Dachshund Homolog 1: Unveiling Its Potential Role in Megakaryopoiesis and Bacillus anthracis Lethal Toxin-Induced Thrombocytopenia
Abstract
1. Introduction
2. Results
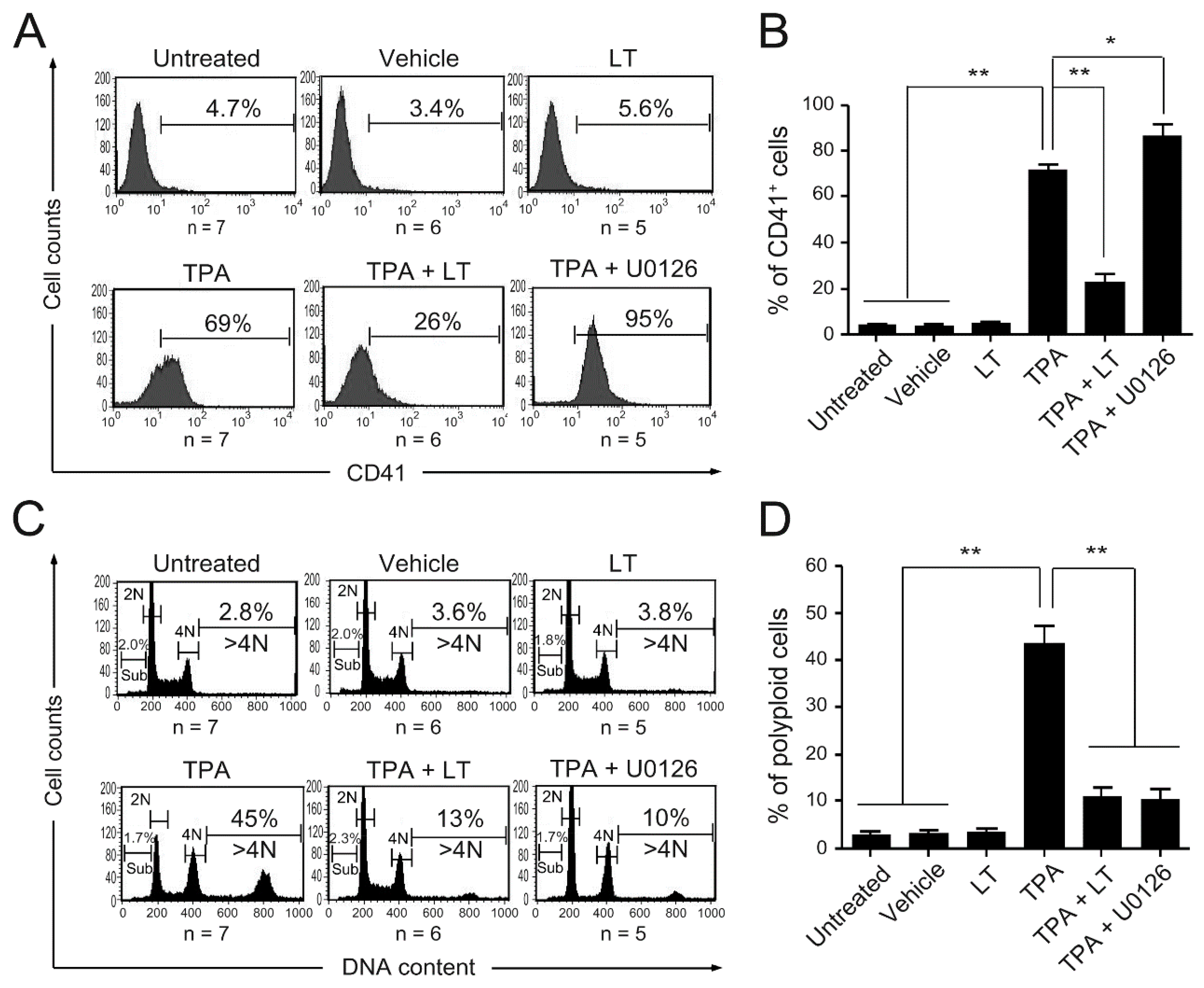
2.1. Bacillus anthracis Lethal Toxin Could Inhibit TPA-Induced Megakaryocytic Differentiation
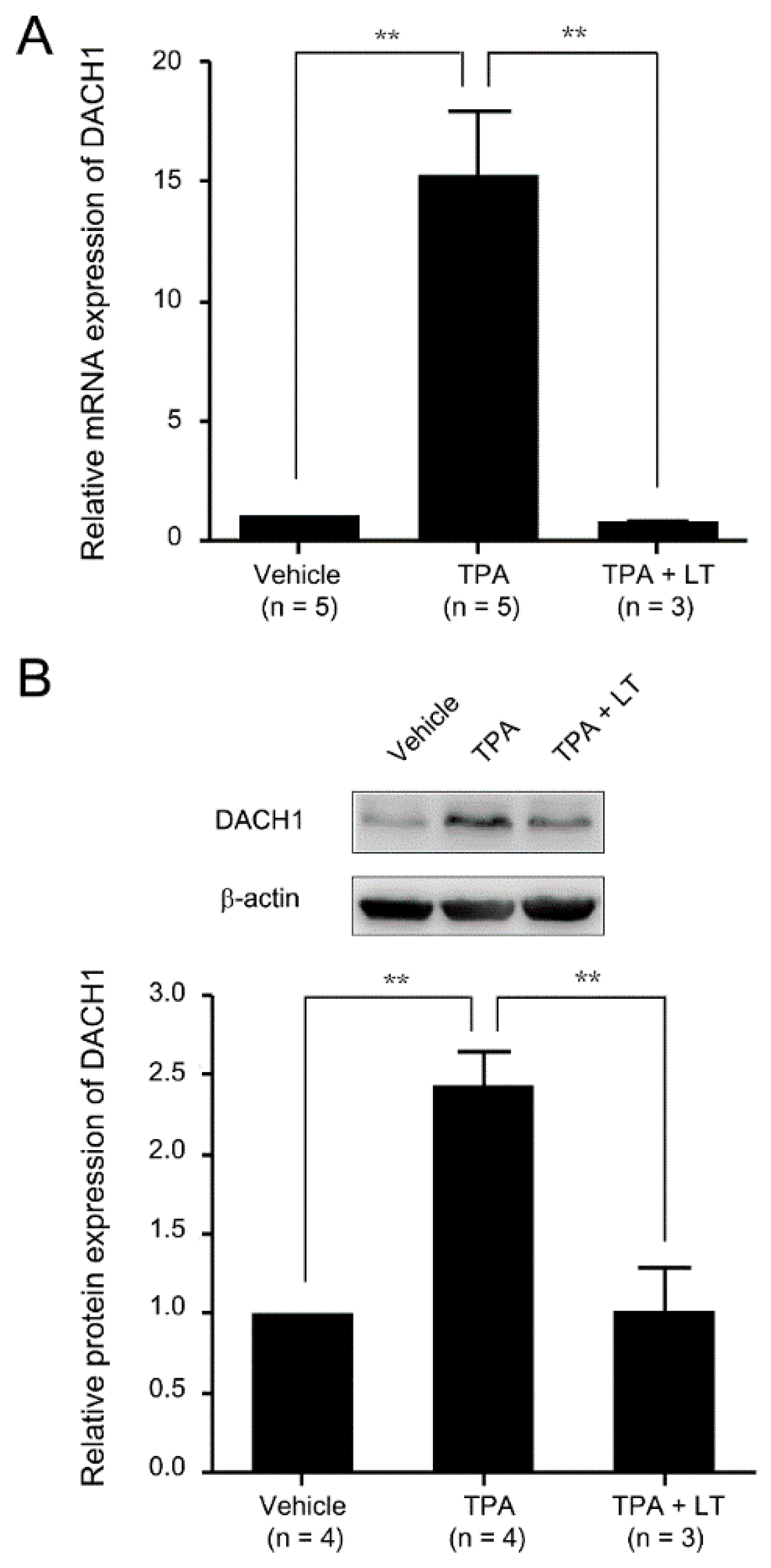
2.2. DACH1 Gene Expression Was Increased during the Process of Megakaryocytic Differentiation
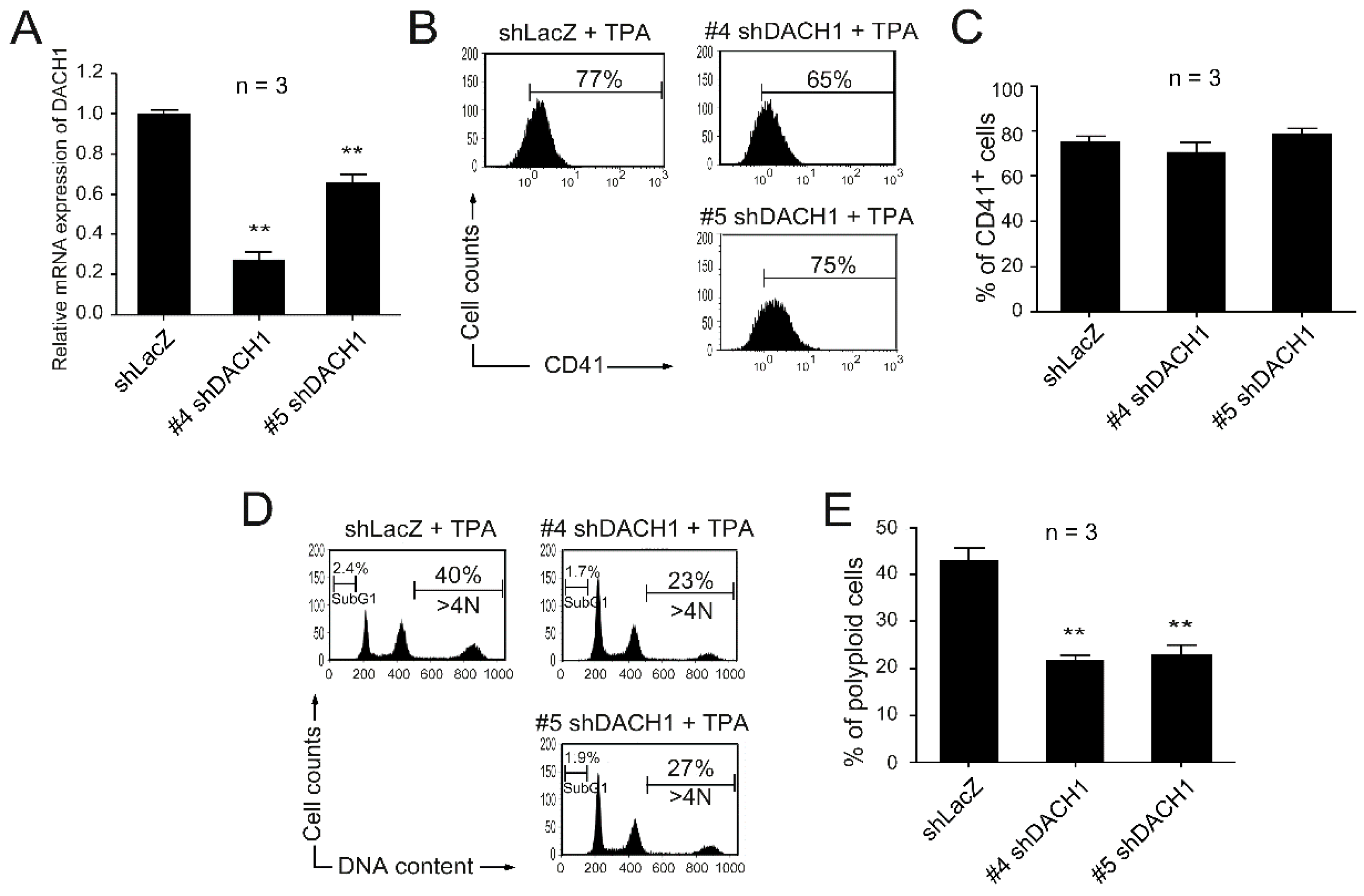
2.3. Knockdown of the DACH1 Gene mRNA Could Block TPA-Induced Megakaryocytic Differentiation
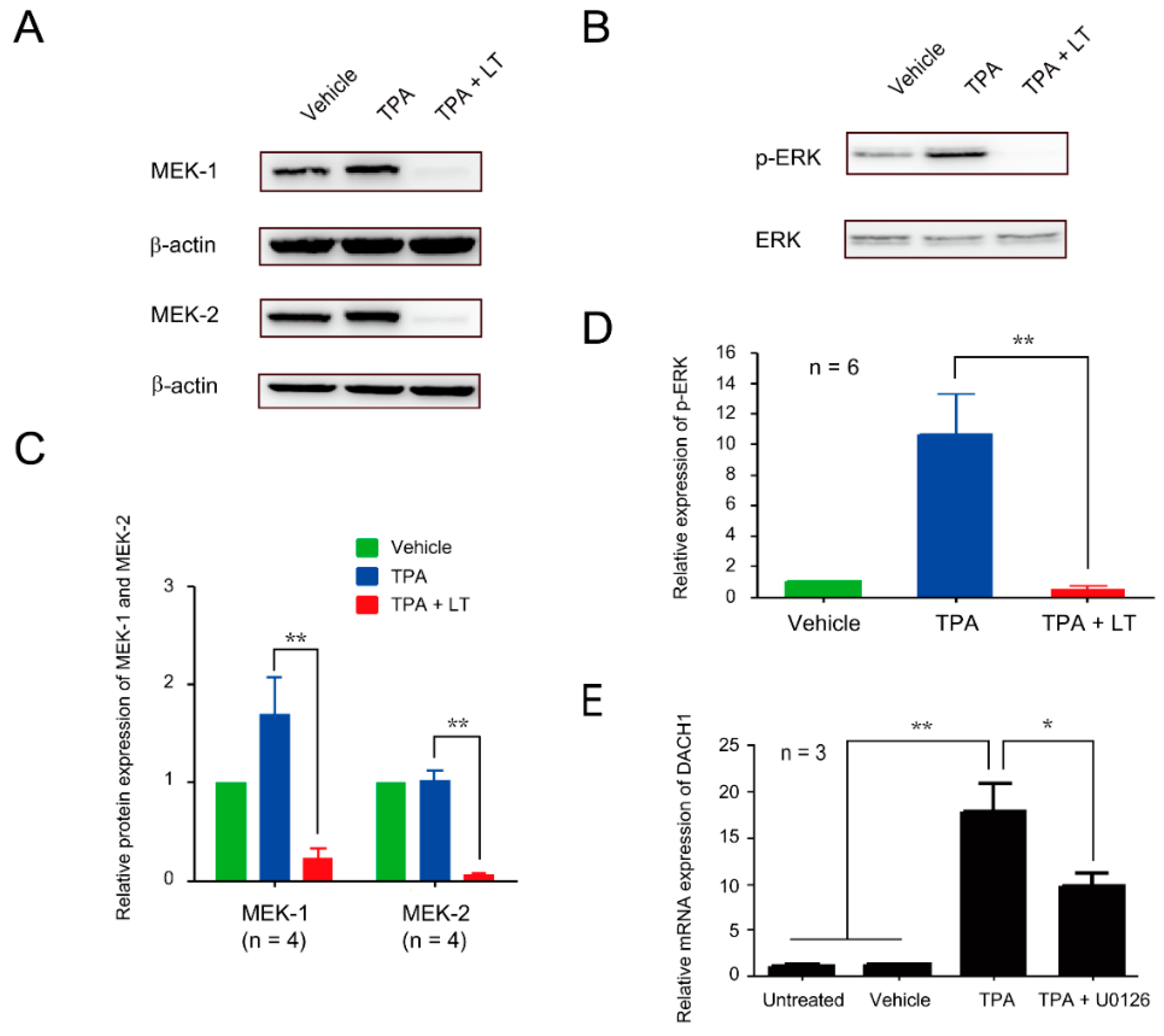
2.4. ERK Signaling Pathway Played a Role in DACH1-Mediated Suppression of Megakaryocytic Differentiation Induced by LT
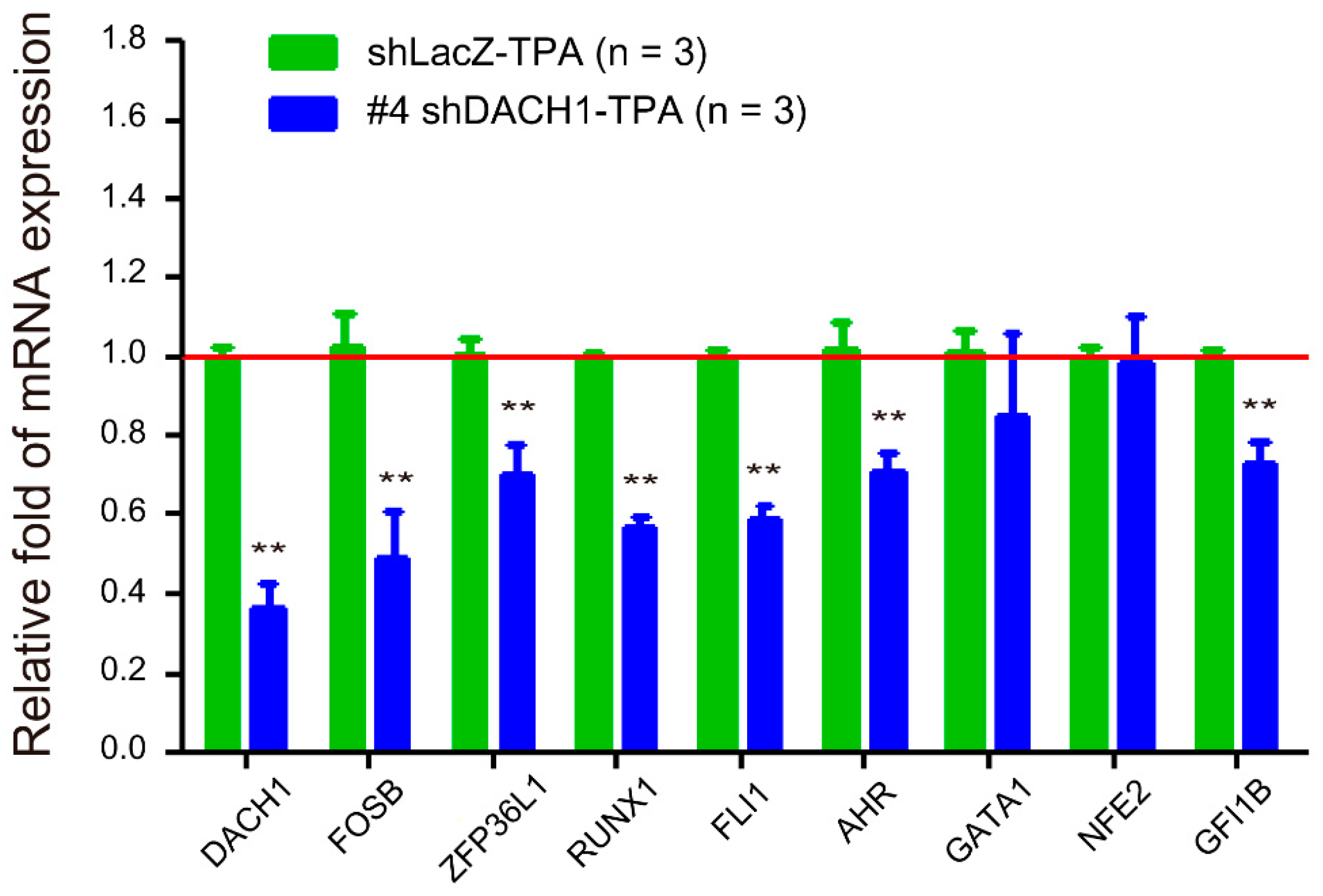
2.5. Relationship between the DACH1 Gene and Other Megakaryopoiesis-Related Genes
2.6. DACH1 Gene Is Upregulated during CD34–MK In Vitro Differentiation
2.7. Suppressed mRNA Expression in Thrombocytopenic Patients
3. Discussion
4. Materials and Methods
4.1. TPA-Induced Megakaryocytic Differentiation in HEL Cells
4.2. Flow Cytometry Assay
4.2.1. CD Marker Characterization
4.2.2. DNA Content Analysis
4.3. RNA Isolation and cDNA Microarray Assay
4.4. Quantitative Reverse Transcription Polymerase Chain Reaction
4.5. Short Hairpin RNA Knockdown Assay
4.6. Western Blot Analysis
4.7. CD34–Megakaryocytes In Vitro Differentiation
4.8. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Mock, M.; Fouet, A. Anthrax. Annu. Rev. Microbiol. 2001, 55, 647–671. [Google Scholar] [CrossRef] [PubMed]
- Booth, M.; Donaldson, L.; Cui, X.; Sun, J.; Cole, S.; Dailsey, S.; Hart, A.; Johns, N.; McConnell, P.; McLennan, T.; et al. Confirmed Bacillus anthracis infection among persons who inject drugs, Scotland, 2009–2010. Emerg. Infect. Dis. 2014, 20, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Jernigan, D.B.; Raghunathan, P.L.; Bell, B.P.; Brechner, R.; Bresnitz, E.A.; Butler, J.C.; Cetron, M.; Cohen, M.; Doyle, T.; Fischer, M.; et al. Investigation of bioterrorism-related anthrax, United States, 2001: Epidemiologic findings. Emerg. Infect. Dis. 2002, 8, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Abramova, F.A.; Grinberg, L.M.; Yampolskaya, O.V.; Walker, D.H. Pathology of inhalational anthrax in 42 cases from the Sverdlovsk outbreak of 1979. Proc. Natl. Acad. Sci. USA 1993, 90, 2291–2294. [Google Scholar] [CrossRef] [PubMed]
- Dixon, T.C.; Meselson, M.; Guillemin, J.; Hanna, P.C. Anthrax. N. Engl. J. Med. 1999, 341, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Shafazand, S.; Doyle, R.; Ruoss, S.; Weinacker, A.; Raffin, T.A. Inhalational anthrax: Epidemiology, diagnosis, and management. Chest 1999, 116, 1369–1376. [Google Scholar] [CrossRef]
- Freedman, A.; Afonja, O.; Chang, M.W.; Mostashari, F.; Blaser, M.; Perez-Perez, G.; Lazarus, H.; Schacht, R.; Guttenberg, J.; Traister, M.; et al. Cutaneous anthrax associated with microangiopathic hemolytic anemia and coagulopathy in a 7-month-old infant. JAMA 2002, 287, 869–874. [Google Scholar] [CrossRef]
- Brossier, F.; Mock, M. Toxins of Bacillus anthracis. Toxicon 2001, 39, 1747–1755. [Google Scholar] [CrossRef]
- Collier, R.J.; Young, J.A. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 2003, 19, 45–70. [Google Scholar] [CrossRef]
- Mourez, M. Anthrax toxins. Rev. Physiol. Biochem. Pharmacol. 2004, 152, 135–164. [Google Scholar]
- Bardwell, A.J.; Abdollahi, M.; Bardwell, L. Anthrax lethal factor-cleavage products of MAPK (mitogen-activated protein kinase) kinases exhibit reduced binding to their cognate MAPKs. Biochem. J. 2004, 378 Pt 2, 569–577. [Google Scholar] [CrossRef]
- Hagemann, C.; Blank, J.L. The ups and downs of MEK kinase interactions. Cell Signal. 2001, 13, 863–875. [Google Scholar] [CrossRef]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef] [PubMed]
- Moayeri, M.; Leppla, S.H. The roles of anthrax toxin in pathogenesis. Curr. Opin. Microbiol. 2004, 7, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Hanna, P.C.; Acosta, D.; Collier, R.J. On the role of macrophages in anthrax. Proc. Natl. Acad. Sci. USA 1993, 90, 10198–10201. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Lingappa, J.; Leppla, S.H.; Agrawal, S.; Jabbar, A.; Quinn, C.; Pulendran, B. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature 2003, 424, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.E. Anthrax lethal toxin induces human endothelial cell apoptosis. Infect. Immun. 2004, 72, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Comer, J.E.; Chopra, A.K.; Peterson, J.W.; Konig, R. Direct inhibition of T-lymphocyte activation by anthrax toxins in vivo. Infect. Immun. 2005, 73, 8275–8281. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Cordoba-Rodriguez, R.; Lankford, C.S.; Frucht, D.M. Anthrax lethal toxin blocks MAPK kinase-dependent IL-2 production in CD4+ T cells. J. Immunol. 2005, 174, 4966–4971. [Google Scholar] [CrossRef] [PubMed]
- Kau, J.H.; Sun, D.S.; Tsai, W.J.; Shyu, H.F.; Huang, H.H.; Lin, H.C.; Chang, H.H. Antiplatelet activities of anthrax lethal toxin are associated with suppressed p42/44 and p38 mitogen-activated protein kinase pathways in the platelets. J. Infect. Dis. 2005, 192, 1465–1474. [Google Scholar] [CrossRef]
- Paccani, S.R.; Tonello, F.; Ghittoni, R.; Natale, M.; Muraro, L.; D’Elios, M.M.; Tang, W.J.; Montecucco, C.; Baldari, C.T. Anthrax toxins suppress T lymphocyte activation by disrupting antigen receptor signaling. J. Exp. Med. 2005, 201, 325–331. [Google Scholar] [CrossRef]
- Crawford, M.A.; Aylott, C.V.; Bourdeau, R.W.; Bokoch, G.M. Bacillus anthracis Toxins Inhibit Human Neutrophil NADPH Oxidase Activity. J. Immunol. 2006, 176, 7557–7565. [Google Scholar] [CrossRef]
- Fang, H.; Xu, L.; Chen, T.Y.; Cyr, J.M.; Frucht, D.M. Anthrax lethal toxin has direct and potent inhibitory effects on B cell proliferation and immunoglobulin production. J. Immunol. 2006, 176, 6155–6161. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.E. Manipulation of host signalling pathways by anthrax toxins. Biochem. J. 2007, 402, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.K.; Chang, H.H.; Lin, G.L.; Wang, T.P.; Lai, Y.L.; Lin, T.K.; Hsieh, M.C.; Kau, J.H.; Huang, H.H.; Hsu, H.L.; et al. Suppressive effects of anthrax lethal toxin on megakaryopoiesis. PLoS ONE 2013, 8, e59512. [Google Scholar] [CrossRef] [PubMed]
- Grinberg, L.M.; Abramova, F.A.; Yampolskaya, O.V.; Walker, D.H.; Smith, J.H. Quantitative pathology of inhalational anthrax I: Quantitative microscopic findings. Mod. Pathol. 2001, 14, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Moayeri, M.; Haines, D.; Young, H.A.; Leppla, S.H. Bacillus anthracis lethal toxin induces TNF-alpha-independent hypoxia-mediated toxicity in mice. J. Clin. Investig. 2003, 112, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, I.; Kanakura, Y. Molecular control of megakaryopoiesis and thrombopoiesis. Int. J. Hematol. 2002, 75, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Weiss, M.J.; Poncz, M. Megakaryocyte biology and related disorders. J. Clin. Investig. 2005, 115, 3332–3338. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, V.R.; Tomer, A. Megakaryocyte development and platelet production. Br. J. Haematol. 2006, 134, 453–466. [Google Scholar] [CrossRef]
- Chang, Y.; Bluteau, D.; Debili, N.; Vainchenker, W. From hematopoietic stem cells to platelets. J. Thromb. Haemost. 2007, 5 (Suppl. S1), 318–327. [Google Scholar] [CrossRef]
- Wen, Q.; Goldenson, B.; Crispino, J.D. Normal and malignant megakaryopoiesis. Expert Rev. Mol. Med. 2011, 13, e32. [Google Scholar] [CrossRef]
- Deutsch, V.R.; Tomer, A. Advances in megakaryocytopoiesis and thrombopoiesis: From bench to bedside. Br. J. Haematol. 2013, 161, 778–793. [Google Scholar] [CrossRef]
- Avanzi, M.P.; Mitchell, W.B. Ex vivo production of platelets from stem cells. Br. J. Haematol. 2014, 165, 237–247. [Google Scholar] [CrossRef]
- Noh, J.Y. Megakaryopoiesis and Platelet Biology: Roles of Transcription Factors and Emerging Clinical Implications. Int. J. Mol. Sci. 2021, 22, 9615. [Google Scholar] [CrossRef] [PubMed]
- Pintado, C.O.; Friend, M.; Llanes, D. Characterisation of a membrane receptor on ruminants and equine platelets and peripheral blood leukocytes similar to the human integrin receptor glycoprotein IIb/IIIa (CD41/61). Vet. Immunol. Immunopathol. 1995, 44, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Furukawa, Y.; Iwase, S.; Terui, Y.; Nakamura, M.; Kitagawa, S.; Kitagawa, M.; Komatsu, N.; Miura, Y. Polyploidization and functional maturation are two distinct processes during megakaryocytic differentiation: Involvement of cyclin-dependent kinase inhibitor p21 in polyploidization. Blood 1997, 89, 3980–3990. [Google Scholar] [CrossRef] [PubMed]
- Cavalloni, G.; Dane, A.; Piacibello, W.; Bruno, S.; Lamas, E.; Brechot, C.; Aglietta, M. The involvement of human-nuc gene in polyploidization of K562 cell line. Exp. Hematol. 2000, 28, 1432–1440. [Google Scholar] [CrossRef]
- Guerriero, R.; Mattia, G.; Testa, U.; Chelucci, C.; Macioce, G.; Casella, I.; Samoggia, P.; Peschle, C.; Hassan, H.J. Stromal cell-derived factor 1alpha increases polyploidization of megakaryocytes generated by human hematopoietic progenitor cells. Blood 2001, 97, 2587–2595. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.J.; Chae, H.C.; Seoh, J.Y.; Ryu, K.H.; Park, H.K.; Kim, Y.J.; Woo, S.Y. Pim-1 induced polyploidy but did not affect megakaryocytic differentiation of K562 cells and CD34+ cells from cord blood. Eur. J. Haematol. 2007, 78, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Zimmet, J.; Ravid, K. Polyploidy: Occurrence in nature, mechanisms, and significance for the megakaryocyte-platelet system. Exp. Hematol. 2000, 28, 3–16. [Google Scholar] [CrossRef]
- Ravid, K.; Lu, J.; Zimmet, J.M.; Jones, M.R. Roads to polyploidy: The megakaryocyte example. J. Cell. Physiol. 2002, 190, 7–20. [Google Scholar] [CrossRef]
- Patel, S.R.; Hartwig, J.H.; Italiano, J.E., Jr. The biogenesis of platelets from megakaryocyte proplatelets. J. Clin. Investig. 2005, 115, 3348–3354. [Google Scholar] [CrossRef]
- Tijssen, M.R.; Ghevaert, C. Transcription factors in late megakaryopoiesis and related platelet disorders. J. Thromb. Haemost. 2013, 11, 593–604. [Google Scholar] [CrossRef]
- Mazzi, S.; Lordier, L.; Debili, N.; Raslova, H.; Vainchenker, W. Megakaryocyte and polyploidization. Exp. Hematol. 2018, 57, 1–13. [Google Scholar] [CrossRef]
- Lindsey, S.; Papoutsakis, E.T. The aryl hydrocarbon receptor (AHR) transcription factor regulates megakaryocytic polyploidization. Br. J. Haematol. 2011, 152, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Melemed, A.S.; Ryder, J.W.; Vik, T.A. Activation of the mitogen-activated protein kinase pathway is involved in and sufficient for megakaryocytic differentiation of CMK cells. Blood 1997, 90, 3462–3470. [Google Scholar] [CrossRef] [PubMed]
- Racke, F.K.; Lewandowska, K.; Goueli, S.; Goldfarb, A.N. Sustained activation of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway is required for megakaryocytic differentiation of K562 cells. J. Biol. Chem. 1997, 272, 23366–23370. [Google Scholar] [CrossRef] [PubMed]
- Rojnuckarin, P.; Drachman, J.G.; Kaushansky, K. Thrombopoietin-induced activation of the mitogen-activated protein kinase (MAPK) pathway in normal megakaryocytes: Role in endomitosis. Blood 1999, 94, 1273–1282. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, R.; Ogata, H.; Kobayashi, Y. Requirement of thrombopoietin-induced activation of ERK for megakaryocyte differentiation and of p38 for erythroid differentiation. Ann. Hematol. 2001, 80, 284–291. [Google Scholar] [CrossRef]
- Jacquel, A.; Herrant, M.; Defamie, V.; Belhacene, N.; Colosetti, P.; Marchetti, S.; Legros, L.; Deckert, M.; Mari, B.; Cassuto, J.P.; et al. A survey of the signaling pathways involved in megakaryocytic differentiation of the human K562 leukemia cell line by molecular and c-DNA array analysis. Oncogene 2006, 25, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Mardon, G.; Solomon, N.M.; Rubin, G.M. dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development 1994, 120, 3473–3486. [Google Scholar] [CrossRef] [PubMed]
- Ayres, J.A.; Shum, L.; Akarsu, A.N.; Dashner, R.; Takahashi, K.; Ikura, T.; Slavkin, H.C.; Nuckolls, G.H. DACH: Genomic characterization, evaluation as a candidate for postaxial polydactyly type A2, and developmental expression pattern of the mouse homologue. Genomics 2001, 77, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Popov, V.M.; Wu, K.; Zhou, J.; Powell, M.J.; Mardon, G.; Wang, C.; Pestell, R.G. The Dachshund gene in development and hormone-responsive tumorigenesis. Trends Endocrinol. Metab. 2010, 21, 41–49. [Google Scholar] [CrossRef]
- Davis, R.J.; Harding, M.; Moayedi, Y.; Mardon, G. Mouse Dach1 and Dach2 are redundantly required for Mullerian duct development. Genesis 2008, 46, 205–213. [Google Scholar] [CrossRef]
- Riggs, M.J.; Lin, N.; Wang, C.; Piecoro, D.W.; Miller, R.W.; Hampton, O.A.; Rao, M.; Ueland, F.R.; Kolesar, J.M. DACH1 mutation frequency in endometrial cancer is associated with high tumor mutation burden. PLoS ONE 2020, 15, e0244558. [Google Scholar] [CrossRef] [PubMed]
- Aman, S.; Li, Y.; Cheng, Y.; Yang, Y.; Lv, L.; Li, B.; Xia, K.; Li, S.; Wu, H. DACH1 inhibits breast cancer cell invasion and metastasis by down-regulating the transcription of matrix metalloproteinase 9. Cell Death Discov. 2021, 7, 351. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Ding, H.; Xu, C.; Kou, X. Methylation status of DACH1 gene in esophageal cancer and its clinical significance. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2021, 38, 1002–1006. [Google Scholar]
- Yu, J.; Jiang, P.; Zhao, K.; Chen, Z.; Zuo, T.; Chen, B. Role of DACH1 on Proliferation, Invasion, and Apoptosis in Human Lung Adenocarcinoma Cells. Curr. Mol. Med. 2021, 21, 806–811. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, W.; Wang, W.; Xu, Y.; Jiang, L.; Gu, Z. Diagnostic and Prognostic Value of DACH1 Methylation in the Sensitivity of Esophageal Cancer to Radiotherapy. Contrast Media Mol. Imaging 2022, 2022, 6857685. [Google Scholar] [CrossRef]
- Gu, Q.; Li, L.; Yao, J.; Dong, F.Y.; Gan, Y.; Zhou, S.; Wang, X.; Wang, X.F. Identification and verification of the temozolomide resistance feature gene DACH1 in gliomas. Front. Oncol. 2023, 13, 1120103. [Google Scholar] [CrossRef] [PubMed]
- Nasirpour, M.H.; Salimi, M.; Majidi, F.; Minuchehr, Z.; Mozdarani, H. Study of DACH1 Expression and its Epigenetic Regulators as Possible Breast Cancer-Related Biomarkers. Avicenna J. Med. Biotechnol. 2023, 15, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, L.; Wang, M. Alteration of DACH1 methylation patterns in lung cancer contributes to cell proliferation and migration. Biochem. Cell Biol. 2018, 96, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Amann-Zalcenstein, D.; Tian, L.; Schreuder, J.; Tomei, S.; Lin, D.S.; Fairfax, K.A.; Bolden, J.E.; McKenzie, M.D.; Jarratt, A.; Hilton, A.; et al. A new lymphoid-primed progenitor marked by Dach1 downregulation identified with single cell multi-omics. Nat. Immunol. 2020, 21, 1574–1584. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Ehrlich, L.I.; Seita, J.; Murakami, P.; Doi, A.; Lindau, P.; Lee, H.; Aryee, M.J.; Irizarry, R.A.; Kim, K.; et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 2010, 467, 338–342. [Google Scholar] [CrossRef]
- Catani, M.V.; Fezza, F.; Baldassarri, S.; Gasperi, V.; Bertoni, A.; Pasquariello, N.; Finazzi-Agro, A.; Sinigaglia, F.; Avigliano, L.; Maccarrone, M. Expression of the endocannabinoid system in the bi-potential HEL cell line: Commitment to the megakaryoblastic lineage by 2-arachidonoylglycerol. J. Mol. Med. 2009, 87, 65–74. [Google Scholar] [CrossRef]
- Garcia, P.; Cales, C. Endoreplication in megakaryoblastic cell lines is accompanied by sustained expression of G1/S cyclins and downregulation of cdc25C. Oncogene 1996, 13, 695–703. [Google Scholar] [PubMed]
- Chang, H.H.; Wang, T.P.; Chen, P.K.; Lin, Y.Y.; Liao, C.H.; Lin, T.K.; Chiang, Y.W.; Lin, W.B.; Chiang, C.Y.; Kau, J.H.; et al. Erythropoiesis suppression is associated with anthrax lethal toxin-mediated pathogenic progression. PLoS ONE 2013, 8, e71718. [Google Scholar] [CrossRef]
- Kau, J.H.; Sun, D.S.; Huang, H.H.; Wong, M.S.; Lin, H.C.; Chang, H.H. Role of visible light-activated photocatalyst on the reduction of anthrax spore-induced mortality in mice. PLoS ONE 2009, 4, e4167. [Google Scholar] [CrossRef]
- Kau, J.H.; Sun, D.S.; Huang, H.S.; Lien, T.S.; Huang, H.H.; Lin, H.C.; Chang, H.H. Sublethal doses of anthrax lethal toxin on the suppression of macrophage phagocytosis. PLoS ONE 2010, 5, e14289. [Google Scholar] [CrossRef]
- Hong, Y.; Martin, J.F.; Vainchenker, W.; Erusalimsky, J.D. Inhibition of protein kinase C suppresses megakaryocytic differentiation and stimulates erythroid differentiation in HEL cells. Blood 1996, 87, 123–131. [Google Scholar] [CrossRef]
- Debili, N.; Robin, C.; Schiavon, V.; Letestu, R.; Pflumio, F.; Mitjavila-Garcia, M.T.; Coulombel, L.; Vainchenker, W. Different expression of CD41 on human lymphoid and myeloid progenitors from adults and neonates. Blood 2001, 97, 2023–2030. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.; Bradley, K.A. Anthrax toxin: Can a little be a good thing? Trends Microbiol. 2004, 12, 143–145. [Google Scholar] [CrossRef] [PubMed]
- Duesbery, N.S.; Webb, C.P.; Leppla, S.H.; Gordon, V.M.; Klimpel, K.R.; Copeland, T.D.; Ahn, N.G.; Oskarsson, M.K.; Fukasawa, K.; Paull, K.D.; et al. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 1998, 280, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Raftrey, B.; Williams, M.; Rios Coronado, P.E.; Fan, X.; Chang, A.H.; Zhao, M.; Roth, R.; Trimm, E.; Racelis, R.; D’Amato, G.; et al. Dach1 Extends Artery Networks and Protects Against Cardiac Injury. Circ. Res. 2021, 129, 702–716. [Google Scholar] [CrossRef]
- Chang, A.H.; Raftrey, B.C.; D’Amato, G.; Surya, V.N.; Poduri, A.; Chen, H.I.; Goldstone, A.B.; Woo, J.; Fuller, G.G.; Dunn, A.R.; et al. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev. 2017, 31, 1308–1324. [Google Scholar] [CrossRef] [PubMed]
- MacGrogan, D.; de la Pompa, J.L. DACH1-Driven Arterialization: Angiogenic Therapy for Ischemic Heart Disease? Circ. Res. 2021, 129, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Vyas, P.; Ault, K.; Jackson, C.W.; Orkin, S.H.; Shivdasani, R.A. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood 1999, 93, 2867–2875. [Google Scholar] [CrossRef] [PubMed]
- Muntean, A.G.; Pang, L.; Poncz, M.; Dowdy, S.F.; Blobel, G.A.; Crispino, J.D. Cyclin D-Cdk4 is regulated by GATA-1 and required for megakaryocyte growth and polyploidization. Blood 2007, 109, 5199–5207. [Google Scholar] [CrossRef]
- Lordier, L.; Bluteau, D.; Jalil, A.; Legrand, C.; Pan, J.; Rameau, P.; Jouni, D.; Bluteau, O.; Mercher, T.; Leon, C.; et al. RUNX1-induced silencing of non-muscle myosin heavy chain IIB contributes to megakaryocyte polyploidization. Nat. Commun. 2012, 3, 717. [Google Scholar] [CrossRef]
- Thomsen, J.S.; Kietz, S.; Strom, A.; Gustafsson, J.A. HES-1, a novel target gene for the aryl hydrocarbon receptor. Mol. Pharmacol. 2004, 65, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Hattori, M.; Hirai, N.; Shinozuka, Y.; Hirata, H.; Kageyama, R.; Sakai, T.; Minato, N. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol. Cell. Biol. 2005, 25, 4262–4271. [Google Scholar] [CrossRef] [PubMed]
- Mikesova, E.; Barankova, L.; Sakmaryova, I.; Tatarkova, I.; Seeman, P. Quantitative multiplex real-time PCR for detection of PLP1 gene duplications in Pelizaeus-Merzbacher patients. Genet. Test. 2006, 10, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Moutsatsou, P.; Psarra, A.M.; Tsiapara, A.; Paraskevakou, H.; Davaris, P.; Sekeris, C.E. Localization of the glucocorticoid receptor in rat brain mitochondria. Arch. Biochem. Biophys. 2001, 386, 69–78. [Google Scholar] [CrossRef]
- Cortin, V.; Pineault, N.; Garnier, A. Ex vivo megakaryocyte expansion and platelet production from human cord blood stem cells. Methods Mol. Biol. 2009, 482, 109–126. [Google Scholar]
| WBC (103/μL) | RBC (106/μL) | Hb (g/dL) | Hct (%) | MCV (fL) | MCH (pg) | MCHC (%) | PLT (103/μL) | |
|---|---|---|---|---|---|---|---|---|
| donor 1 | 6.22 | 4.21 | 13.4 | 39.9 | 94.8 | 31.8 | 33.6 | 420 |
| donor 2 | 5.06 | 4.81 | 14.1 | 41.2 | 85.7 | 29.3 | 34.2 | 281 |
| donor 3 | 7.01 | 4.70 | 14.1 | 40.5 | 86.2 | 30.0 | 34.8 | 241 |
| patient 1 | 10.01 | 3.07 ↓ | 8.7 ↓ | 26.2 ↓ | 85.3 | 28.3 | 33.2 | 44 ↓ |
| patient 2 | 4.40 | 3.16 ↓ | 10.0 ↓ | 30.2 ↓ | 95.6 | 31.6 | 33.1 | 143 ↓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, G.-L.; Chang, H.-H.; Lin, W.-T.; Liou, Y.-S.; Lai, Y.-L.; Hsieh, M.-H.; Chen, P.-K.; Liao, C.-Y.; Tsai, C.-C.; Wang, T.-F.; et al. Dachshund Homolog 1: Unveiling Its Potential Role in Megakaryopoiesis and Bacillus anthracis Lethal Toxin-Induced Thrombocytopenia. Int. J. Mol. Sci. 2024, 25, 3102. https://doi.org/10.3390/ijms25063102
Lin G-L, Chang H-H, Lin W-T, Liou Y-S, Lai Y-L, Hsieh M-H, Chen P-K, Liao C-Y, Tsai C-C, Wang T-F, et al. Dachshund Homolog 1: Unveiling Its Potential Role in Megakaryopoiesis and Bacillus anthracis Lethal Toxin-Induced Thrombocytopenia. International Journal of Molecular Sciences. 2024; 25(6):3102. https://doi.org/10.3390/ijms25063102
Chicago/Turabian StyleLin, Guan-Ling, Hsin-Hou Chang, Wei-Ting Lin, Yu-Shan Liou, Yi-Ling Lai, Min-Hua Hsieh, Po-Kong Chen, Chi-Yuan Liao, Chi-Chih Tsai, Tso-Fu Wang, and et al. 2024. "Dachshund Homolog 1: Unveiling Its Potential Role in Megakaryopoiesis and Bacillus anthracis Lethal Toxin-Induced Thrombocytopenia" International Journal of Molecular Sciences 25, no. 6: 3102. https://doi.org/10.3390/ijms25063102
APA StyleLin, G.-L., Chang, H.-H., Lin, W.-T., Liou, Y.-S., Lai, Y.-L., Hsieh, M.-H., Chen, P.-K., Liao, C.-Y., Tsai, C.-C., Wang, T.-F., Chu, S.-C., Kau, J.-H., Huang, H.-H., Hsu, H.-L., & Sun, D.-S. (2024). Dachshund Homolog 1: Unveiling Its Potential Role in Megakaryopoiesis and Bacillus anthracis Lethal Toxin-Induced Thrombocytopenia. International Journal of Molecular Sciences, 25(6), 3102. https://doi.org/10.3390/ijms25063102






