Low-Toxicity Self-Photosensitized Biohybrid Systems for Enhanced Light-Driven H2 Production
Abstract
1. Introduction
2. Results
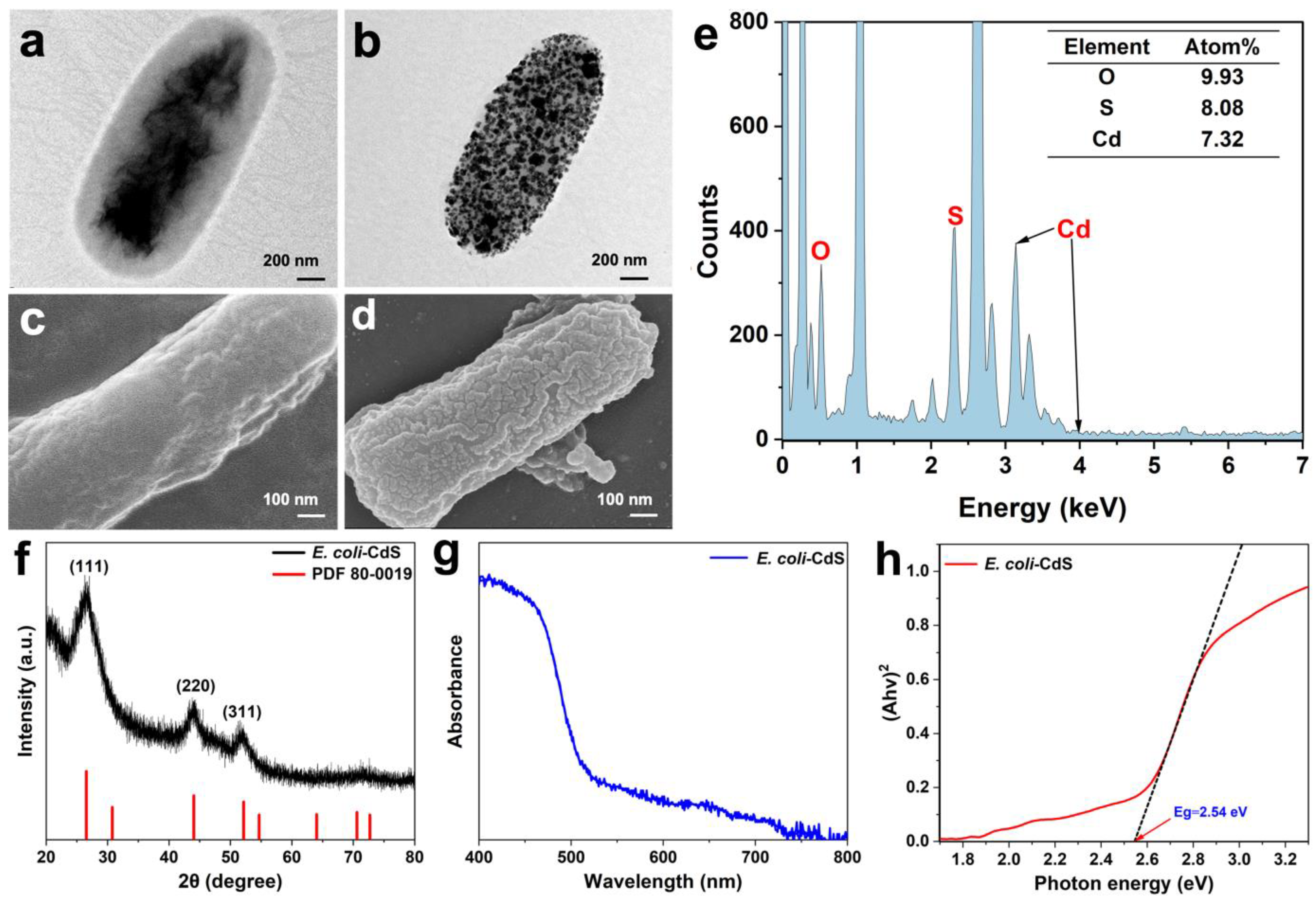
2.1. Construction and Characterization of the E. coli–CdS Hybrid System
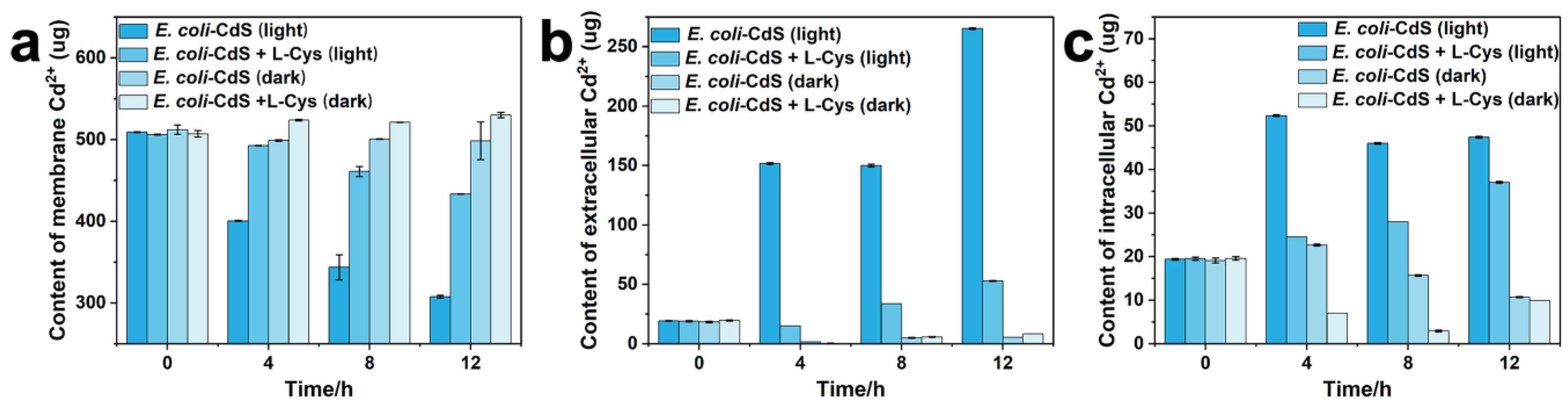
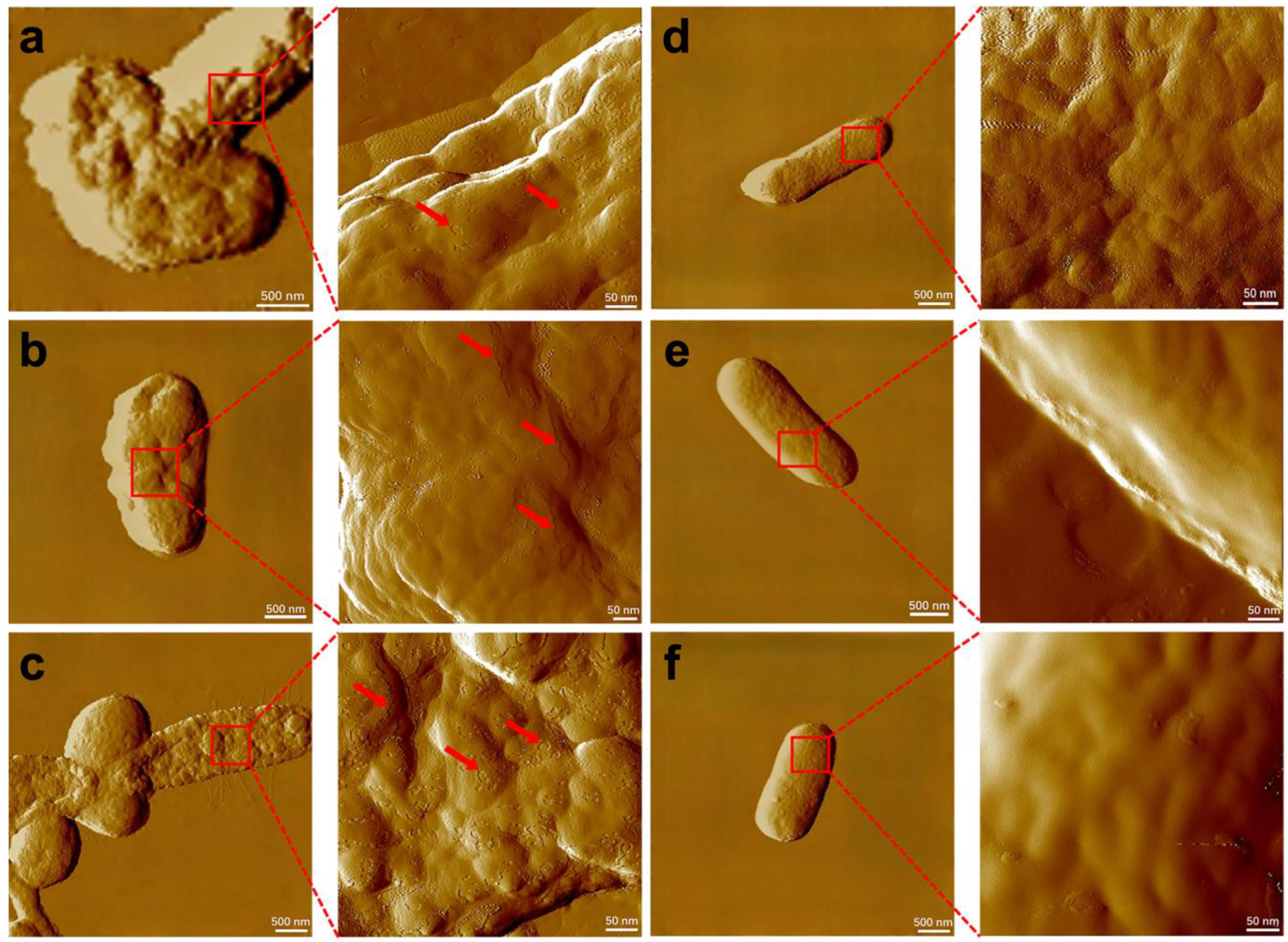
2.2. Toxicity Assessment of Cd2+ and CdS NPs on Whole Bacterial Cells
2.3. Construction of the Engineered efeB/OE–CdS System
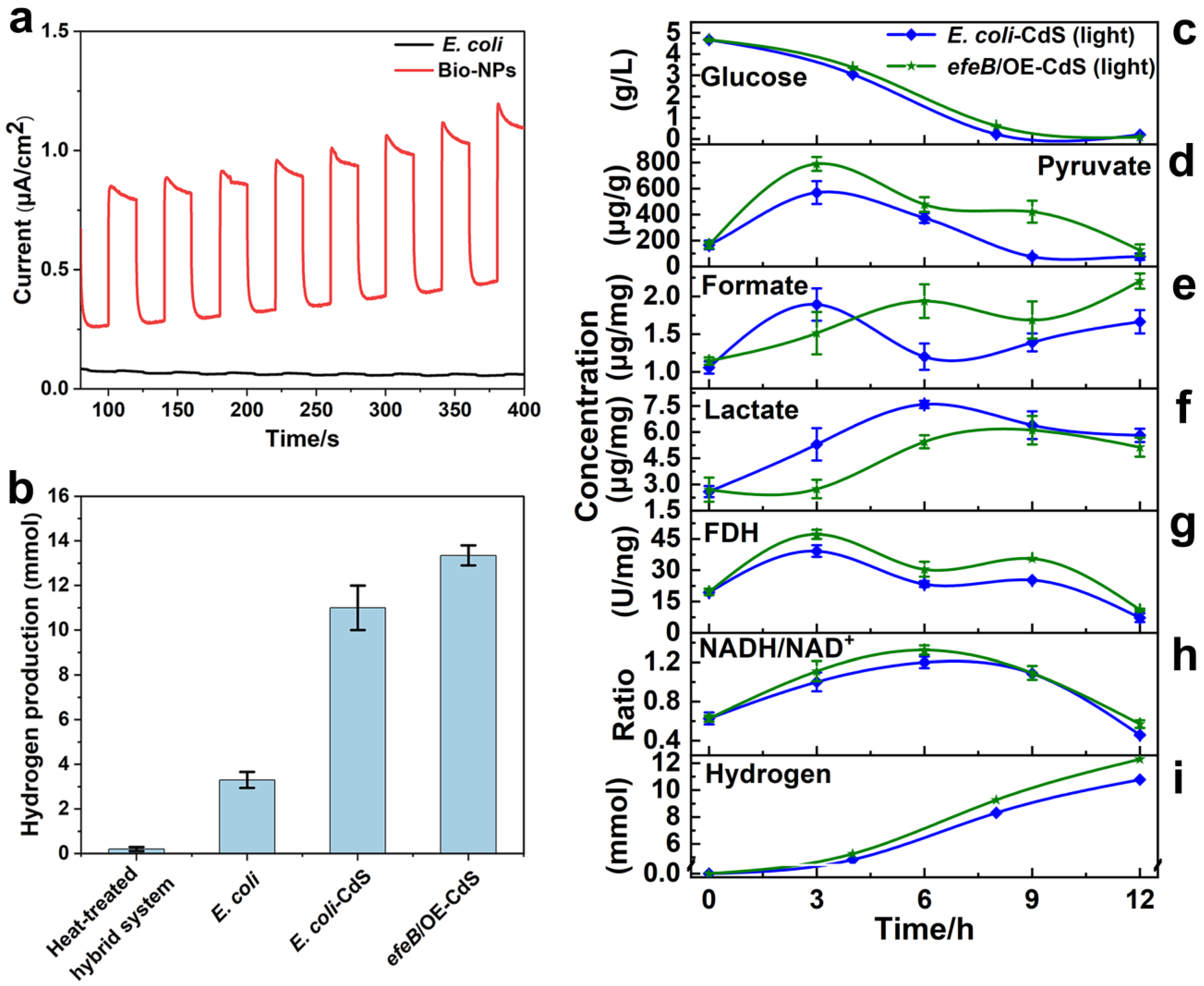
2.4. Contribution of EfeB to Light-Driven Hydrogen Production
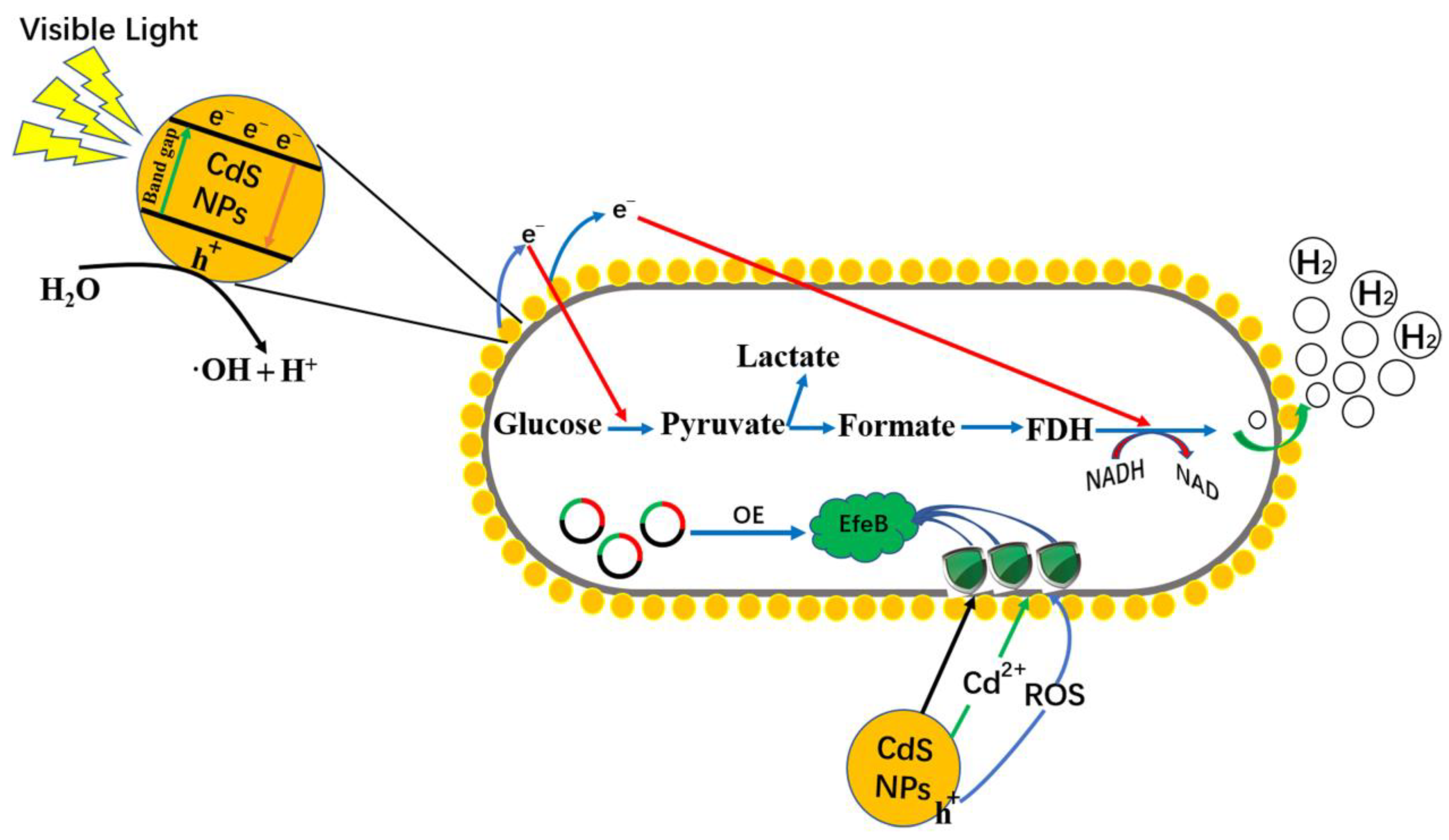
2.5. Mechanism Underlying Hydrogen Production in Light-Assisted Hybrid Systems
3. Discussion
4. Materials and Methods
4.1. Wild-Type and Genetically Engineered E. coli Strains
4.2. Construction and Characterization of Biohybrid Systems
4.3. Cd2+ and CdS Assay during Light Irradiation
4.4. Cell Enzyme Activity and MDA Assay under Oxidative Stress
4.5. Quantitative Genetic Testing
4.6. Cell PI Staining and Atomic Force Microscopy Analyses of Cell Surfaces
4.7. Light-Assisted Fermentative Hydrogen Generation
4.8. Metabolite Assay in Biohybrid Systems during Hydrogen Production
4.9. Analytical Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- King, D. Global clean energy in 2017. Science 2017, 355, 111. [Google Scholar] [CrossRef]
- Turner, J.M. The matter of a clean energy future. Science 2022, 376, 1361. [Google Scholar] [CrossRef]
- Sun, X.D.; Jiang, S.Y.; Huang, H.W.; Li, H.; Jia, B.H.; Ma, T.Y. Solar Energy Catalysis. Angew. Chem. Int. Ed. 2022, 61, 2204880. [Google Scholar] [CrossRef]
- Zhuang, C.; Li, L.; Ban, C.; Liu, Y.; Liu, X. Synthesis, analysis and electrical properties of silicon doped BN nanowires. J. Alloys Compd. 2018, 731, 84–89. [Google Scholar] [CrossRef]
- Sakimoto, K.K.; Wong, A.B.; Yang, P. Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 2016, 351, 74–77. [Google Scholar] [CrossRef]
- Luo, B.F.; Wang, Y.Z.; Li, D.; Shen, H.Q.; Xu, L.X.; Fang, Z.; Xia, Z.L.; Ren, J.L.; Shi, W.D.; Yong, Y.C. A Periplasmic Photosensitized Biohybrid System for Solar Hydrogen Production. Adv. Energy Mater. 2021, 11, 2100256. [Google Scholar] [CrossRef]
- He, Y.; Wang, S.R.; Han, X.Y.; Shen, J.Y.; Lu, Y.W.; Zhao, J.Z.; Shen, C.P.; Qiao, L. Photosynthesis of Acetate by Sporomusa ovata-CdS Biohybrid System. ACS Appl. Mater. Interfaces 2022, 14, 23364–23374. [Google Scholar] [CrossRef]
- Wang, B.; Xiao, K.M.; Jiang, Z.F.; Wang, J.F.; Yu, J.C.; Wong, P.K. Biohybrid photoheterotrophic metabolism for significant enhancement of biological nitrogen fixation in pure microbial cultures. Energy Environ. Sci. 2019, 12, 2185–2191. [Google Scholar] [CrossRef]
- Martins, M.; Toste, C.; Pereira, I.A.C. Enhanced Light-Driven Hydrogen Production by Self-Photosensitized Biohybrid Systems. Angew. Chem. Int. Ed. 2021, 60, 9055–9062. [Google Scholar] [CrossRef]
- Han, H.-X.; Tian, L.-J.; Liu, D.-F.; Yu, H.-Q.; Sheng, G.-P.; Xiong, Y. Reversing Electron Transfer Chain for Light-Driven Hydrogen Production in Biotic–Abiotic Hybrid Systems. J. Am. Chem. Soc. 2022, 144, 6434–6441. [Google Scholar] [CrossRef]
- Honda, Y.; Watanabe, M.; Hagiwara, H.; Ida, S.; Ishihara, T. Inorganic/whole-cell biohybrid photocatalyst for highly efficient hydrogen production from water. Appl. Catal. B-Environ. 2017, 210, 400–406. [Google Scholar] [CrossRef]
- Wang, B.; Zeng, C.; Chu, K.H.; Wu, D.; Yip, H.Y.; Ye, L.; Wong, P.K. Enhanced Biological Hydrogen Production from Escherichia coli with Surface Precipitated Cadmium Sulfide Nanoparticles. Adv. Energy Mater. 2017, 7, 1700611. [Google Scholar] [CrossRef]
- Utterback, J.K.; Wilker, M.B.; Brown, K.A.; King, P.W.; Eaves, J.D.; Dukovic, G. Competition between electron transfer, trapping, and recombination in CdS nanorod-hydrogenase complexes. Phys. Chem. Chem. Phys. 2015, 17, 5538–5542. [Google Scholar] [CrossRef]
- Reisner, E.; Powell, D.J.; Cavazza, C.; Fontecilla-Camps, J.C.; Armstrong, F.A. Visible Light-Driven H2 Production by Hydrogenases Attached to Dye-Sensitized TiO2 Nanoparticles. J. Am. Chem. Soc. 2009, 131, 18457–18466. [Google Scholar] [CrossRef]
- Lee, C.-Y.; Park, H.S.; Fontecilla-Camps, J.C.; Reisner, E. Photoelectrochemical H2 Evolution with a Hydrogenase Immobilized on a TiO2-Protected Silicon Electrode. Angew. Chem. 2016, 128, 6075–6078. [Google Scholar] [CrossRef]
- Chen, M.; Zhou, X.-F.; Yu, Y.-Q.; Liu, X.; Zeng, R.J.-X.; Zhou, S.-G.; He, Z. Light-driven nitrous oxide production via autotrophic denitrification by self-photosensitized Thiobacillus denitrificans. Environ. Int. 2019, 127, 353–360. [Google Scholar] [CrossRef]
- Huang, S.; Tang, J.; Liu, X.; Dong, G.; Zhou, S. Fast Light-Driven Biodecolorization by a Geobacter sulfurreducens-CdS Biohybrid. ACS Sustain. Chem. Eng. 2019, 7, 15427–15433. [Google Scholar] [CrossRef]
- Liu, P.-C.; Ma, X.-L.; Li, T.-T.; Yan, F.; Wu, L.-J.; Xiao, X. Elucidation of photodegradation of p-chlorophenol in a biophotoelectric reductive degradation system by density functional theory calculations. Int. Biodeterior. Biodegrad. 2020, 151, 104969. [Google Scholar] [CrossRef]
- Zuo, W.L.; Yu, Y.D.; Huang, H. Making waves: Microbe-photocatalyst hybrids may provide new opportunities for treating heavy metal polluted wastewater. Water Res. 2021, 195, 116984. [Google Scholar] [CrossRef]
- Chen, X.Y.; Feng, Q.Y.; Cai, Q.H.; Huang, S.F.; Yu, Y.Q.; Zeng, R.J.; Chen, M.; Zhou, S.G. Mn3O4 Nanozyme Coating Accelerates Nitrate Reduction and Decreases N2O Emission during Photoelectrotrophic Denitrification by Thiobacillus denitrificans-CdS. Environ. Sci. Technol. 2020, 54, 10820–10830. [Google Scholar] [CrossRef]
- Cui, D.; Wang, J.; Wang, H.; Yang, Y.; Zhao, M. The cytotoxicity of endogenous CdS and Cd2+ ions during CdS NPs biosynthesis. J. Hazard. Mater. 2021, 409, 124485. [Google Scholar] [CrossRef]
- Newman, D.K.; Beveridge, T.J.; Morel, F. Precipitation of Arsenic Trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 1997, 63, 2022–2028. [Google Scholar] [CrossRef]
- Suresh, A.K.; Doktycz, M.J.; Wang, W.; Moon, J.-W.; Gu, B.; Meyer, H.M.; Hensley, D.K.; Allison, D.P.; Phelps, T.J.; Pelletier, D.A. Monodispersed biocompatible silver sulfide nanoparticles: Facile extracellular biosynthesis using the γ-proteobacterium, Shewanella oneidensis. Acta Biomater. 2011, 7, 4253–4258. [Google Scholar] [CrossRef]
- Yu, Z.; Hao, R.; Zhang, L.; Zhu, Y. Effects of TiO2, SiO2, Ag and CdTe/CdS quantum dots nanoparticles on toxicity of cadmium towards Chlamydomonas reinhardtii. Ecotoxicol. Environ. Saf. 2018, 156, 75–86. [Google Scholar] [CrossRef]
- Bellanger, X.; Schneider, R.; Dezanet, C.; Arroua, B.; Balan, L.; Billard, P.; Merlin, C. Zn2+ leakage and photo-induced reactive oxidative species do not explain the full toxicity of ZnO core Quantum Dots. J. Hazard. Mater. 2020, 396, 122616. [Google Scholar] [CrossRef]
- Chen, N.; He, Y.; Su, Y.; Li, X.; Huang, Q.; Wang, H.; Zhang, X.; Tai, R.; Fan, C. The cytotoxicity of cadmium-based quantum dots. Biomaterials 2012, 33, 1238–1244. [Google Scholar] [CrossRef]
- Zhi, B.; Mishra, S.; Hudson-Smith, N.V.; Kortshagen, U.R.; Haynes, C.L. Toxicity Evaluation of Boron- and Phosphorus-Doped Silicon Nanocrystals toward Shewanella oneidensis MR-1. ACS Appl. Nano Mater. 2018, 1, 4884–4893. [Google Scholar] [CrossRef]
- Sakimoto, K.K.; Kornienko, N.; Cestellos-Blanco, S.; Lim, J.; Liu, C.; Yang, P.D. Physical Biology of the Materials-Microorganism Interface. J. Am. Chem. Soc. 2018, 140, 1978–1985. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, H.; Tian, Z.; Lu, D.; Yu, Y.; Cestellos-Blanco, S.; Sakimoto, K.K.; Yang, P. Bacteria photosensitized by intracellular gold nanoclusters for solar fuel production. Nat. Nanotechnol. 2018, 13, 900–905. [Google Scholar] [CrossRef]
- Meyer, J.; Hamwi, S.; Kroger, M.; Kowalsky, W.; Riedl, T.; Kahn, A. Transition Metal Oxides for Organic Electronics: Energetics, Device Physics and Applications. Adv. Mater. 2012, 24, 5408–5427. [Google Scholar] [CrossRef]
- Cui, S.; Tian, L.-J.; Li, J.; Wang, X.-M.; Liu, H.-Q.; Fu, X.-Z.; He, R.-L.; Lam, P.K.S.; Huang, T.-Y.; Li, W.-W. Light-assisted fermentative hydrogen production in an intimately-coupled inorganic-bio hybrid with self-assembled nanoparticles. Chem. Eng. J. 2022, 428, 131254. [Google Scholar] [CrossRef]
- Sun, P.Q.; Li, K.L.; Lin, K.; Wei, W.; Zhao, J. Ag2S Quantum Dots for Use in Whole-Cell Biohybrid Catalyst for Visible-Light-Driven Photocatalytic Organic Pollutant Degradation. ACS Appl. Nano Mater. 2022, 5, 9754–9760. [Google Scholar] [CrossRef]
- Wang, C.L.; Lum, A.M.; Ozuna, S.C.; Clark, D.S.; Keasling, J.D. Aerobic sulfide production and cadmium precipitation by Escherichia coli expressing the Treponema denticola cysteine desulfhydrase gene. Appl. Microbiol. Biotechnol. 2001, 56, 425–430. [Google Scholar] [CrossRef]
- Wang, Y.L.; Liu, Y.Q.; Zhao, N.; Wang, J.Y.; Yang, Y.; Cui, D.Z.; Zhao, M. Fe3O4 nanozyme coating enhances light-driven biohydrogen production in self-photosensitized Shewanella oneidensis-CdS hybrid systems. Biotechnol. J. 2023, 18, e2300084. [Google Scholar] [CrossRef]
- Hossain, S.T.; Mukherjee, S.K. Toxicity of cadmium sulfide (CdS) nanoparticles against Escherichia coli and HeLa cells. J. Hazard. Mater. 2013, 260, 1073–1082. [Google Scholar] [CrossRef]
- Derfus, A.M.; Chan, W.C.W.; Bhatia, S.N. Probing the Cytotoxicity Of Semiconductor Quantum Dots. Nano Lett. 2004, 4, 11–18. [Google Scholar] [CrossRef]
- Pechstedt, K.; Whittle, T.; Baumberg, J.; Melvin, T. Photoluminescence of Colloidal CdSe/ZnS Quantum Dots: The Critical Effect of Water Molecules. J. Phys. Chem. C 2010, 114, 12069–12077. [Google Scholar] [CrossRef]
- Sobhanan, J.; Jones, P.; Kohara, R.; Sugino, S.; Vacha, M.; Subrahmanyam, C.; Takano, Y.; Lacy, F.; Biju, V. Toxicity of nanomaterials due to photochemical degradation and the release of heavy metal ions. Nanoscale 2020, 12, 22049–22058. [Google Scholar] [CrossRef]
- Khan, F.U.; Chen, Y.; Khan, N.U.; Ahmad, A.; Tahir, K.; Khan, Z.U.; Khan, A.U.; Khan, S.U.; Raza, M.; Wan, P. Visible light inactivation of E. coli, Cytotoxicity and ROS determination of biochemically capped gold nanoparticles. Microb. Pathog. 2017, 107, 419–424. [Google Scholar] [CrossRef]
- Kurokawa, H.; Taninaka, A.; Yoshitomi, T.; Shigekawa, H.; Matsui, H. Near-Infrared Light Irradiation of Porphyrin-Modified Gold Nanoparticles Promotes Cancer-Cell-Specific Cytotoxicity. Molecules 2022, 27, 27041238. [Google Scholar] [CrossRef]
- Semchyshyn, H. Hydrogen peroxide-induced response in E-coli and S-cerevisiae: Different stages of the flow of the genetic information. Cent. Eur. J. Biol. 2009, 4, 142–153. [Google Scholar] [CrossRef]
- Wang, Y.; Li, H.; Li, T.; He, H.; Du, X.; Zhang, X.; Kong, J. Cytoprotective effect of Streptococcus thermophilus against oxidative stress mediated by a novel peroxidase (EfeB). J. Dairy Sci. 2018, 101, 6955–6963. [Google Scholar] [CrossRef]
- Wei, W.; Sun, P.; Li, Z.; Song, K.; Su, W.; Wang, B.; Liu, Y.; Zhao, J. A surface-display biohybrid approach to light-driven hydrogen production in air. Sci. Adv. 2018, 4, eaap9253. [Google Scholar] [CrossRef]
- Wroblewska-Wolna, A.M.; Harvie, A.J.; Rowe, S.F.; Critchley, K.; Butt, J.N.; Jeuken, L.J.C. Quantum dot interactions with and toxicity to Shewanella oneidensis MR-1. Nanotechnology 2020, 31, 134005. [Google Scholar] [CrossRef]
- Ghonimi, N.A.M.; Elsharkawi, K.A.; Khyal, D.S.M.; Abdelghani, A.A. Serum malondialdehyde as a lipid peroxidation marker in multiple sclerosis patients and its relation to disease characteristics. Mult. Scler. Relat. Disord. 2021, 51, 102941. [Google Scholar] [CrossRef]
- Wang, Z.; Zhu, X.; Su, Y.; Xu, W.; Liu, H.; Liu, Z.; Chen, W.; Wang, J. Dimethyl phthalate damaged the cell membrane of Escherichia coli K12. Ecotoxicol. Environ. Saf. 2019, 180, 208–214. [Google Scholar] [CrossRef]
- Semerád, J.; Čvančarová, M.; Filip, J.; Kašlík, J.; Zlotá, J.; Soukupová, J.; Cajthaml, T. Novel assay for the toxicity evaluation of nanoscale zero-valent iron and derived nanomaterials based on lipid peroxidation in bacterial species. Chemosphere 2018, 213, 568–577. [Google Scholar] [CrossRef]
- Foster, H.A.; Ditta, I.B.; Varghese, S.; Steele, A. Photocatalytic disinfection using titanium dioxide: Spectrum and mechanism of antimicrobial activity. Appl. Microbiol. Biotechnol. 2011, 90, 1847–1868. [Google Scholar] [CrossRef]
- Abdel-Rahman, M.A.; Tashiro, Y.; Sonomoto, K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 2013, 31, 877–902. [Google Scholar] [CrossRef]
- Li, C.; Wang, H.; Naghadeh, S.B.; Zhang, J.Z.; Fang, P. Visible light driven hydrogen evolution by photocatalytic reforming of lignin and lactic acid using one-dimensional NiS/CdS nanostructures. Appl. Catal. B-Environ. 2018, 227, 229–239. [Google Scholar] [CrossRef]
- Saanchez, A.M.; Bennett, G.N.; San, K.Y. Effect of different levels of NADH availability on metabolic fluxes of Escherichia coli chemostat cultures in defined medium. J. Biotechnol. 2005, 117, 395–405. [Google Scholar] [CrossRef]
- Huang, L.; Liu, X.; Zhang, Z.; Ye, J.; Rensing, C.; Zhou, S.; Nealson, K.H. Light-driven carbon dioxide reduction to methane by Methanosarcina barkeri in an electric syntrophic coculture. ISME J. 2022, 16, 370–377. [Google Scholar] [CrossRef]
- Wang, B.; Jiang, Z.; Yu, J.C.; Wang, J.; Wong, P.K. Enhanced CO2 reduction and valuable C2+ chemical production by a CdS-photosynthetic hybrid system. Nanoscale 2019, 11, 9296–9301. [Google Scholar] [CrossRef]
- Ye, J.; Yu, J.; Zhang, Y.Y.; Chen, M.; Liu, X.; Zhou, S.G.; He, Z. Light-driven carbon dioxide reduction to methane by Methanosarcina barkeri-CdS biohybrid. Appl. Catal. B 2019, 257, 117916. [Google Scholar] [CrossRef]
- Zhang, J.; Nanjaraj Urs, A.N.; Lin, L.; Zhou, Y.; Hu, Y.; Hua, G.; Gao, Q.; Yuchi, Z.; Zhang, Y. Structure of glycerol dehydrogenase (GldA) from Escherichia coli. Acta Crystallogr. F-Struct. Biol. Commun. 2019, 75, 176–183. [Google Scholar] [CrossRef]
- Breuil, C.; Saddler, J.N. Comparison of the 3,5-dinitrosalicylic acid and Nelson-Somogyi methods of assaying for reducing sugars and determining cellulase activity. Enzym. Microb. Technol. 1985, 7, 327–332. [Google Scholar] [CrossRef]
- Jiang, Z.F.; Wang, B.; Yu, J.C.; Wang, J.F.; An, T.C.; Zhao, H.J.; Li, H.M.; Yuan, S.Q.; Wong, P.K. AglnS2/In2S3 heterostructure sensitization of Escherichia coli for sustainable hydrogen production. Nano Energy 2018, 46, 234–240. [Google Scholar] [CrossRef]
- Honda, Y.; Shinohara, Y.; Watanabe, M.; Ishihara, T.; Fujii, H. Photo-biohydrogen Production by Photosensitization with Biologically Precipitated Cadmium Sulfide in Hydrogen-Forming Recombinant Escherichia coli. ChemBioChem 2020, 21, 3389–3397. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Liu, Y.; Bai, L.; Wang, J.; Zhao, N.; Cui, D.; Zhao, M. Low-Toxicity Self-Photosensitized Biohybrid Systems for Enhanced Light-Driven H2 Production. Int. J. Mol. Sci. 2024, 25, 3085. https://doi.org/10.3390/ijms25063085
Wang Y, Liu Y, Bai L, Wang J, Zhao N, Cui D, Zhao M. Low-Toxicity Self-Photosensitized Biohybrid Systems for Enhanced Light-Driven H2 Production. International Journal of Molecular Sciences. 2024; 25(6):3085. https://doi.org/10.3390/ijms25063085
Chicago/Turabian StyleWang, Yuelei, Yuqi Liu, Long Bai, Jueyu Wang, Na Zhao, Daizong Cui, and Min Zhao. 2024. "Low-Toxicity Self-Photosensitized Biohybrid Systems for Enhanced Light-Driven H2 Production" International Journal of Molecular Sciences 25, no. 6: 3085. https://doi.org/10.3390/ijms25063085
APA StyleWang, Y., Liu, Y., Bai, L., Wang, J., Zhao, N., Cui, D., & Zhao, M. (2024). Low-Toxicity Self-Photosensitized Biohybrid Systems for Enhanced Light-Driven H2 Production. International Journal of Molecular Sciences, 25(6), 3085. https://doi.org/10.3390/ijms25063085




