.; Hwang, D.J. Expression Analysis of the Metacaspase Gene Family in Arabidopsis. J. Plant Biol. 2013, 56, 391–398. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Van Breusegem, F.; Gallois, P.; Zavialov, A.; Lam, E.; Bozhkov, P.V. Metacaspases. Cell Death Differ. 2011, 18, 1279–1288. [Google Scholar] [CrossRef]
- Glazebrook, J. Genes Controlling Expression of Defense Responses in Arabidopsis—2001 Status. Curr. Opin. Plant Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Penninckx, I.A.; Cammue, B.P.; Broekaert, W.F. The Complexity of Disease Signaling in Arabidopsis. Curr. Opin. Immunol. 2001, 13, 63–68. [Google Scholar] [CrossRef]
- Burke, R.; Schwarze, J.; Sherwood, O.L.; Jnaid, Y.; McCabe, P.F.; Kacprzyk, J. Stressed to Death: The Role of Transcription Factors in Plant Programmed Cell Death Induced by Abiotic and Biotic Stimuli. Front. Plant Sci. 2020, 11, 1235. [Google Scholar] [CrossRef]
- Bernacki, M.J.; Czarnocka, W.; Zaborowska, M.; Różańska, E.; Labudda, M.; Rusaczonek, A.; Witoń, D.; Karpiński, S. EDS1-Dependent Cell Death and the Antioxidant System in Arabidopsis Leaves Is Deregulated by the Mammalian Bax. Cells 2020, 9, 2454. [Google Scholar] [CrossRef]
- Lacomme, C.; Cruz, S.S. Bax-Induced Cell Death in Tobacco Is Similar to the Hypersensitive Response. Proc. Natl. Acad. Sci. USA 1999, 96, 7956–7961. [Google Scholar] [CrossRef]
- Danon, A.; Delorme, V.; Mailhac, N.; Gallois, P. Plant Programmed Cell Death: A Common Way to Die. Plant Physiol. Biochem. 2000, 38, 647–655. [Google Scholar] [CrossRef]
- Williams, B.; Dickman, M. Plant Programmed Cell Death: Can’t Live with It; Can’t Live without It. Mol. Plant Pathol. 2008, 9, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Aarts, N.; Metz, M.; Holub, E.; Staskawicz, B.J.; Daniels, M.J.; Parker, J.E. Different Requirements for EDS1 and NDR1 by Disease Resistance Genes Define at Least Two R Gene-Mediated Signaling Pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 1998, 95, 10306–10311. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Gobbato, E.; Kracher, B.; Qiu, J.; Bautor, J.; Parker, J.E. A Core Function of EDS1 with PAD4 Is to Protect the Salicylic Acid Defense Sector in Arabidopsis Immunity. New Phytol. 2017, 213, 1802–1817. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Koshino, Y.; Sawa, S.; Ishiguro, S.; Okada, K.; Tanaka, A. Overexpression of Chlorophyllide a Oxygenase (CAO) Enlarges the Antenna Size of Photosystem II in Arabidopsis Thaliana. Plant J. 2001, 26, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Lv, R.; Li, Z.; Li, M.; Dogra, V.; Lv, S.; Liu, R.; Lee, K.P.; Kim, C. Uncoupled Expression of Nuclear and Plastid Photosynthesis-Associated Genes Contributes to Cell Death in a Lesion Mimic Mutant. Plant Cell 2019, 31, 210–230. [Google Scholar] [CrossRef] [PubMed]
- Szechyńska-Hebda, M.; Ghalami, R.Z.; Kamran, M.; Van Breusegem, F.; Karpiński, S. To Be or Not to Be? Are Reactive Oxygen Species, Antioxidants, and Stress Signalling Universal Determinants of Life or Death? Cells 2022, 11, 4105. [Google Scholar] [CrossRef] [PubMed]
- Ahn, G.; Jung, I.J.; Cha, J.-Y.; Jeong, S.Y.; Shin, G.-I.; Ji, M.G.; Kim, M.G.; Lee, S.Y.; Kim, W.-Y. Phytochrome B Positively Regulates Red Light-Mediated ER Stress Response in Arabidopsis. Front. Plant Sci. 2022, 13, 846294. [Google Scholar] [CrossRef] [PubMed]
- Fichman, Y.; Myers, R.J.; Grant, D.G.; Mittler, R. Plasmodesmata-Localized Proteins and ROS Orchestrate Light-Induced Rapid Systemic Signaling in Arabidopsis. Sci. Signal. 2021, 14, eabf0322. [Google Scholar] [CrossRef] [PubMed]
- Kawai-Yamada, M.; Jin, L.; Yoshinaga, K.; Hirata, A.; Uchimiya, H. Mammalian Bax-Induced Plant Cell Death Can Be down-Regulated by Overexpression of Arabidopsis Bax Inhibitor-1 (AtBI-1). Proc. Natl. Acad. Sci. USA 2001, 98, 12295–12300. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Métraux, J.-P. Salicylic Acid Induction–Deficient Mutants of Arabidopsis Express PR-2 and PR-5 and Accumulate High Levels of Camalexin after Pathogen Inoculation. Plant Cell 1999, 11, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Kalbina, I.; Strid, Å. Supplementary Ultraviolet-B Irradiation Reveals Differences in Stress Responses between Arabidopsis Thaliana Ecotypes. Plant Cell Environ. 2006, 29, 754–763. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, A.; Satapathy, K.B.; Panigrahi, G.K. Ectopic Expression of Disease Resistance Protein Promotes Resistance against Pathogen Infection and Drought Stress in Arabidopsis. Physiol. Mol. Plant Pathol. 2023, 124, 101949. [Google Scholar] [CrossRef]
- Czégény, G.; Mátai, A.; Hideg, É. UV-B Effects on Leaves—Oxidative Stress and Acclimation in Controlled Environments. Plant Sci. 2016, 248, 57–63. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Deng, H.; Sun, X.; Wang, A.; Tang, X.; Gao, Y.; Zhang, N.; Wang, L.; Yang, S.; et al. Tomato UV-B Receptor SlUVR8 Mediates Plant Acclimation to UV-B Radiation and Enhances Fruit Chloroplast Development via Regulating SlGLK2. Sci. Rep. 2018, 8, 6097. [Google Scholar] [CrossRef]
- Nasibova, A.N. UV-B Radiation Effects on Electron-Transport Reactions in Biomaterials. Adv. Biol. Earth Sci. 2022, 7, 13–18. [Google Scholar]
- Piccini, C.; Cai, G.; Dias, M.C.; Romi, M.; Longo, R.; Cantini, C. UV-B Radiation Affects Photosynthesis-Related Processes of Two Italian Olea Europaea (L.) Varieties Differently. Plants 2020, 9, 1712. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS Signaling: The New Wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef]
- Shah, J. The Salicylic Acid Loop in Plant Defense. Curr. Opin. Plant Biol. 2003, 6, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Straus, M.R.; Rietz, S.; Ver Loren van Themaat, E.; Bartsch, M.; Parker, J.E. Salicylic Acid Antagonism of EDS1-Driven Cell Death Is Important for Immune and Oxidative Stress Responses in Arabidopsis. Plant J. 2010, 62, 628–640. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Kenton, P.; Lloyd, A.J.; Ougham, H.; Prats, E. The Hypersensitive Response; the Centenary Is upon Us but How Much Do We Know? J. Exp. Bot. 2008, 59, 501–520. [Google Scholar] [CrossRef] [PubMed]
- Velikova, V.; Sharkey, T.D.; Loreto, F. Stabilization of Thylakoid Membranes in Isoprene-Emitting Plants Reduces Formation of Reactive Oxygen Species. Plant Signal. Behav. 2012, 7, 139–141. [Google Scholar] [CrossRef]
- Meuwly, P.; Métraux, J.P. Ortho-Anisic Acid as Internal Standard for the Simultaneous Quantitation of Salicylic Acid and Its Putative Biosynthetic Precursors in Cucumber Leaves. Anal. Biochem. 1993, 214, 500–505. [Google Scholar] [CrossRef]
- Górecka, M.; Lewandowska, M.; Dąbrowska-Bronk, J.; Białasek, M.; Barczak-Brzyżek, A.; Kulasek, M.; Mielecki, J.; Kozłowska-Makulska, A.; Gawroński, P.; Karpiński, S. Photosystem II 22kDa Protein Level—A Prerequisite for Excess Light-Inducible Memory, Cross-Tolerance to UV-C and Regulation of Electrical Signalling. Plant Cell Environ. 2020, 43, 649–661. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.; Harper, S.; Speicher, D.W. Detection of Proteins on Blot Membranes. Curr. Protoc. Protein Sci. 2016, 86, 10.8.1–10.8.11. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernacki, M.J.; Rusaczonek, A.; Gołębiewska, K.; Majewska-Fala, A.B.; Czarnocka, W.; Karpiński, S.M. METACASPASE8 (MC8) Is a Crucial Protein in the LSD1-Dependent Cell Death Pathway in Response to Ultraviolet Stress. Int. J. Mol. Sci. 2024, 25, 3195. https://doi.org/10.3390/ijms25063195
Bernacki MJ, Rusaczonek A, Gołębiewska K, Majewska-Fala AB, Czarnocka W, Karpiński SM. METACASPASE8 (MC8) Is a Crucial Protein in the LSD1-Dependent Cell Death Pathway in Response to Ultraviolet Stress. International Journal of Molecular Sciences. 2024; 25(6):3195. https://doi.org/10.3390/ijms25063195
Chicago/Turabian StyleBernacki, Maciej Jerzy, Anna Rusaczonek, Kinga Gołębiewska, Agata Barbara Majewska-Fala, Weronika Czarnocka, and Stanisław Mariusz Karpiński. 2024. "METACASPASE8 (MC8) Is a Crucial Protein in the LSD1-Dependent Cell Death Pathway in Response to Ultraviolet Stress" International Journal of Molecular Sciences 25, no. 6: 3195. https://doi.org/10.3390/ijms25063195
APA StyleBernacki, M. J., Rusaczonek, A., Gołębiewska, K., Majewska-Fala, A. B., Czarnocka, W., & Karpiński, S. M. (2024). METACASPASE8 (MC8) Is a Crucial Protein in the LSD1-Dependent Cell Death Pathway in Response to Ultraviolet Stress. International Journal of Molecular Sciences, 25(6), 3195. https://doi.org/10.3390/ijms25063195










