Integrated Analysis of DNA Methylation and Gene Expression Profiles in a Rat Model of Osteoarthritis
Abstract
1. Introduction
2. Results
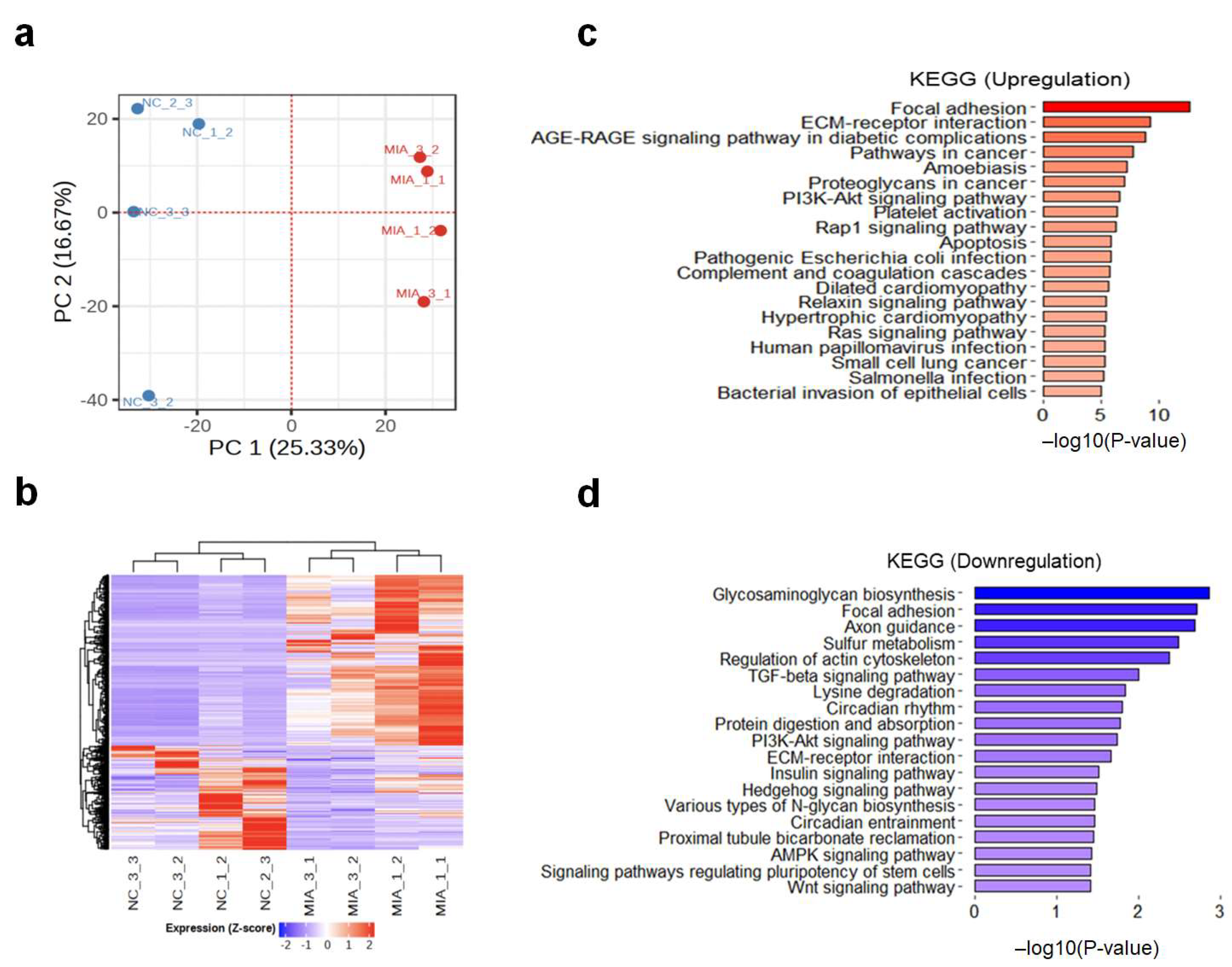
2.1. Transcriptomic Changes in the MIA-Induced Rat Model of OA
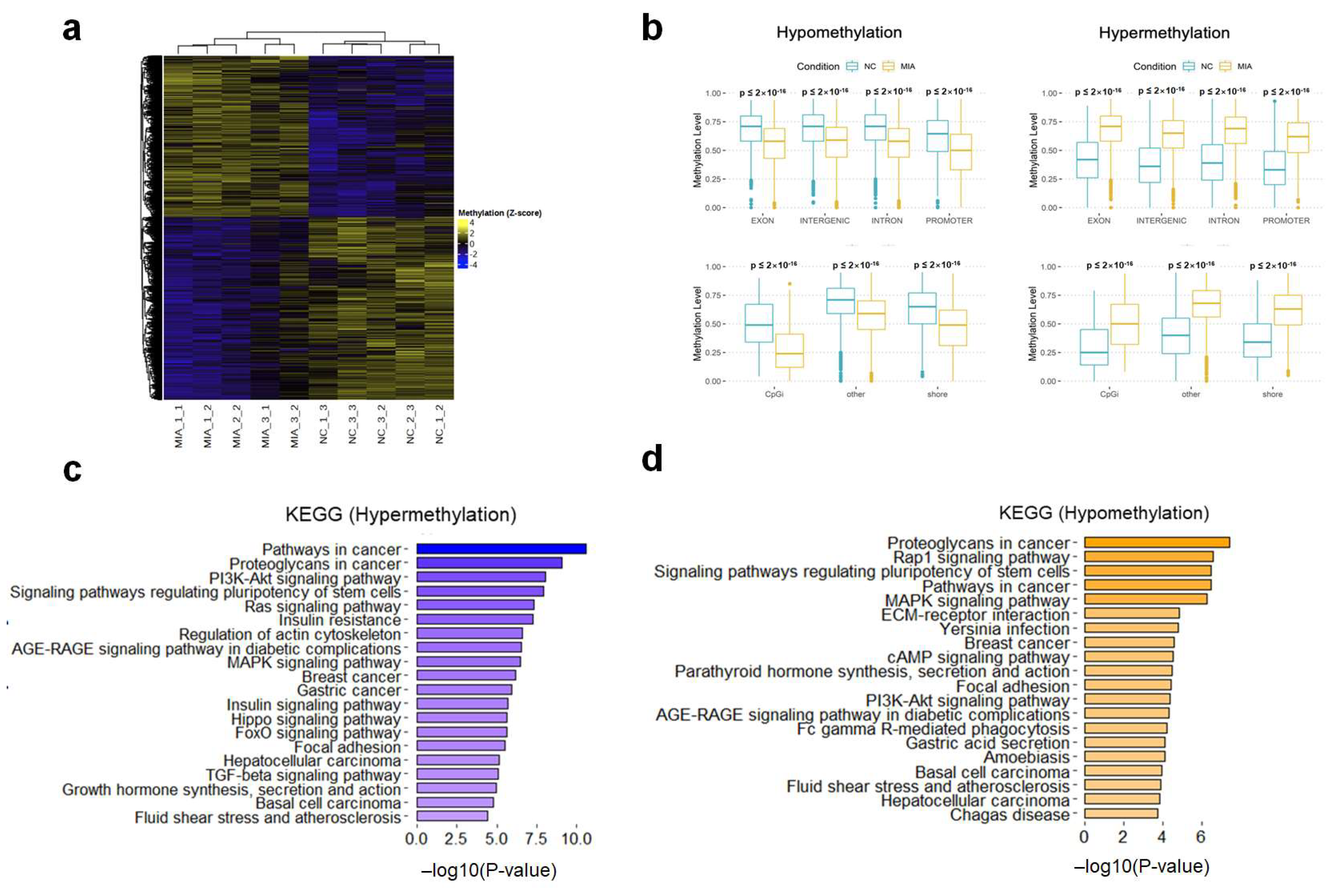
2.2. Genome-Wide DNA Methylation Profiling of OA
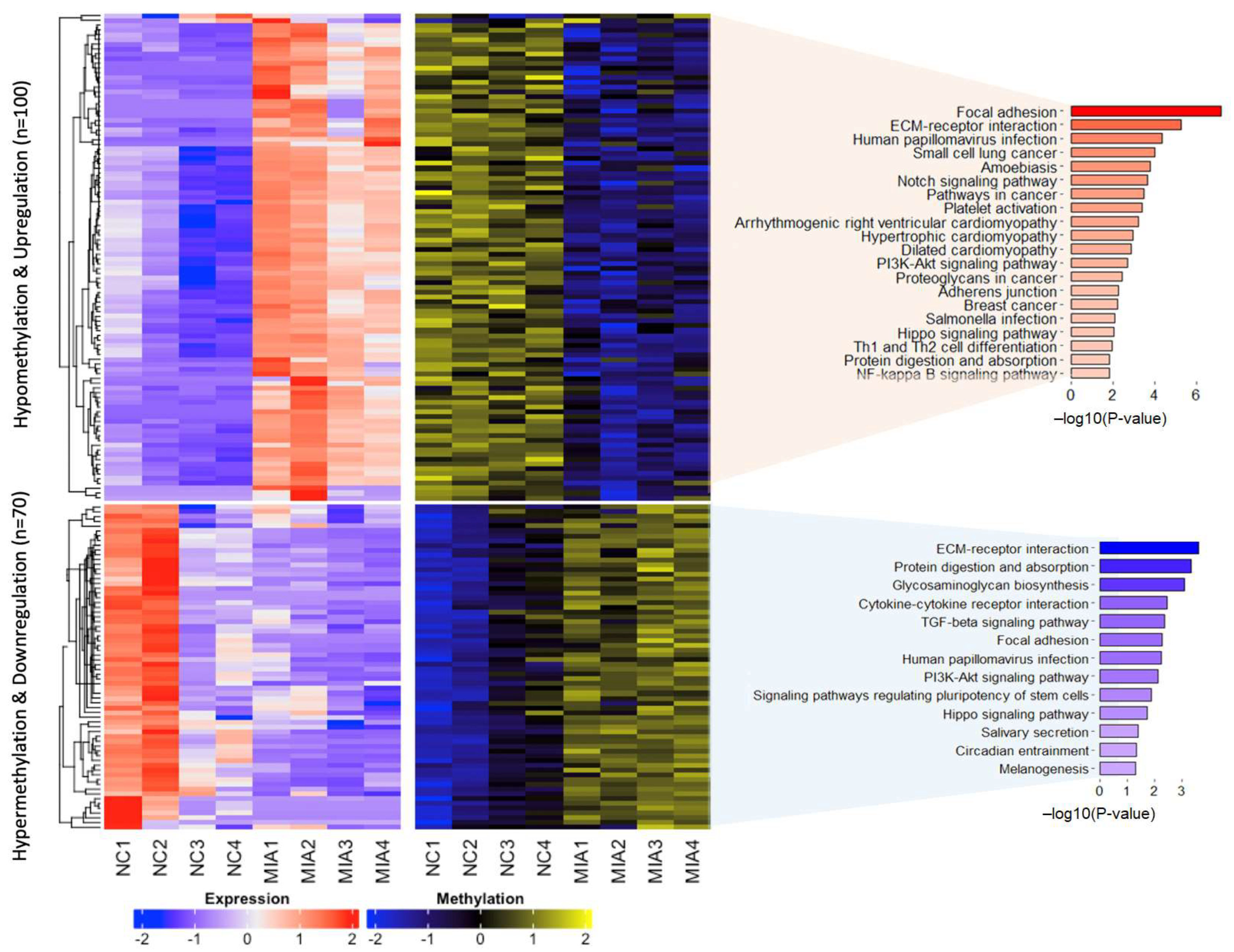
2.3. Dysregulated Gene Expression Induced by DNA Methylation
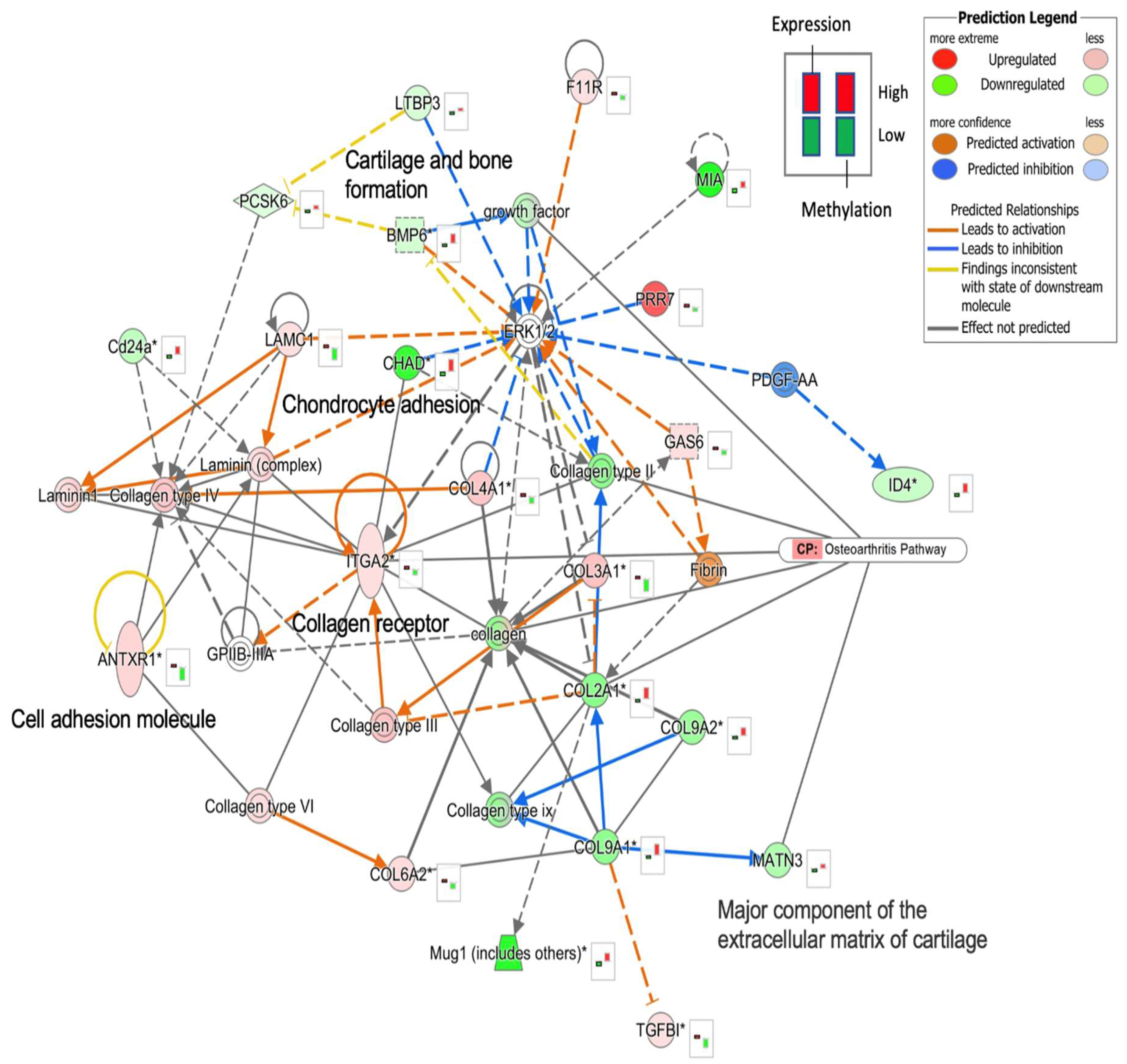
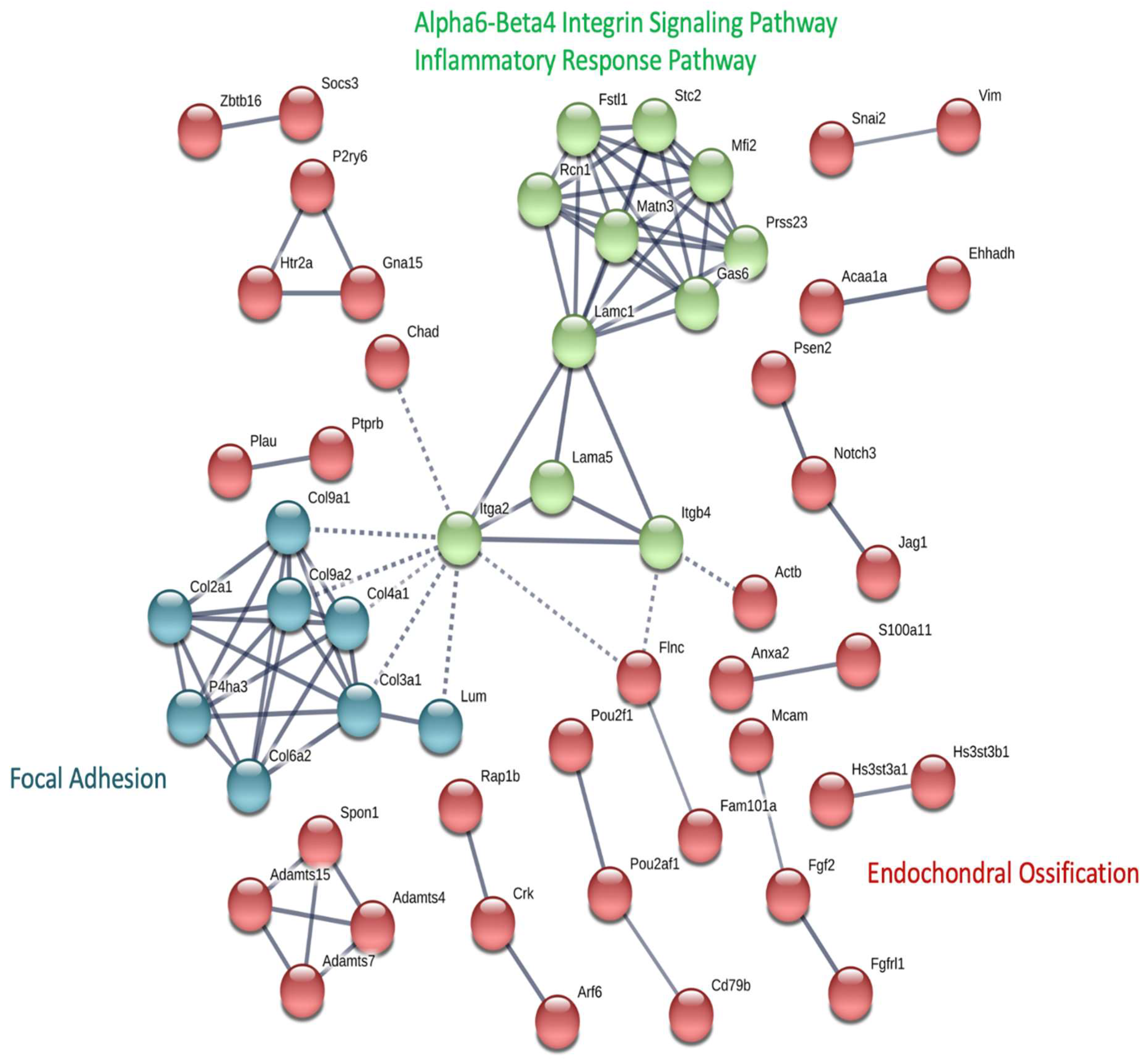
2.4. Functional Network Analysis of OA Candidate Gene
3. Discussion
4. Materials and Methods
4.1. Monosodium Iodoacetate (MIA)-Induced Rat Model
4.2. RNA Extraction and Quality Control
4.3. RNA-Sequencing Analysis
4.4. DNA Extraction and Methyl-Sequencing Analysis
4.5. Functional Annotation and Enrichment Analysis
4.6. Selection of Candidate Genes Based on DNA Methylation and Expression Levels
4.7. Network Inference Using Ingenuity Pathway Analysis (IPA) and STRING
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johnson, V.L.; Hunter, D.J. The epidemiology of osteoarthritis. Best. Pract. Res. Clin. Rheumatol. 2014, 28, 5–15. [Google Scholar] [CrossRef]
- Mora, J.C.; Przkora, R.; Cruz-Almeida, Y. Knee osteoarthritis: Pathophysiology and current treatment modalities. J. Pain Res. 2018, 11, 2189–2196. [Google Scholar] [CrossRef] [PubMed]
- Neogi, T. The epidemiology and impact of pain in osteoarthritis. Osteoarthr. Cartil. 2013, 21, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, D.; Cho, Y.; Kim, J.H. Epigenetic Regulation of Chondrocyte Catabolism and Anabolism in Osteoarthritis. Mol. Cells 2015, 38, 677–684. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, Z.; Alexander, P.G.; Ocasio-Nieves, B.D.; Yocum, L.; Lin, H.; Tuan, R.S. Pathogenesis of Osteoarthritis: Risk Factors, Regulatory Pathways in Chondrocytes, and Experimental Models. Biology 2020, 9, 194. [Google Scholar] [CrossRef] [PubMed]
- Krakowski, P.; Karpiński, R.; Maciejewski, R.; Jonak, J.; Jurkiewicz, A. Short-Term Effects of Arthroscopic Microfracturation of Knee Chondral Defects in Osteoarthritis. Appl. Sci. 2020, 10, 8312. [Google Scholar] [CrossRef]
- Karpiński, R. Knee joint osteoarthritis diagnosis based on selected acoustic signal discriminants using machine learning. Appl. Comput. Sci. 2022, 18, 71–85. [Google Scholar] [CrossRef]
- Almhdie-Imjabbar, A.; Toumi, H.; Lespessailles, E. Radiographic Biomarkers for Knee Osteoarthritis: A Narrative Review. Life 2023, 13, 237. [Google Scholar] [CrossRef]
- Tong, L.; Yu, H.; Huang, X.; Shen, J.; Xiao, G.; Chen, L.; Wang, H.; Xing, L.; Chen, D. Current understanding of osteoarthritis pathogenesis and relevant new approaches. Bone Res. 2022, 10, 60. [Google Scholar] [CrossRef]
- Yao, Q.; Wu, X.; Tao, C.; Gong, W.; Chen, M.; Qu, M.; Zhong, Y.; He, T.; Chen, S.; Xiao, G. Osteoarthritis: Pathogenic signaling pathways and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 56. [Google Scholar] [CrossRef]
- Xia, B.; Di, C.; Zhang, J.; Hu, S.; Jin, H.; Tong, P. Osteoarthritis pathogenesis: A review of molecular mechanisms. Calcif. Tissue Int. 2014, 95, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Saberi Hosnijeh, F.; Bierma-Zeinstra, S.M.; Bay-Jensen, A.C. Osteoarthritis year in review 2018: Biomarkers (biochemical markers). Osteoarthr. Cartil. 2019, 27, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Hu, Y.; Shen, J.; Liu, X.; Wang, T.; Li, L.; Li, J. Integrative analysis of DNA methylation and gene expression reveals key molecular signatures in acute myocardial infarction. Clin. Epigenet. 2022, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xiao, X.; Zhang, L.; Wang, B.; Li, P.; Cheng, B.; Liang, C.; Ma, M.; Guo, X.; Zhang, F.; et al. An integrative analysis of DNA methylation and transcriptome showed the dysfunction of MAPK pathway was involved in the damage of human chondrocyte induced by T-2 toxin. BMC Mol. Cell Biol. 2022, 23, 4. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lee, J.; Ding, J.; Sun, S. Integrative analysis of gene expression and methylation data for breast cancer cell lines. BioData Min. 2018, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Chen, J.; Yang, Y.; Lin, J.; Wu, Z. Genome-wide expression and methylation profiling reveal candidate genes in osteoarthritis. Clin. Exp. Rheumatol. 2017, 35, 983–990. [Google Scholar]
- Rushton, M.D.; Reynard, L.N.; Barter, M.J.; Refaie, R.; Rankin, K.S.; Young, D.A.; Loughlin, J. Characterization of the cartilage DNA methylome in knee and hip osteoarthritis. Arthritis Rheumatol. 2014, 66, 2450–2460. [Google Scholar] [CrossRef]
- Rushton, M.D.; Young, D.A.; Loughlin, J.; Reynard, L.N. Differential DNA methylation and expression of inflammatory and zinc transporter genes defines subgroups of osteoarthritic hip patients. Ann. Rheum. Dis. 2015, 74, 1778–1782. [Google Scholar] [CrossRef]
- Wang, X.; Tang, D.; Shen, P.; Xu, H.; Qiu, H.; Wu, T.; Gao, X. Analysis of DNA methylation in chondrocytes in rats with knee osteoarthritis. BMC Musculoskelet. Disord. 2017, 18, 377. [Google Scholar] [CrossRef][Green Version]
- Thysen, S.; Luyten, F.P.; Lories, R.J. Targets, models and challenges in osteoarthritis research. Dis. Model. Mech. 2015, 8, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.-S.; Hsu, F.-M.; Chen, P.-Y. Profiling genome-wide DNA methylation. Epigenet. Chromatin 2016, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Duarte, A. DNA Methylation in Osteoarthritis: Current Status and Therapeutic Implications. Open Rheumatol. J. 2018, 12, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarasimhan, R.; Liang, G. The Role of DNA Methylation in Cancer. Adv. Exp. Med. Biol. 2016, 945, 151–172. [Google Scholar]
- Baek, S.J.; Ban, H.J.; Park, S.M.; Kim, S.Y.; Lee, S.; Jin, H.J. Genome-wide DNA methylation profiling reveals candidate biomarkers and probable molecular mechanism of metabolic syndrome. Genes. Dis. 2022, 9, 833–836. [Google Scholar] [CrossRef]
- Chun, J.M.; Lee, A.Y.; Nam, J.Y.; Lim, K.S.; Choe, M.S.; Lee, M.Y.; Kim, C.; Kim, J.S. Effects of Dipsacus asperoides Extract on Monosodium Iodoacetate-Induced Osteoarthritis in Rats Based on Gene Expression Profiling. Front. Pharmacol. 2021, 12, 615157. [Google Scholar] [CrossRef]
- Chun, J.M.; Lee, A.Y.; Nam, J.Y.; Lee, M.Y.; Choe, M.S.; Lim, K.S.; Kim, C.; Kim, J.S. Protective effects of Phlomis umbrosa extract on a monosodium iodoacetate-induced osteoarthritis model and prediction of molecular mechanisms using transcriptomics. Phytomedicine 2021, 81, 153429. [Google Scholar] [CrossRef]
- Poole, A.R.; Kobayashi, M.; Yasuda, T.; Laverty, S.; Mwale, F.; Kojima, T.; Sakai, T.; Wahl, C.; El-Maadawy, S.; Webb, G.; et al. Type II collagen degradation and its regulation in articular cartilage in osteoarthritis. Ann. Rheum. Dis. 2002, 61 (Suppl. S2), ii78–ii81. [Google Scholar] [CrossRef]
- Alvarez-Garcia, O.; Fisch, K.M.; Wineinger, N.E.; Akagi, R.; Saito, M.; Sasho, T.; Su, A.I.; Lotz, M.K. Increased DNA Methylation and Reduced Expression of Transcription Factors in Human Osteoarthritis Cartilage. Arthritis Rheumatol. 2016, 68, 1876–1886. [Google Scholar] [CrossRef]
- Song, D.; Qi, W.; Lv, M.; Yuan, C.; Tian, K.; Zhang, F. Combined bioinformatics analysis reveals gene expression and DNA methylation patterns in osteoarthritis. Mol. Med. Rep. 2018, 17, 8069–8078. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.; Zeggini, E. Functional genomics in osteoarthritis: Past, present, and future. J. Orthop. Res. 2016, 34, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Song, D.H.; Kim, S.H.; Jung, Y.; Kim, S.J. Development and characterization of various osteoarthritis models for tissue engineering. PLoS ONE 2018, 13, e0194288. [Google Scholar] [CrossRef] [PubMed]
- Combe, R.; Bramwell, S.; Field, M.J. The monosodium iodoacetate model of osteoarthritis: A model of chronic nociceptive pain in rats? Neurosci. Lett. 2004, 370, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, N. Genome-wide expression and methylation profiles reveal candidate genes and biological processes underlying synovial inflammatory tissue of patients with osteoarthritis. Int. J. Rheum. Dis. 2015, 18, 783–790. [Google Scholar] [CrossRef] [PubMed]
- De Santis, M.; Di Matteo, B.; Chisari, E.; Cincinelli, G.; Angele, P.; Lattermann, C.; Filardo, G.; Vitale, N.D.; Selmi, C.; Kon, E. The Role of Wnt Pathway in the Pathogenesis of OA and Its Potential Therapeutic Implications in the Field of Regenerative Medicine. Biomed. Res. Int. 2018, 2018, 7402947. [Google Scholar] [CrossRef]
- Hosseinzadeh, A.; Kamrava, S.K.; Joghataei, M.T.; Darabi, R.; Shakeri-Zadeh, A.; Shahriari, M.; Reiter, R.J.; Ghaznavi, H.; Mehrzadi, S. Apoptosis signaling pathways in osteoarthritis and possible protective role of melatonin. J. Pineal Res. 2016, 61, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Rigoglou, S.; Papavassiliou, A.G. The NF-κB signalling pathway in osteoarthritis. Int. J. Biochem. Cell Biol. 2013, 45, 2580–2584. [Google Scholar] [CrossRef]
- Shen, J.; Li, S.; Chen, D. TGF-β signaling and the development of osteoarthritis. Bone Res. 2014, 2, 14002. [Google Scholar] [CrossRef]
- Sun, K.; Luo, J.; Guo, J.; Yao, X.; Jing, X.; Guo, F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: A narrative review. Osteoarthr. Cartil. 2020, 28, 400–409. [Google Scholar] [CrossRef]
- Wang, X.; Li, F.; Fan, C.; Wang, C.; Ruan, H. Effects and relationship of ERK1 and ERK2 in interleukin-1β-induced alterations in MMP3, MMP13, type II collagen and aggrecan expression in human chondrocytes. Int. J. Mol. Med. 2011, 27, 583–589. [Google Scholar]
- Liu, Z.; Cai, H.; Zheng, X.; Zhang, B.; Xia, C. The Involvement of Mutual Inhibition of ERK and mTOR in PLCγ1-Mediated MMP-13 Expression in Human Osteoarthritis Chondrocytes. Int. J. Mol. Sci. 2015, 16, 17857–17869. [Google Scholar] [CrossRef] [PubMed]
- Loeser, R.F.; Erickson, E.A.; Long, D.L. Mitogen-activated protein kinases as therapeutic targets in osteoarthritis. Curr. Opin. Rheumatol. 2008, 20, 581–586. [Google Scholar] [CrossRef]
- Jeffries, M.A.; Donica, M.; Baker, L.W.; Stevenson, M.E.; Annan, A.C.; Beth Humphrey, M.; James, J.A.; Sawalha, A.H. Genome-Wide DNA Methylation Study Identifies Significant Epigenomic Changes in Osteoarthritic Subchondral Bone and Similarity to Overlying Cartilage. Arthritis Rheumatol. 2016, 68, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Miyamoto, S.; Mekada, E. Integrin alpha 2 beta 1-dependent EGF receptor activation at cell-cell contact sites. J. Cell Sci. 2000, 113 Pt 12, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Noris, P.; Guidetti, G.F.; Conti, V.; Ceresa, I.F.; Di Pumpo, M.; Pecci, A.; Torti, M.; Savoia, A.; Balduini, C.L. Autosomal dominant thrombocytopenias with reduced expression of glycoprotein Ia. Thromb. Haemost. 2006, 95, 483–489. [Google Scholar] [CrossRef]
- Peters, M.A.; Wendholt, D.; Strietholt, S.; Frank, S.; Pundt, N.; Korb-Pap, A.; Joosten, L.A.; van den Berg, W.B.; Kollias, G.; Eckes, B.; et al. The loss of α2β1 integrin suppresses joint inflammation and cartilage destruction in mouse models of rheumatoid arthritis. Arthritis Rheum. 2012, 64, 1359–1368. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, H.; Yang, X.; Li, Z.; Zhang, D.; Li, B.; Zhang, D.; Li, Q.; Xiong, Y. Potential candidate biomarkers associated with osteoarthritis: Evidence from a comprehensive network and pathway analysis. J. Cell Physiol. 2019, 234, 17433–17443. [Google Scholar] [CrossRef]
- Gong, J.; Lu, X.; Xu, J.; Xiong, W.; Zhang, H.; Yu, X. Coexpression of UCA1 and ITGA2 in pancreatic cancer cells target the expression of miR-107 through focal adhesion pathway. J. Cell Physiol. 2019, 234, 12884–12896. [Google Scholar] [CrossRef]
- Baek, S.J.; Chun, J.M.; Kang, T.W.; Seo, Y.S.; Kim, S.B.; Seong, B.; Jang, Y.; Shin, G.H.; Kim, C. Identification of Epigenetic Mechanisms Involved in the Anti-Asthmatic Effects of Descurainia sophia Seed Extract Based on a Multi-Omics Approach. Molecules 2018, 23, 2879. [Google Scholar] [CrossRef]
- Shin, Y.; Subramaniyam, S.; Chun, J.M.; Jeon, J.H.; Hong, J.M.; Jung, H.; Seong, B.; Kim, C. Genome-Wide Differential Methylation Profiles from Two Terpene-Rich Medicinal Plant Extracts Administered in Osteoarthritis Rats. Plants 2021, 10, 1132. [Google Scholar] [CrossRef] [PubMed]
- Schuelert, N.; McDougall, J.J. Grading of monosodium iodoacetate-induced osteoarthritis reveals a concentration-dependent sensitization of nociceptors in the knee joint of the rat. Neurosci. Lett. 2009, 465, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chun, J.M.; Kim, J.-S.; Kim, C. Integrated Analysis of DNA Methylation and Gene Expression Profiles in a Rat Model of Osteoarthritis. Int. J. Mol. Sci. 2024, 25, 594. https://doi.org/10.3390/ijms25010594
Chun JM, Kim J-S, Kim C. Integrated Analysis of DNA Methylation and Gene Expression Profiles in a Rat Model of Osteoarthritis. International Journal of Molecular Sciences. 2024; 25(1):594. https://doi.org/10.3390/ijms25010594
Chicago/Turabian StyleChun, Jin Mi, Joong-Sun Kim, and Chul Kim. 2024. "Integrated Analysis of DNA Methylation and Gene Expression Profiles in a Rat Model of Osteoarthritis" International Journal of Molecular Sciences 25, no. 1: 594. https://doi.org/10.3390/ijms25010594
APA StyleChun, J. M., Kim, J.-S., & Kim, C. (2024). Integrated Analysis of DNA Methylation and Gene Expression Profiles in a Rat Model of Osteoarthritis. International Journal of Molecular Sciences, 25(1), 594. https://doi.org/10.3390/ijms25010594






