1. Introduction
Osteoarthritis (OA) is the most common form of arthritis and one of the most prevalent diseases in middle-aged and older people. Knee osteoarthritis is one of the leading causes of physical disability in adults [
1,
2,
3,
4]. Initially, it was seen as a disease in which only mechanical degradation of the cartilage occurred, but nowadays, it is considered a very complex disease involving different tissues [
5,
6]. Thus, alterations in the joint happen at different levels, namely, in the metabolism and architecture of the subchondral bone and in the morphology and metabolism of the articular cartilage, presenting periarticular osteophytosis, inflammation, meniscus degeneration, and fibrosis of the synovial membrane in different degrees. In addition, it is related to changes in other tissues, such as ligaments, tendons, and the sand surrounding the musculature [
6,
7,
8,
9,
10]. Changes in the morphometric characteristics of the infrapatellar fat pad have also been observed, supporting an important role in the pathogenesis and progression of OA [
6,
11].
The epidemiology of this disorder is complex and multifactorial, with genetic, biological, and biomechanical components [
6,
7,
9,
10]. The main risk factors are age, obesity, sex, abnormal mechanical joint loading, altered joint morphology, and previous joint injury, especially previous knee injuries [
7,
11,
12,
13,
14,
15,
16].
Despite its worldwide importance, there is no official treatment that cures, reverses, or slows down the development of OA. This could be because its pathogenesis is not yet fully understood [
1,
17]. For this reason, treatment of OA has traditionally consisted of management of the primary cause, followed by treatment of pain, control of clinical signs, and surgical intervention in some late stages [
1,
7,
18].
For OA diagnosis, a new tool called metabolomics has emerged, and it might provide more valuable information, given that current imaging techniques offer a late diagnosis lacking information on the functional adaptation of cartilage. Metabolomics consists of the study of small biological molecules in a system and holds great potential for early diagnosis, monitoring therapies, and the understanding of the pathogenesis of many diseases [
1,
3,
4,
7,
19].
For this reason, it has become an ideal method for the identification of OA biomarkers in a variety of biological samples. Different studies have reported several metabolites and metabolic pathways that can be altered in OA, such as amino acid metabolism, fatty acid and lipid metabolism, phospholipids, arginine, phosphatidylcholine, L-tryptophan, tyrosine, carnitine, and arachidonic acid [
1,
2,
4,
10,
20,
21,
22,
23,
24,
25]. In order to identify biomarkers, one must go to the metabolic pathways that affect amino acid metabolism. These include the biomarkers branched-chain amino acids (BCAAs), arginine, and phospholipid metabolism related to the conversion of phosphatidylcholine (PC) to lysophosphatidylcholine (lysoPC) [
3]. In the study by Zhang et al., they associated six metabolites with knee OA: arginine, sphingomyelin, and different PC [
4].
This current study has been performed through non-targeted metabolome and gene expression detection in samples obtained from rats using a monoiodoacetate (MIA)-induced OA model. The main objective was to evaluate the serum metabolome and identify the main altered metabolites in a patient with osteoarthritis of inflammatory origin. The hypothesis of the present study was that metabolomics could allow for the detection of changes in the serum metabolome in a patient with early osteoarthritis that have not yet been described.
3. Discussion
OA induced in rats by intra-articular MIA produces changes in the serum metabolome, as observed in the results of the study described herein. Furthermore, these variations are associated with the time elapsed since the induction of OA.
The pathophysiology of the OA complex remains unknown nowadays, so it was decided to investigate metabolomics as an advanced diagnostic method to see whether it could provide further insights into the disease. This technique allows us to observe the progress at a biochemical level and monitor the evolution of the treatment [
4,
10,
17,
19]. In addition, as the study period was relatively short, we needed a sampling technique that would allow us to make an early diagnosis compared to other techniques, such as radiography or MRI, where the diagnosis is made in more advanced stages of the disease [
1,
7,
10,
29,
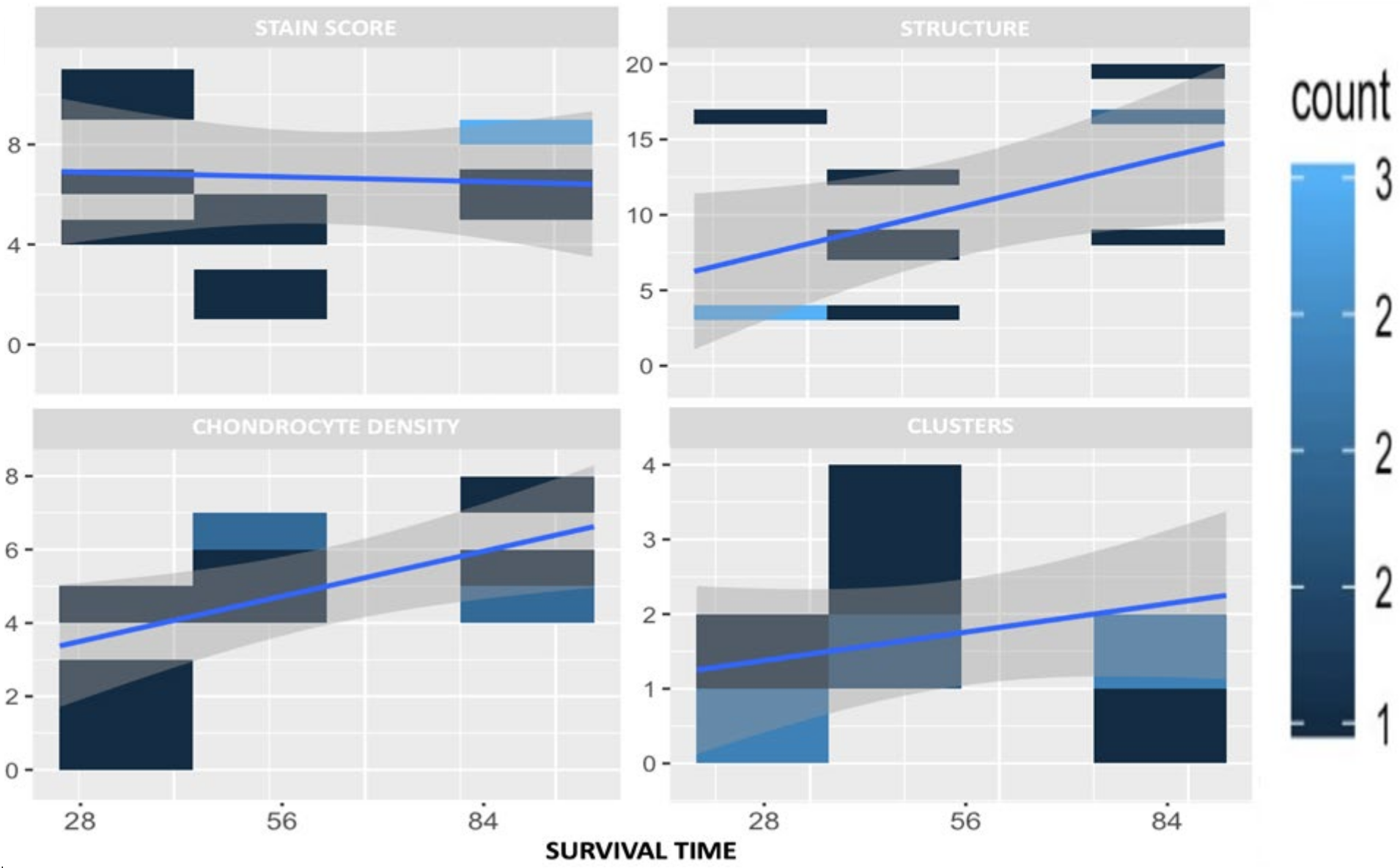
30]. The usefulness of an adequate OA model was verified by histological examinations.
Metabolomics is supported by several studies in which significant differences in serum metabolites were observed between healthy rats and OA rats, as observed in this study [
1,
2,
10,
17,
19,
20]. In the study by Chen et al. in rats with OA, plasma samples were analyzed by metabolomics, and it was shown that density labeling mass spectrometry, the same technique used in this study, had a high sensitivity for detecting metabolites in rat plasma [
1].
On the other hand, one of the factors to consider was the use of serum to analyze the metabolites present in the animal. The study carried out by Zhang et al. compared blood and synovial fluid (SF), observing how the range of metabolites varied considerably, and out of 168, only 8 were consistently related. This study suggests that metabolic changes are joint-specific and other inflammatory processes may influence the concentration of metabolites in serum [
4], as does the study carried out by Guma et al., which states that there is a fairly modest correlation between plasma and SF [
19].
Thus, as discussed in several articles, it is necessary to know which metabolites are altered for a better understanding of the joint status [
3,
19,
31,
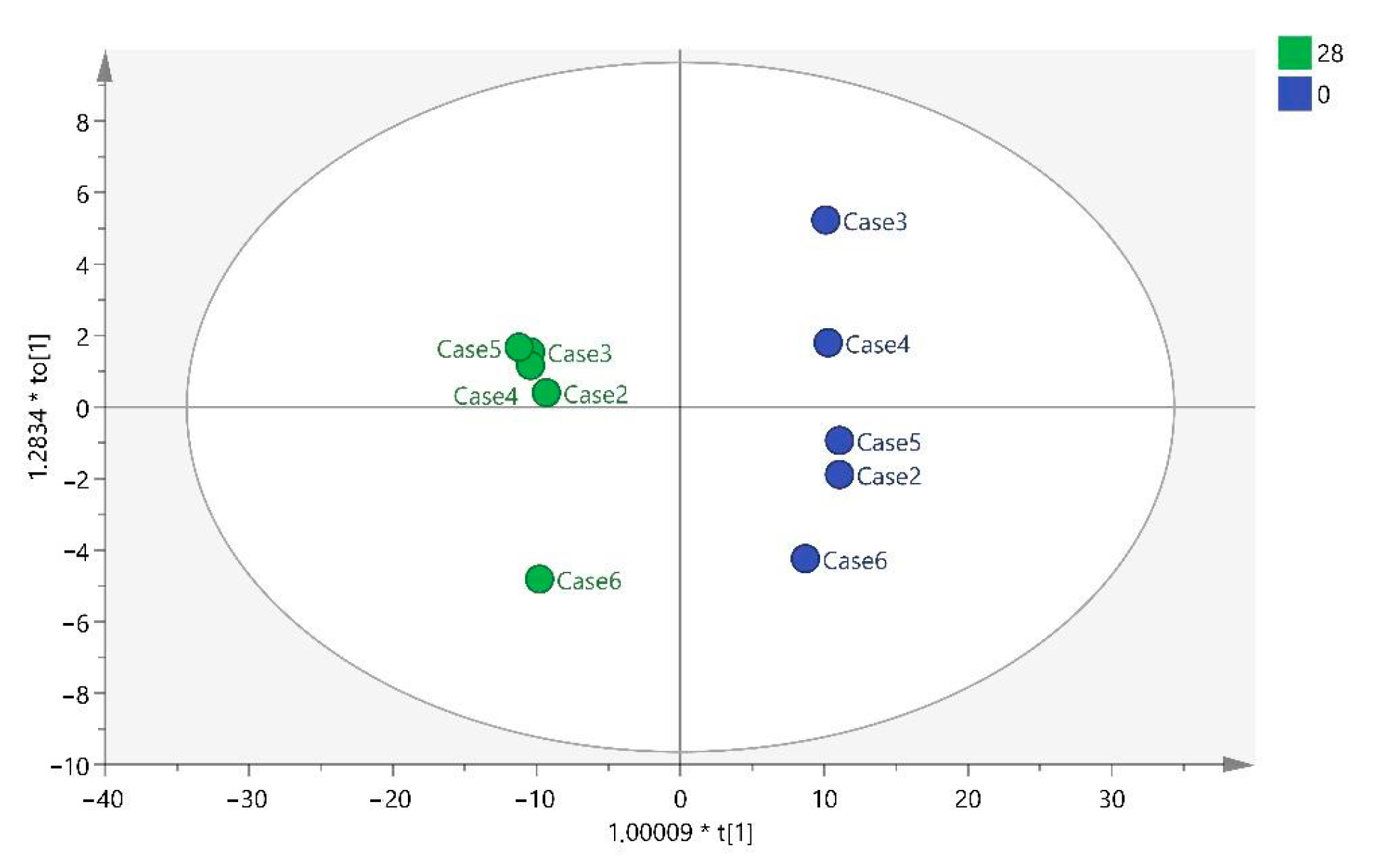
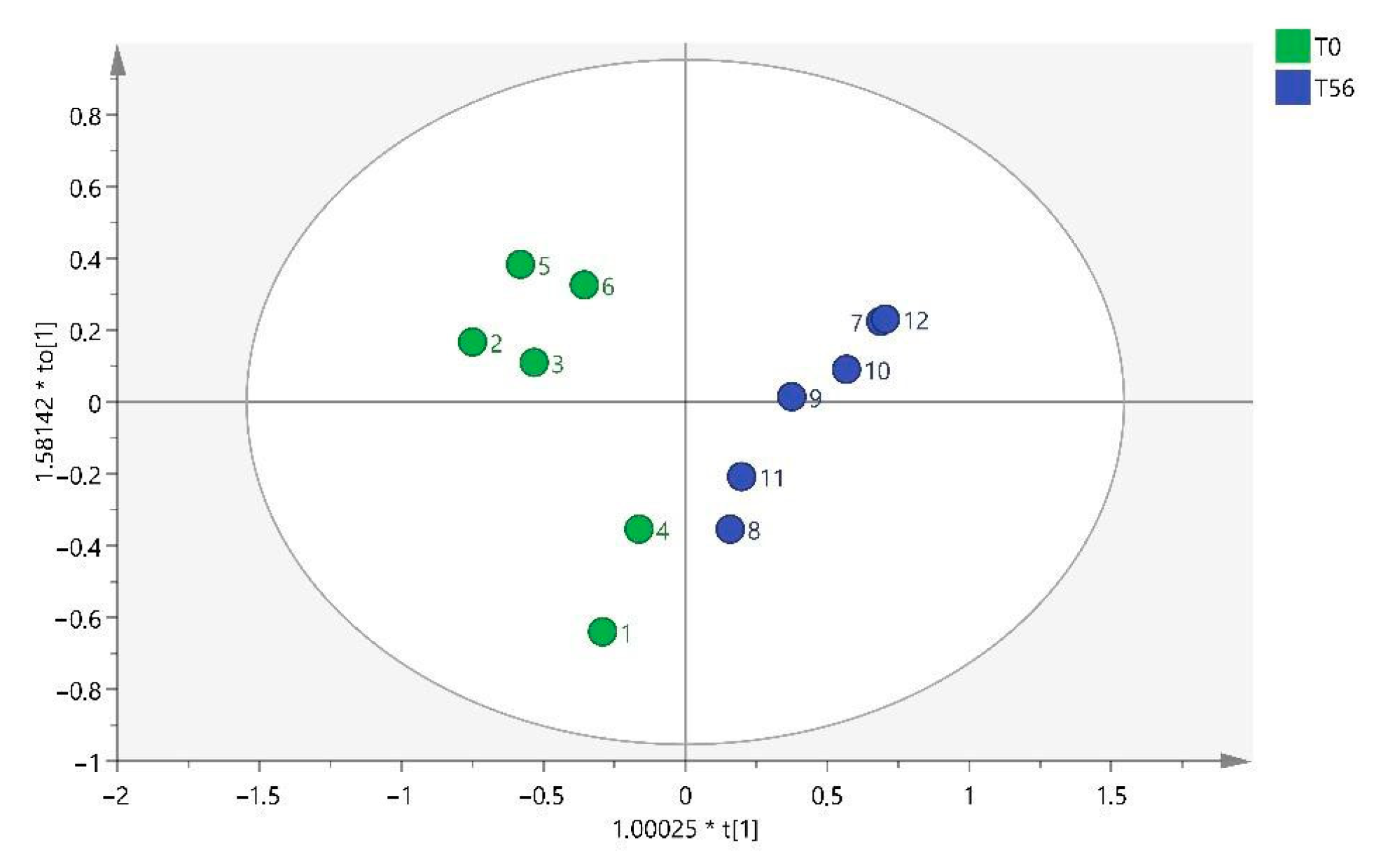
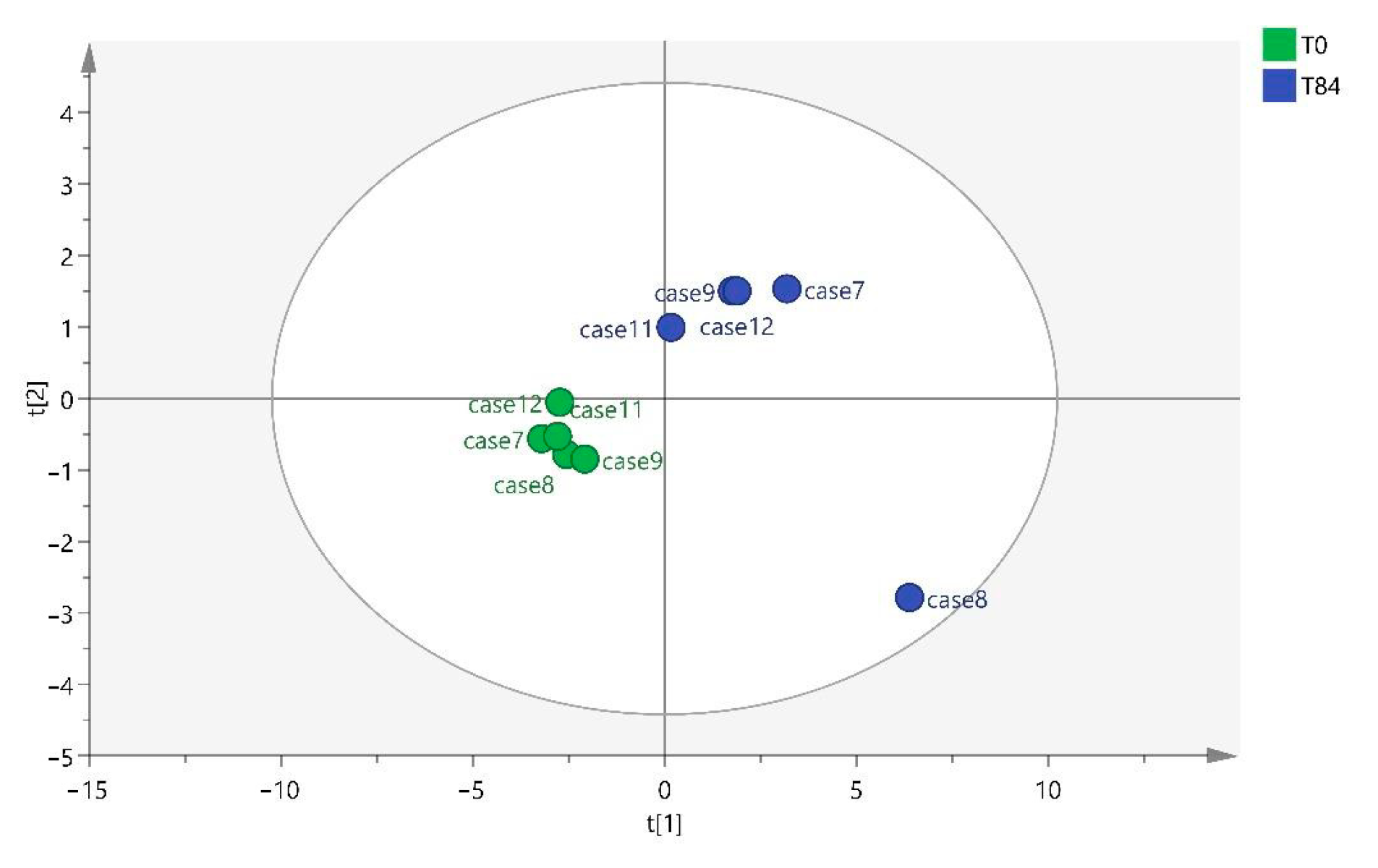
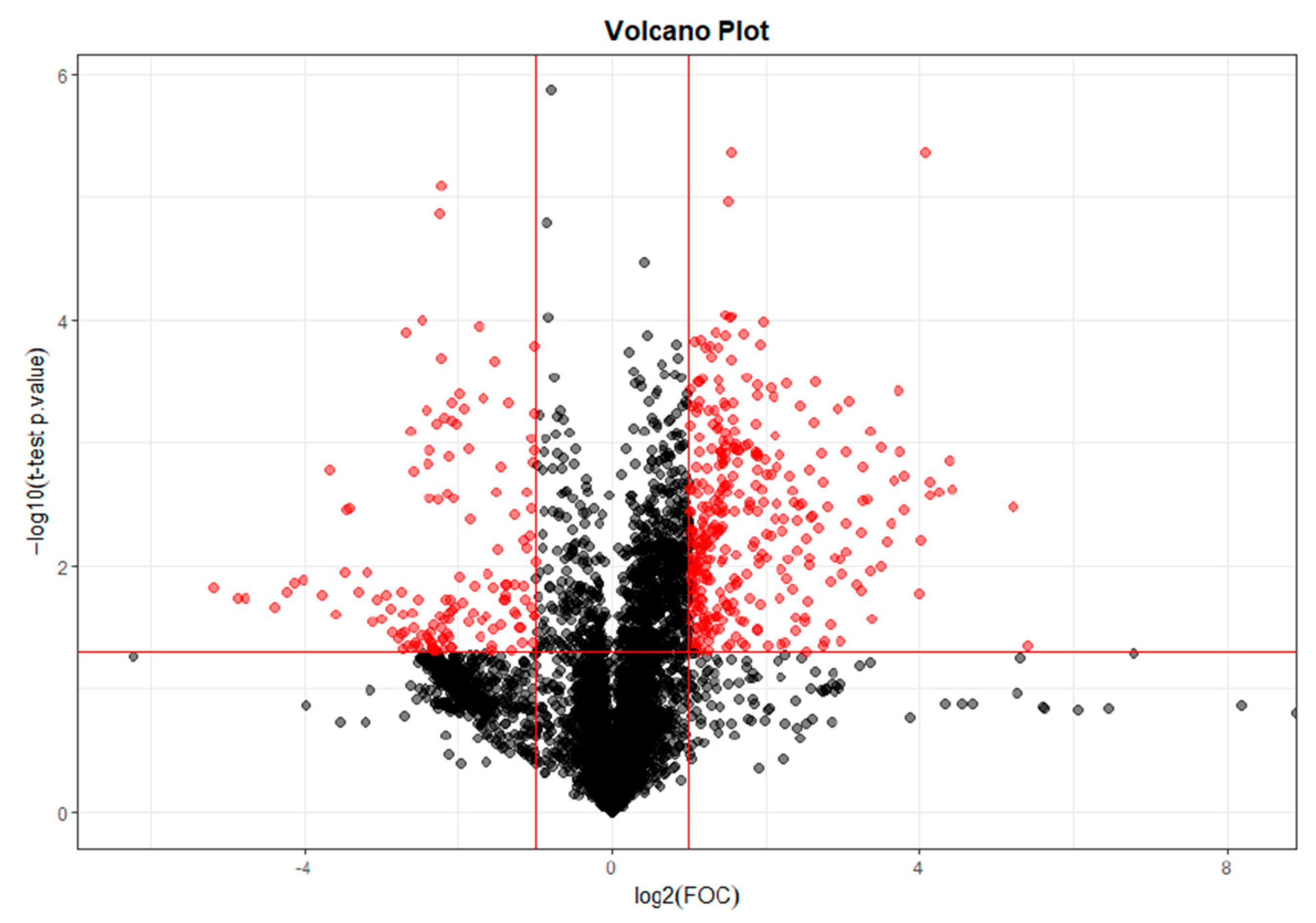
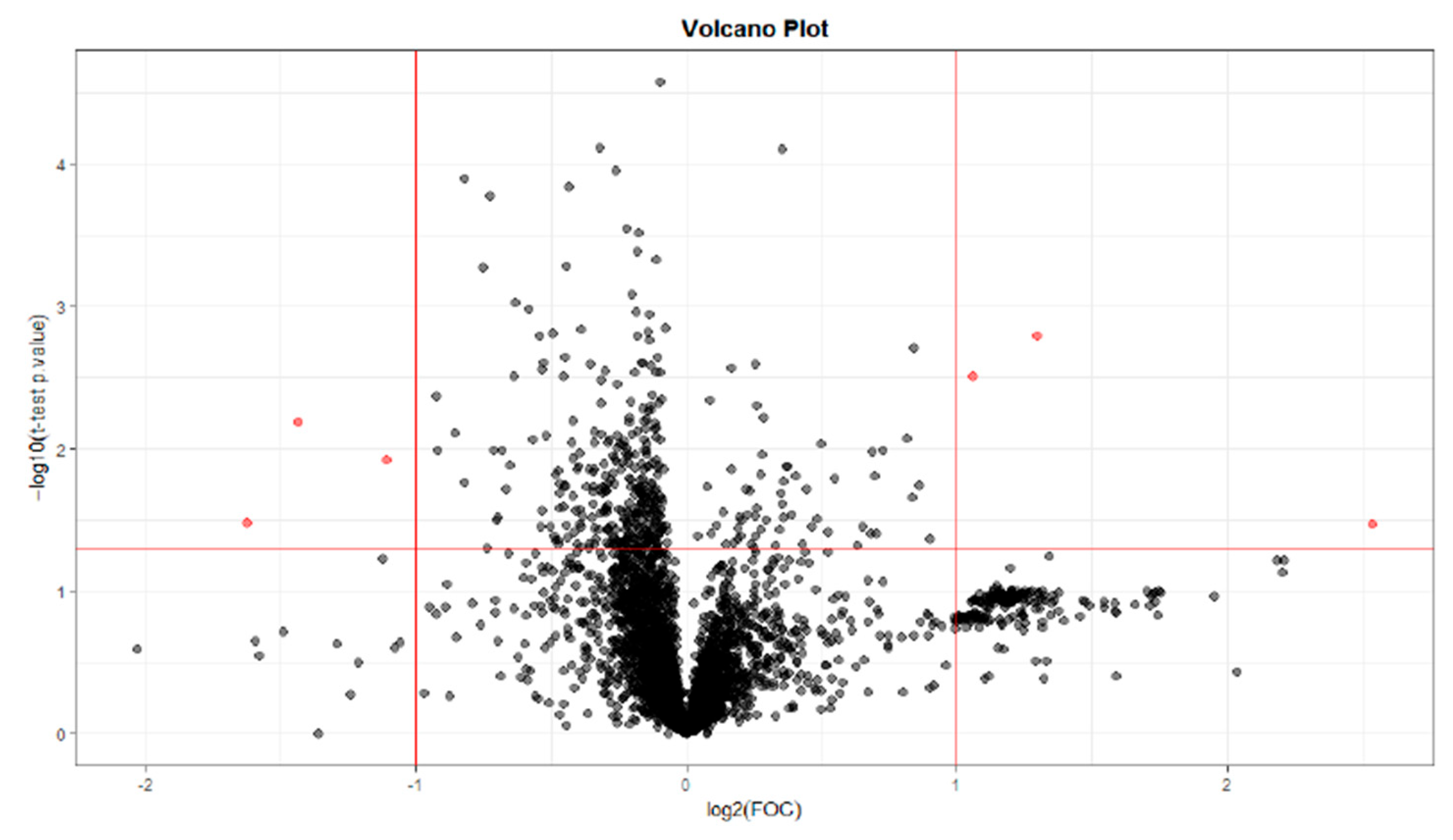
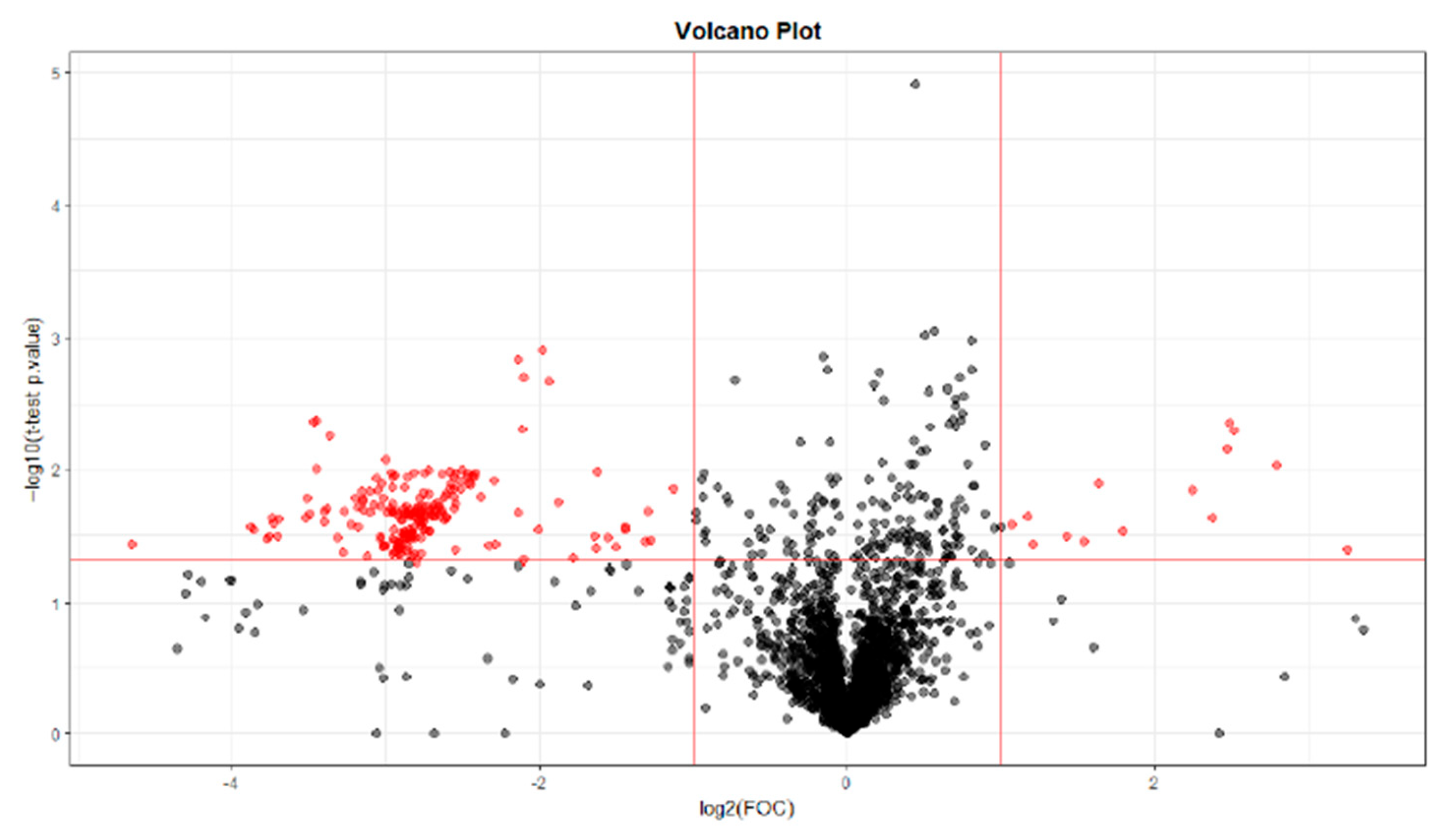
32]. Regarding the metabolites found in this study, they differ depending on whether the sample has been collected at T28, T56, or T84. Different benzenoids, organoheterocyclic compounds, organic acids, and lipid molecules were detected at T28, whereas only organic acids were observed at T56, and at T84, they were mainly lipid molecules.
The differential benzenoid classes found in this study were benzene and substituted derivatives. The organoheterocyclic compound classes found in this study were tetrahydroisoquinolines, pyridines, and derivatives. Previous studies had not observed any association of these metabolites with OA.
Likewise, differences in organic acids and derivatives were detected at T0 vs. T28 and T0 vs. T56. The main organic acid classes found in this study were carboxylic acids and derivatives (T28 and T56) and organic sulfonic acids and derivatives (T28). The organic sulfonic acid detected in the analysis at T0 vs. T28 was taurine. This result correlates with other studies where taurine metabolism was found to be one of the metabolic pathways most involved in OA, as taurine is implicated in the pathophysiology of OA, correlating with subchondral bone sclerosis and playing a vital regulatory role [
21,
29,
30,
33,
34,
35]. Anderson et al. observed that elevated taurine in OA could indicate increased subchondral bone sclerosis [
35]. Yang et al. showed that taurine levels in sclerotic subchondral bone were positively regulated [
30]. Taken together, these studies in synovial fluid revealed that altered taurine metabolism in subchondral bone has a direct correlation with subchondral bone sclerosis in osteoarthritis. In this study, taurine was analyzed from blood samples, so this metabolite could be used as an early biomarker of subchondral bone sclerosis in OA, but further blood studies are required to confirm this hypothesis.
Within the organic acid group, we have also detected carboxylic acids at T28 and T56, just as Swank et al. detected hippurate carboxylic acid in urine after 18 months of OA progression [
21]. Within this group, we found that at T56, the metabolite acetyl citrate was detected. With regard to this metabolite, different studies have detected the presence of citrate in urine and synovial fluid related to OA [
35,
36,
37]. Citrate is an intermediate in the tricarboxylic acid cycle, and its increase indicates an OA-related alteration in the cycle [
33,
35,
36,
37,
38]. Therefore, it would be interesting to further investigate the presence of acetyl citrate in the blood as a biomarker of OA.
Lipid molecules were found at T0 vs. T28 and T0 vs. T84. These possible biomarkers were related to the findings of several studies, where there is an alteration of lipid metabolism associated with OA due to its pro-inflammatory properties [
22,
23,
24,
39,
40]. Kosinska et al. detected alterations at the level of phospholipids and sphingolipids at different stages of the disease in synovial fluid [
41,
42,
43]. Thus, understanding the relationship between OA and lipid molecule analysis may be helpful in future treatments [
5].
The main lipid classes in which differences were found in this study are sphingolipids at T28, prenol lipids at T28 and T84, and glycerophospholipids, steroids, and fatty acyls at T84. These data are consistent with the study by Pousinis et al. that describes the presence of glycerophospholipids, sphingolipids, and fatty acyls in plasma from rats subjected to an OA model for 112 days [
40].
With regard to prenol lipids, their most biologically relevant classes are fat-soluble vitamins (vitamins E, A, and K) [
44]. Neogi et al. observed an association between low plasma levels of vitamin K and an increased prevalence of OA manifestations in the hand and knee. They found the relationship because vitamin K supports calcium homeostasis and facilitates bone mineralization [
45,
46,
47]. Regarding vitamin E, different studies have demonstrated its potent anti-inflammatory properties as well as its role in the prevention and regulation of the progression of age-related diseases [
48,
49]. Therefore, further research on the relationship between OA and prenol lipids would be desirable.
Sphingolipids detected at T28 had been previously detected in other studies and were related to subchondral bone sclerosis in OA. This fact suggests that sphingolipids play an important regulatory role in the pathological process of sclerotic subchondral bone [
24,
30,
40,
42]. Tootsi et al. found changes in serum sphingolipid levels in humans with OA, confirming their involvement in the pathogenesis of OA [
24]. Kosinska et al. found that sphingolipids could alter synovial inflammation and repair responses in damaged joints [
42]. Thus, it would be interesting to use sphingolipids as blood biomarkers for OA.
Phospholipids are molecules associated with inflammation and increased cartilage damage at the synovial fluid level and may be associated with the pathogenesis of OA [
41]. The glycerophospholipids form the essential lipid bilayer of all biological membranes, and changes in glycerophospholipid concentrations and composition are associated with OA development, as shown in a multitude of studies [
4,
22,
24,
40,
50]. Therefore, changes in the concentration of lipid molecules, more specifically glycerophospholipids, may indicate a risk of OA.
The biomarker ursodeoxycholic acid detected in this study belongs to the class of steroids, superclass lipids, and lipid-like molecules (HMDB). Ursodeoxycholic acid is a naturally occurring dihydroxy hydrophilic bile acid, and Moon et al. demonstrated that this bile acid has a preventive potential as a treatment in a model of induced OA by reducing pain and ameliorating cartilage destruction [
51]. On the other hand, Carlson et al. detected metabolites from steroid hormone biosynthesis in the synovial fluid of people with rheumatoid arthritis [
52]. No studies have been found on the detection of alterations in ursodeoxycholic acid at the metabolomic level in animals with OA, so this metabolite should be considered in future investigations.
4-Hydroperoxy-H4-neuroprostane, also known as 14-H4-NeuroP, is a member of the class fatty acyls, superclass lipids, and lipid-like molecules and the direct parent of prostaglandins and related compounds (HMDB) [
26]. Similarly, in the study by Zhao et al., it was observed that serum levels of prostaglandin estradiol2 were significantly increased in the OA group. In addition, several metabolites of the class fatty acyls and superclass lipids, such as aminobutyric acid, stearic acid, or L-carnitine, were increased [
10]. Attur et al. examined plasma lipid prostaglandin E2 (PGE2) and found PGE2 elevated in symptomatic knee OA patients [
53]. Similarly, Gierman et al. associated changes in PGE2 levels with the development of OA [
54]. On the other hand, the study by Shi et al. and Pausinis et al. also shows changes in arachidonic acid or linoleic acid, metabolites within the same classification [
2,
40]. Regarding acylcarnitines of the fatty acyls class, several studies have shown changes in their concentration in the serum of animal models with OA [
50,
55]. Thus, changes in prostaglandin concentrations or fatty acyls could indicate the presence of OA.
There are some limitations to this study. Firstly, the sample size was small and did not allow strong validation of these potential biomarkers. Therefore, a larger sample size would be necessary in future research. Secondly, only blood samples were used, and in the future, it would be interesting to correlate serum with synovial fluid measurements to better understand the observed metabolic changes. Thirdly, the possible relationship between OA and arthritic diseases and whether these biomarkers are useful or not for identifying other forms of arthritis were not explored. Fourthly, variables other than time were not considered, and there is intra-group variability in these results; for this reason, other variables will need to be considered in the future. Lastly, it should be considered that the identified metabolites in this study should be directly evaluated in subsequent targeted studies, aiming to confirm or rule out their role in the modification of serum metabolomes in an inflammatory model of osteoarthritis.