Genome-Wide Identification, Characterization, and Expression Analysis of the BES1 Family Genes under Abiotic Stresses in Phoebe bournei
Abstract
1. Introduction
2. Results
2.1. Analysis of Physicochemical Properties of the BES1 Gene Family in Phoebe bournei
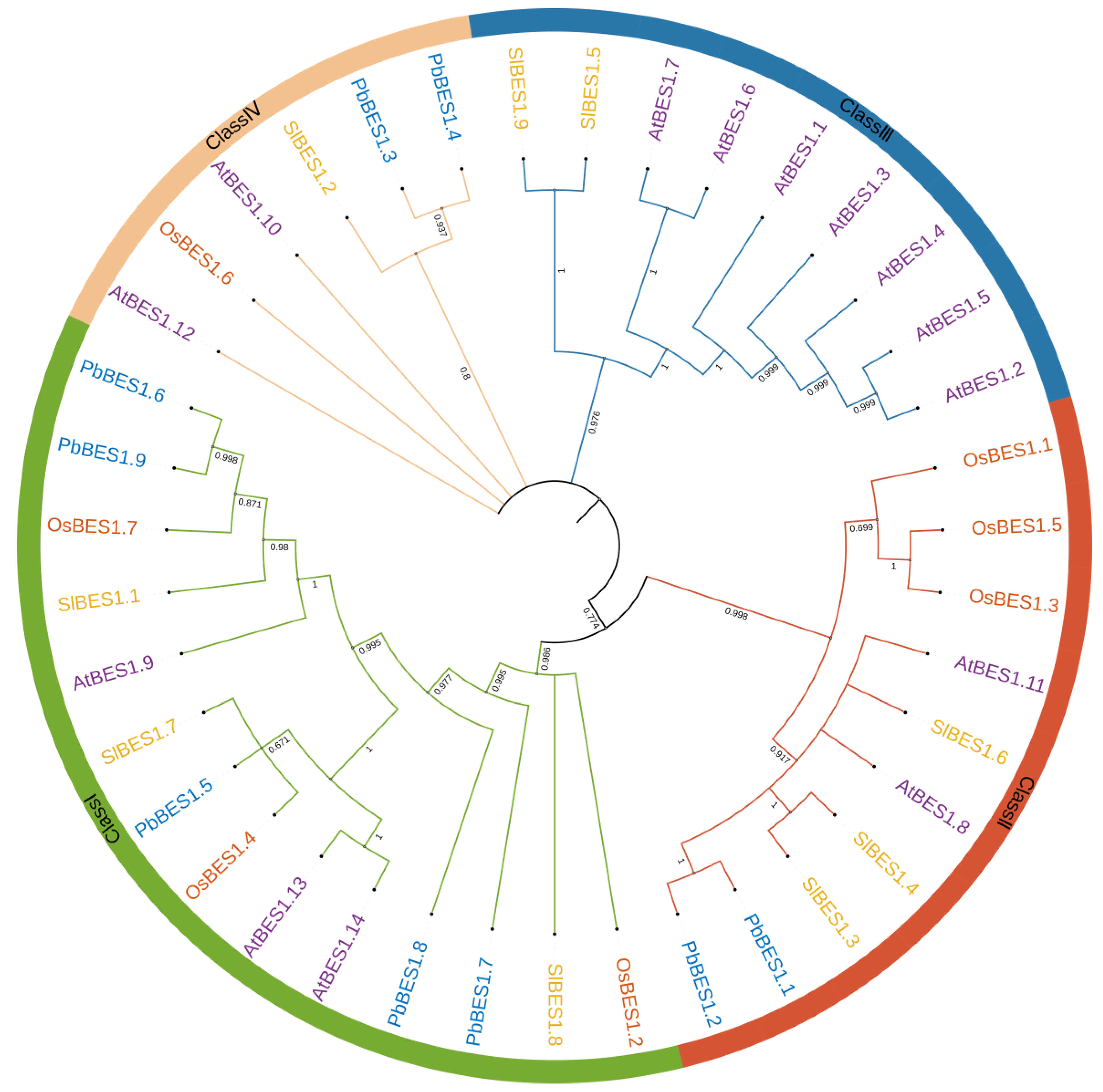
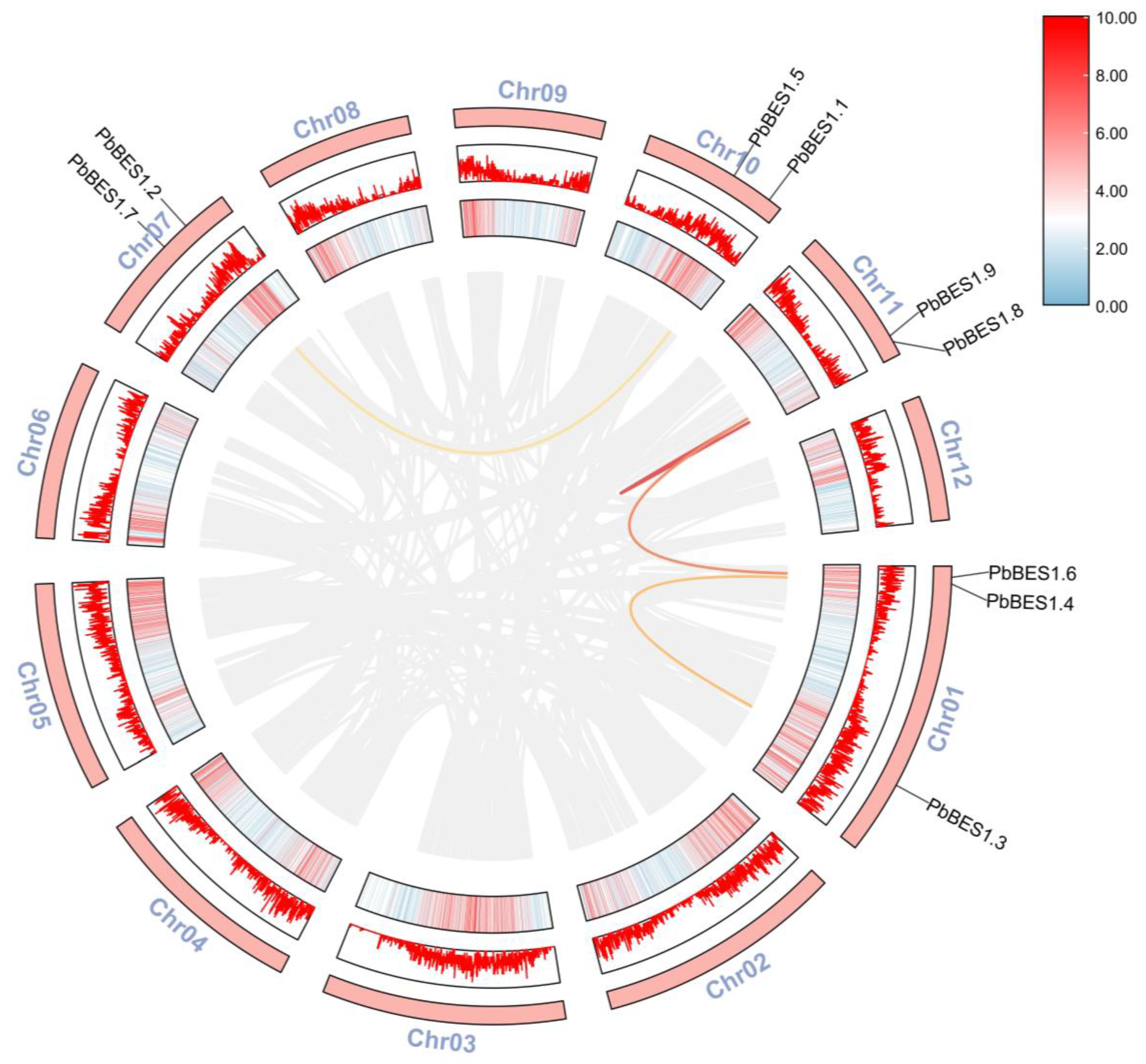
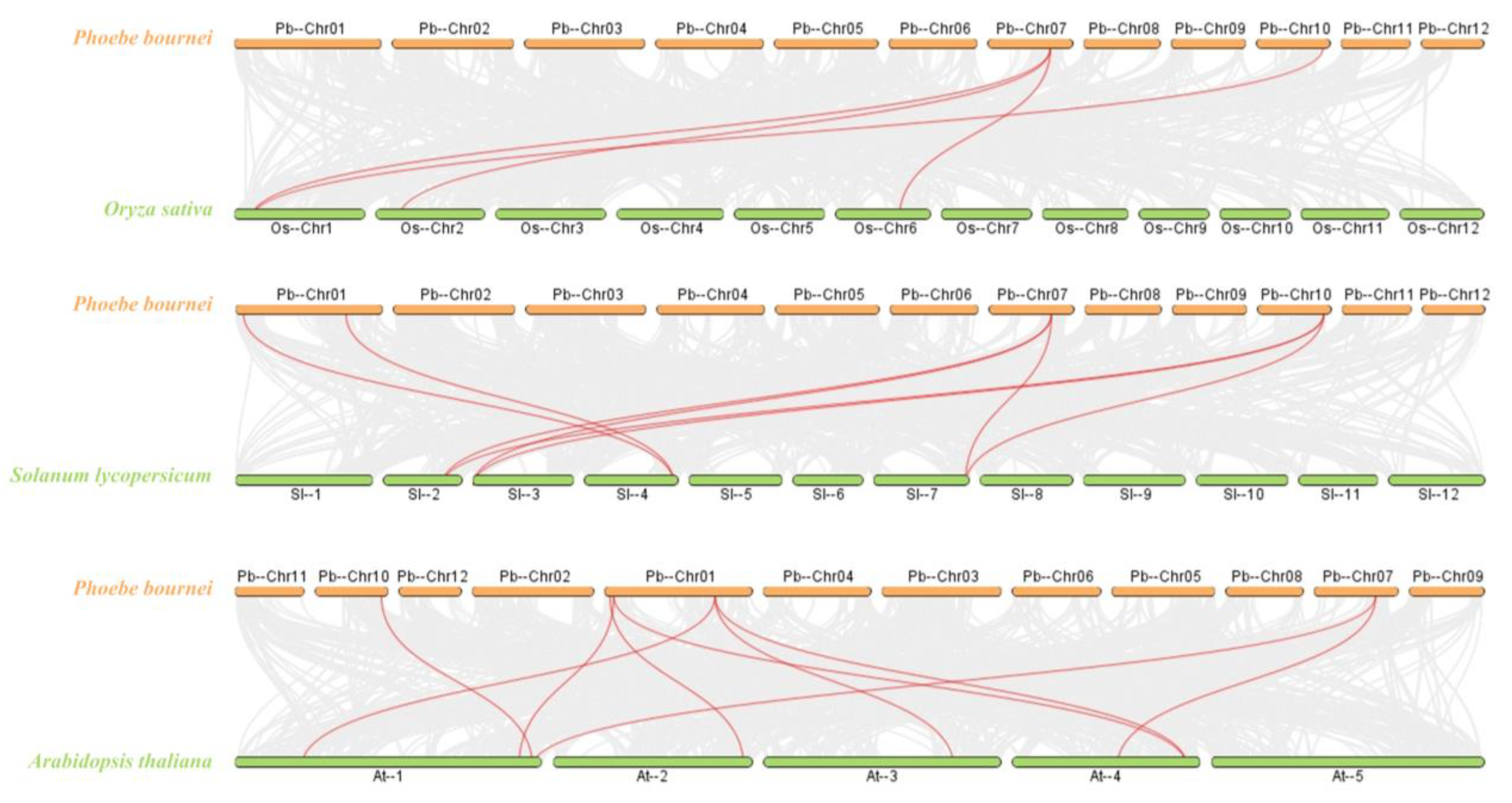
2.2. Protein Evolution and Collinearity Analysis of PbBES1 Genes
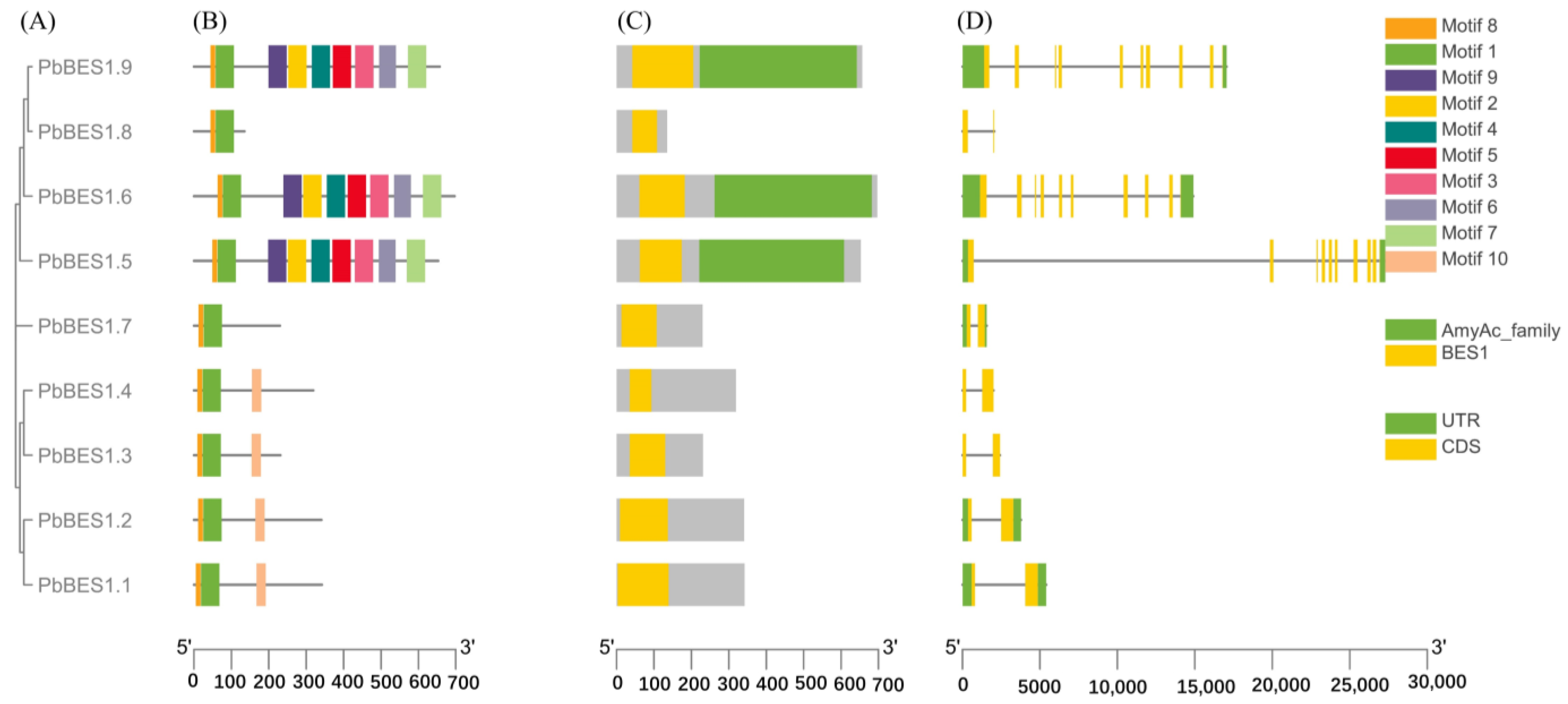
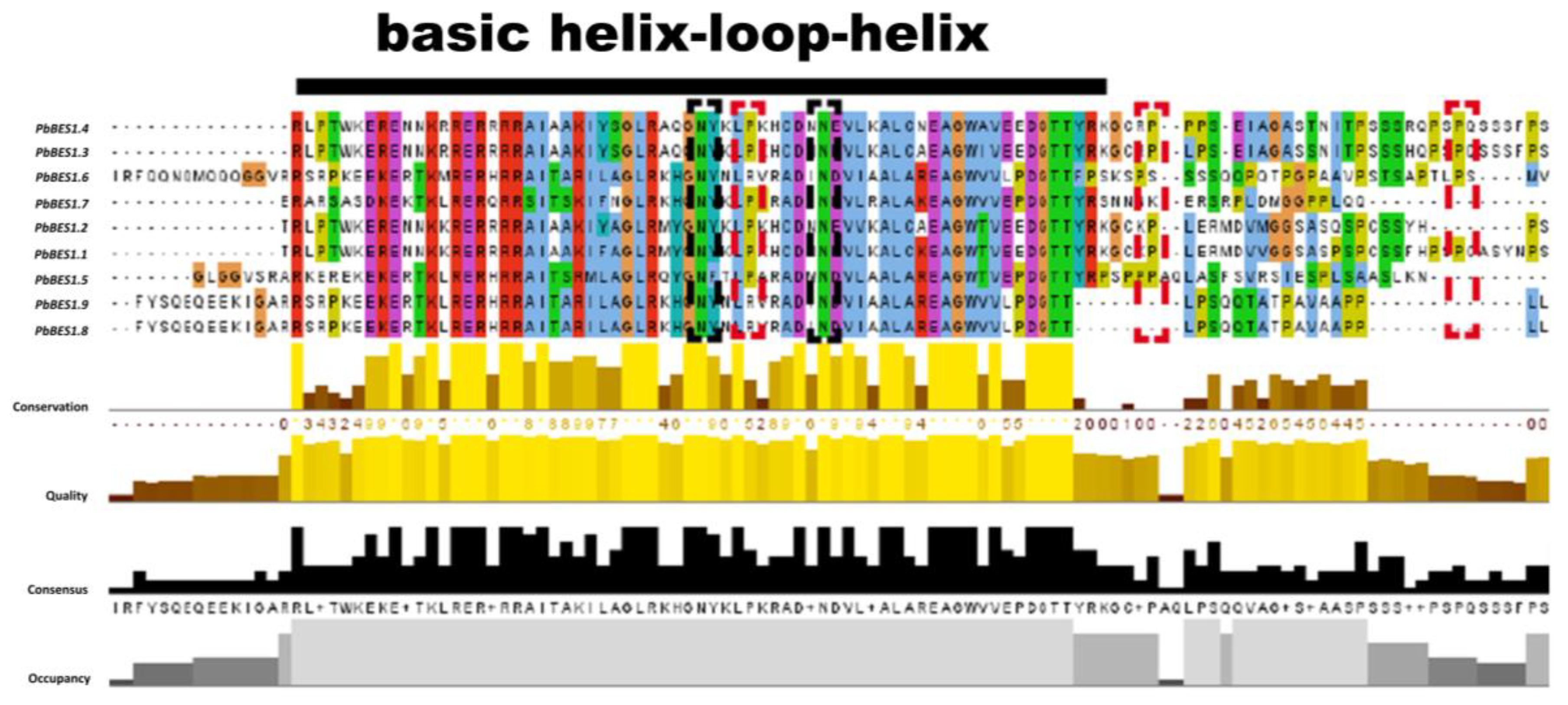
2.3. Analysis of Gene Structure and Conserved Motifs in Phoebe bournei
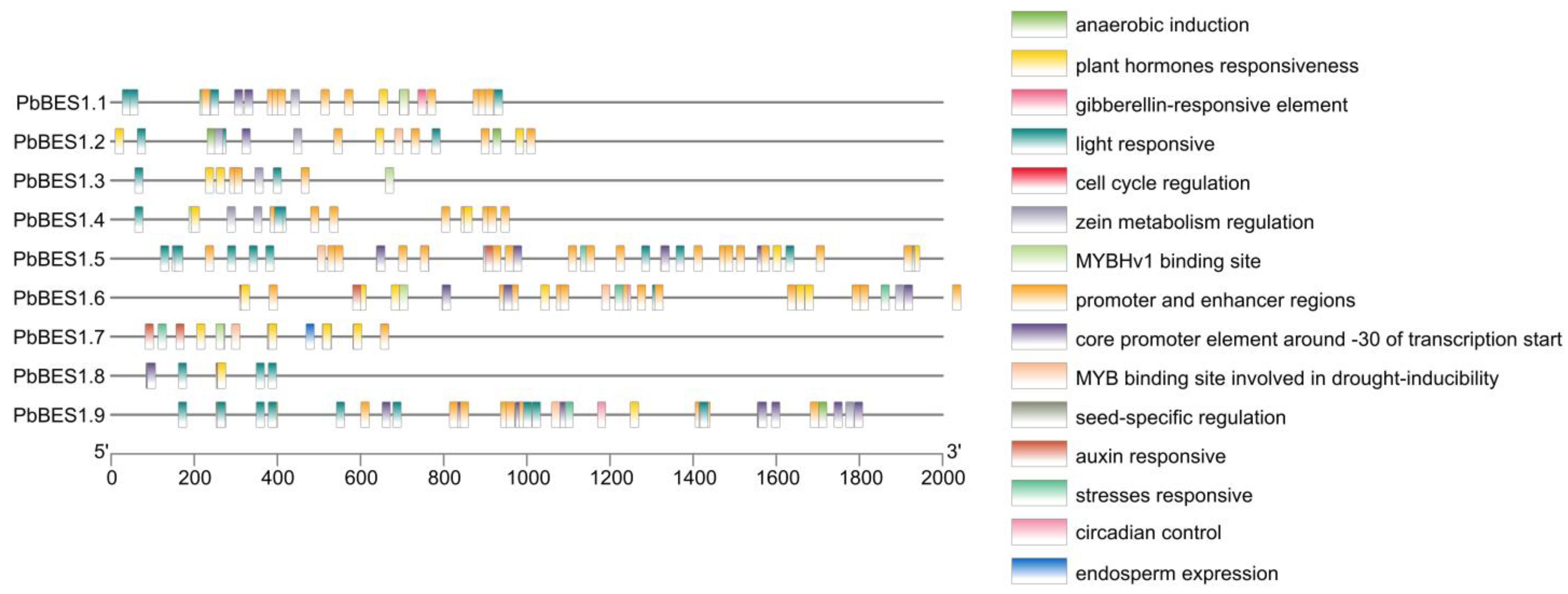
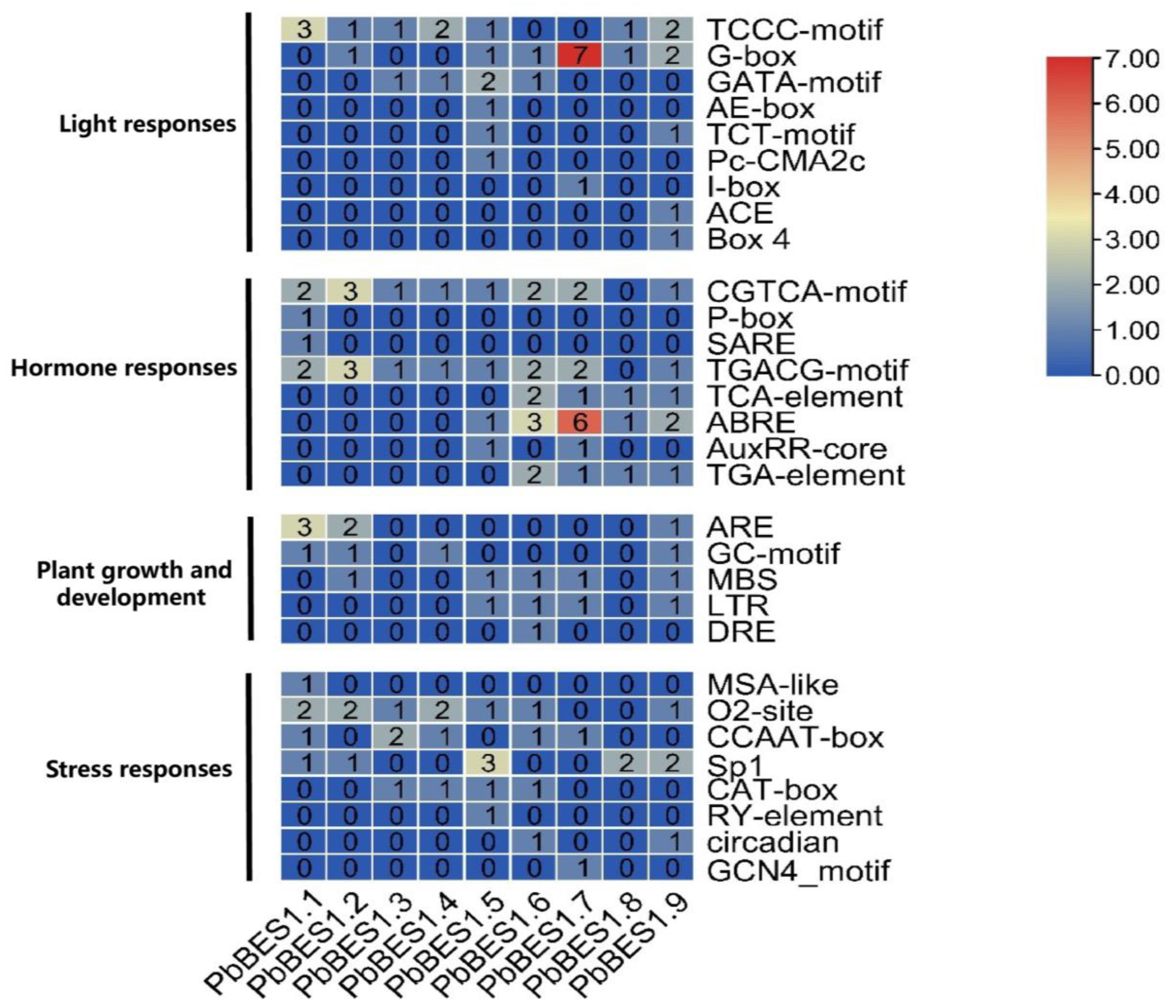
2.4. Cis-Acting Analysis and Structure of the PbBES1 Promoter
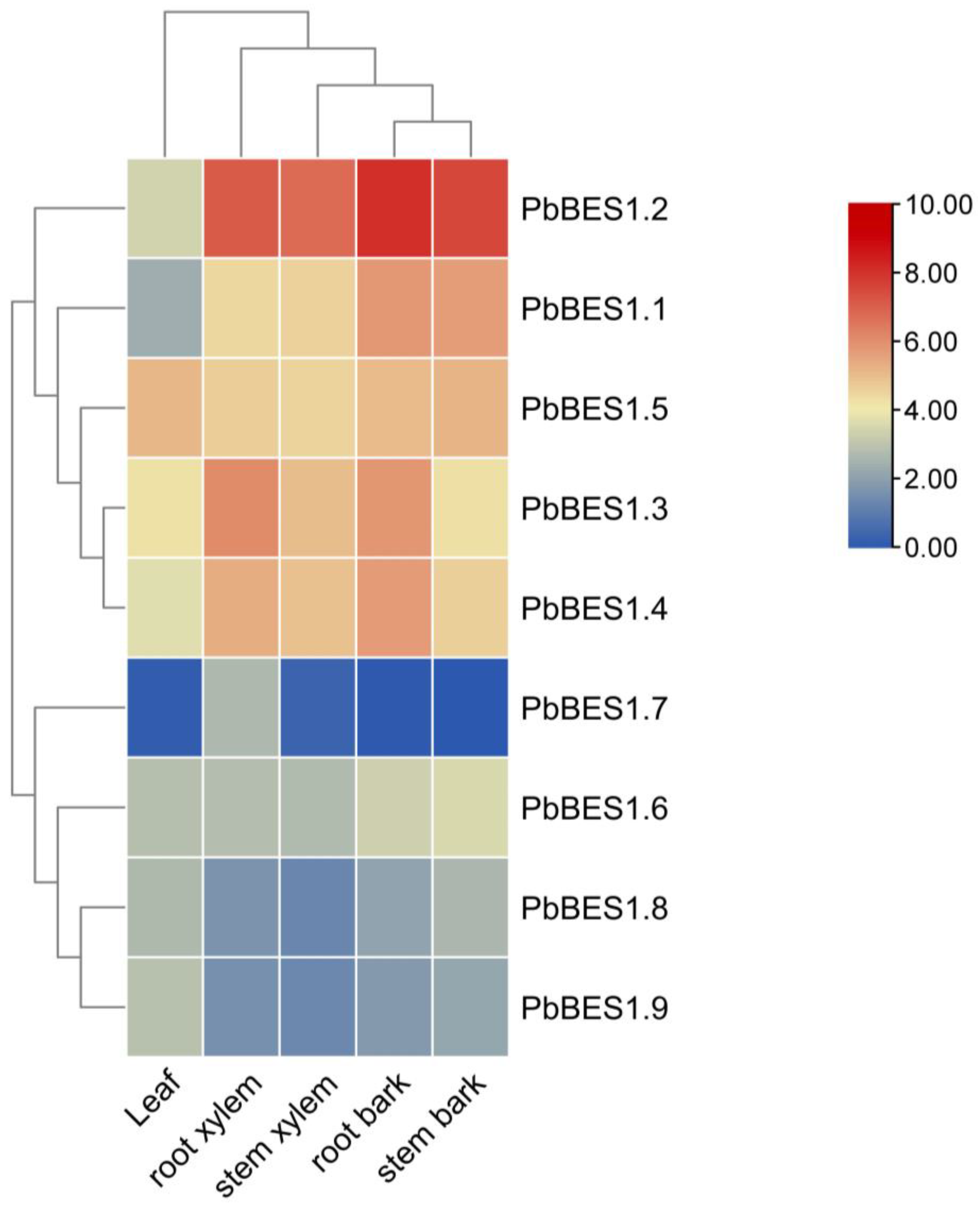
2.5. Heat Map of PbBES1 Gene Expression in Different Tissues
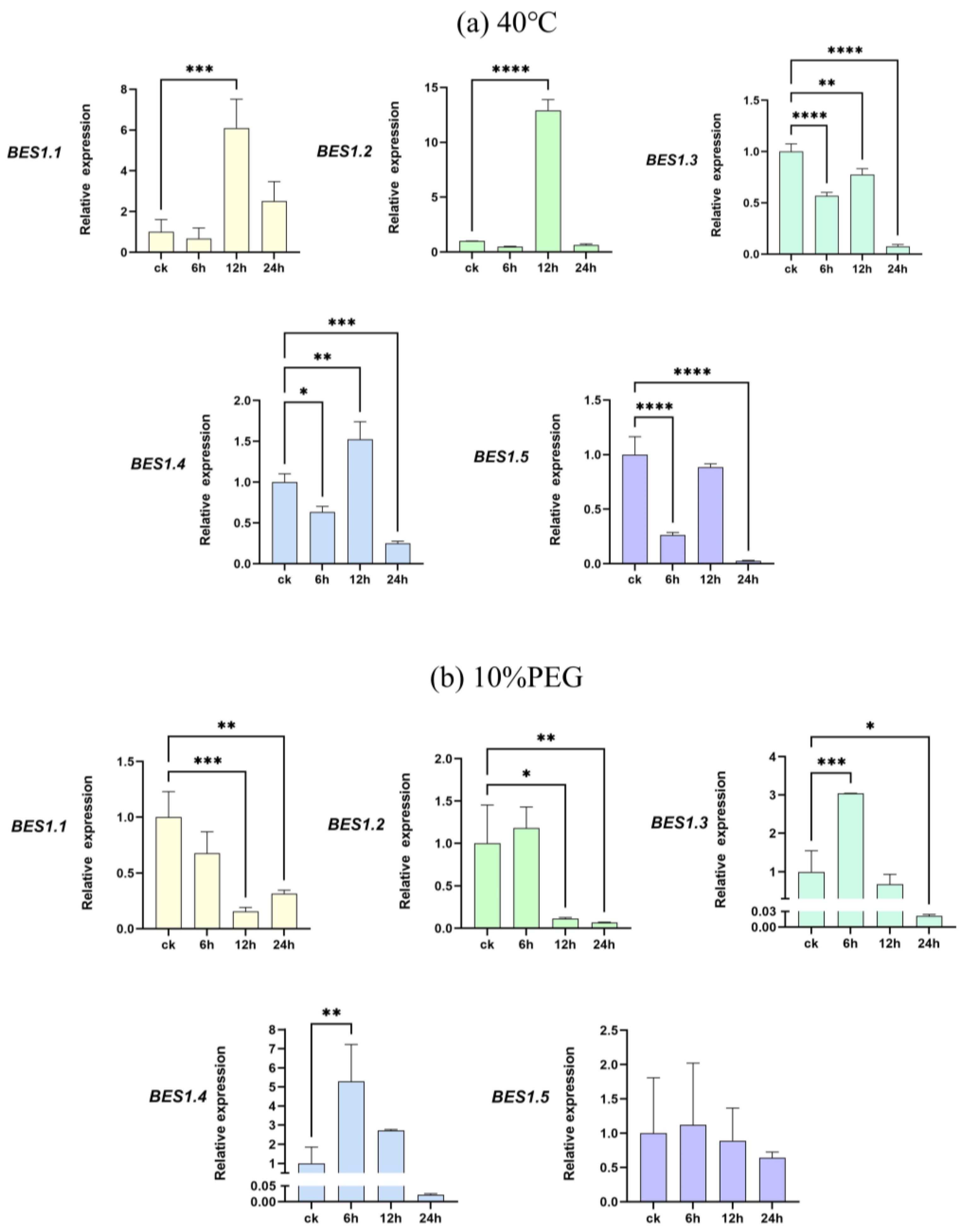
2.6. Abiotic Stress Experiments on the BES1 Gene Family of Phoebe bournei
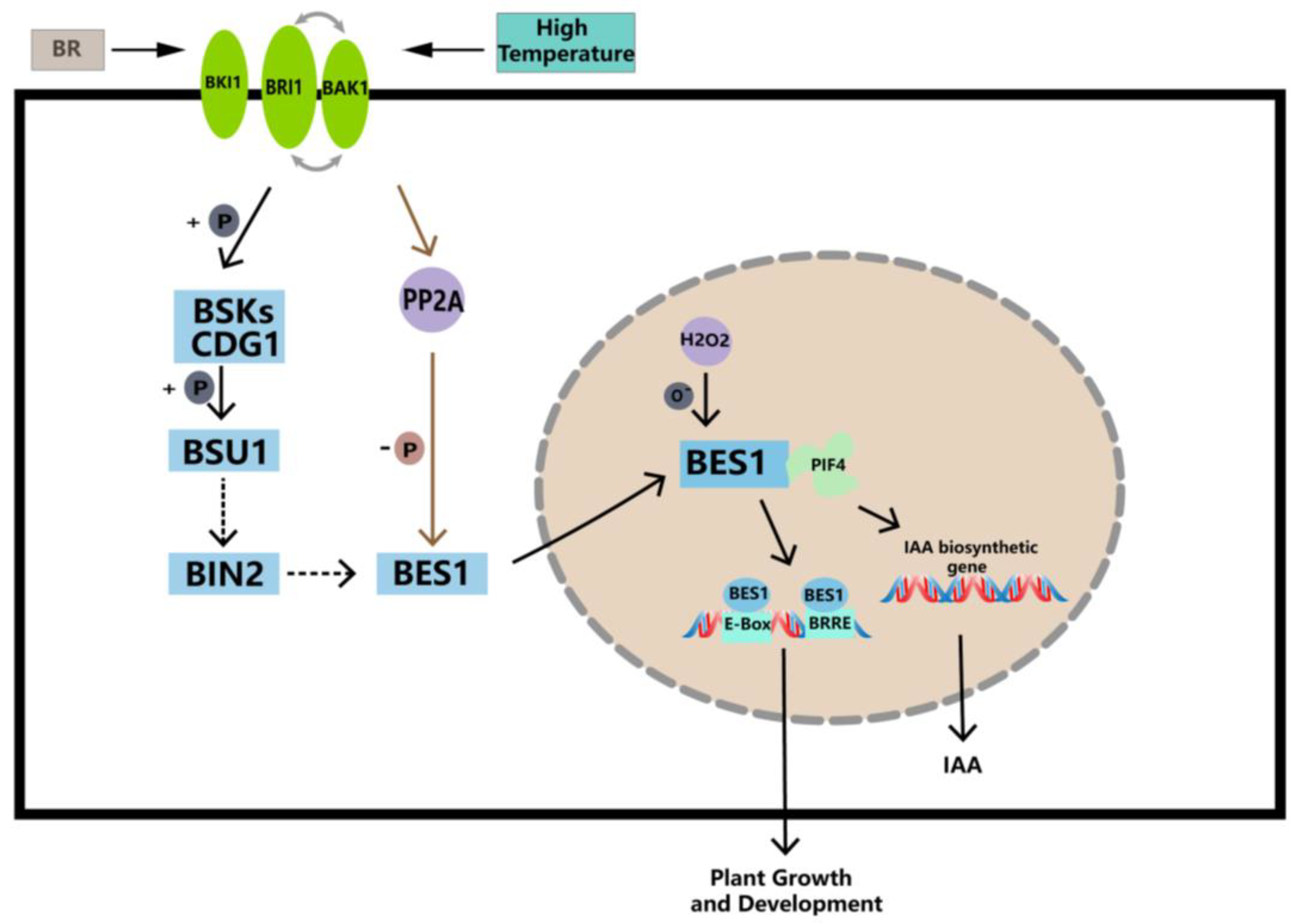
3. Discussion
4. Materials and Methods
4.1. Genomic Data
4.2. Plant Material Sources and Abiotic Stress Treatment
4.2.1. Plant Material Sources
4.2.2. Drought Stress Treatment
4.2.3. Temperature Handling
4.3. Identification and Physical and Chemical Property Analysis
4.4. Phylogenetic Tree Construction
4.5. Chromosome Distribution and Covariance Analysis
4.6. Gene Family Conserved Motifs, Gene Structure Analysis
4.7. Multiple Sequence Comparison and Promoter Cis-Element Analysis of the PbBES1 Gene
4.8. The Expression Profiles of PbBES1 Genes
4.9. RNA Extraction and qRT-PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, J.T.; Lofgren, S.; Farrel, A. Structure-based prediction of transcription factor binding sites. Tsinghua Sci. Technol. 2014, 19, 568–577. [Google Scholar]
- Century, K.; Reuber, T.L.; Ratcliffe, O.J. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008, 147, 20–29. [Google Scholar] [CrossRef]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef]
- Kono, A.; Yin, Y. Updates on BES1/BZR1 Regulatory Networks Coordinating Plant Growth and Stress Responses. Front. Plant Sci. 2020, 11, 617162. [Google Scholar] [CrossRef]
- Wang, R.; Wang, R.; Liu, M.; Yuan, W.; Zhao, Z.; Liu, X.; Peng, Y.; Yang, X.; Sun, Y.; Tang, W. Nucleocytoplasmic trafficking and turnover mechanisms of BRASSINAZOLE RESISTANT1 in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2021, 118, e2101838118. [Google Scholar] [CrossRef]
- Guedes, F.R.C.M.; Maia, C.F.; da Silva, B.R.S.; Batista, B.L.; Alyemeni, M.N.; Ahmad, P.; Lobato, A.K.d.S. Exogenous 24-Epibrassinolide stimulates root protection, and leaf antioxidant enzymes in lead stressed rice plants: Central roles to minimize Pb content and oxidative stress. Environ. Pollut. 2021, 280, 116992. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.W.; Mandava, N.; Worley, J.F.; Plimmer, J.R.; Smith, M.V. Brassins—A New Family of Plant Hormones from Rape Pollen. Nature 1970, 225, 1065–1066. [Google Scholar] [CrossRef] [PubMed]
- Peres, A.L.G.L.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.T.; Mandava, N.B.; Menossi, M. Brassinosteroids, the Sixth Class of Phytohormones: A Molecular View from the Discovery to Hormonal Interactions in Plant Development and Stress Adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef] [PubMed]
- González-García, M.-P.; Vilarrasa-Blasi, J.; Zhiponova, M.; Divol, F.; Mora-García, S.; Russinova, E.; Caño-Delgado, A.I. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 2011, 138, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Russinova, E. Brassinosteroid signalling. Curr. Biol. 2020, 30, R294–R298. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Vukašinović, N.; Hsu, C.-W.; Zhang, J.; Vanhoutte, I.; Shahan, R.; Taylor, I.W.; Greenstreet, L.; Heitz, M.; Afanassiev, A.; et al. Brassinosteroid gene regulatory networks at cellular resolution in the Arabidopsis root. Science 2023, 379, eadf4721. [Google Scholar] [CrossRef] [PubMed]
- Bulgakov, V.P.; Avramenko, T.V. Linking Brassinosteroid and ABA Signaling in the Context of Stress Acclimation. Int. J. Mol. Sci. 2020, 21, 5108. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolomé, J.; Minguet, E.G.; Grau-Enguix, F.; Abbas, M.; Locascio, A.; Thomas, S.G.; Alabadí, D.; Blázquez, M.A. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 13446–13451. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photo-morphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Tang, B.; Xie, Z.; Nolan, T.; Ye, H.; Song, G.; Walley, J.; Yin, Y. GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 2019, 100, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, S.; Tang, B.; Chen, J.; Xie, Z.; Nolan, T.M.; Jiang, H.; Guo, H.; Lin, H.Y.; Li, L.; et al. RD26 mediates crosstalk be-tween drought and brassinosteroid signalling pathways. Nat. Commun. 2017, 8, 14573. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Yang, Y.; Li, J.; Wang, Z.Y. The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2002, 99, 10185–10190. [Google Scholar] [CrossRef]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinoster-oid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef]
- Guo, R.; Qian, H.; Shen, W.; Liu, L.; Zhang, M.; Cai, C.; Zhao, Y.; Qiao, J.; Wang, Q. BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J. Exp. Bot. 2013, 64, 2401–2412. [Google Scholar] [CrossRef]
- Ryu, H.; Cho, H.; Bae, W.; Hwang, I. Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 2014, 5, 4138. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, C.; Wang, X. A recently evolved isoform of the transcription factor BES1 pro-motes brassinosteroid signaling and development in Arabidopsis thaliana. Plant Cell 2015, 27, 361–374. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Mueller, A.K.; Labaied, M.; Kappe, S.H.; Matuschewski, K. Genetically modified Plasmodium para-sites as a protective experimental malaria vaccine. Nature 2005, 433, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR Receptor-like Protein Kinase, Interacts with BRI1 and Modulates Brassinosteroid Signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-W.; Guan, S.; Burlingame, A.L.; Wang, Z.-Y. The CDG1 Kinase Mediates Brassinosteroid Signal Transduction from BRI1 Receptor Kinase to BSU1 Phosphatase and GSK3-like Kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Ortiz, G.; Huq, E.; Quail, P.H. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 2003, 15, 1749–1770. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Park, Y.J.; Son, S.-H.; Roh, J.; Youn, J.H.; Kim, S.Y.; Kim, S.-K. Brassinosteroids signaling via BZR1 down-regulates expression of ACC oxidase 4 to control growth of Arabidopsis thaliana seedlings. Plant Signal. Behav. 2020, 15, 1734333. [Google Scholar] [CrossRef]
- Yu, Z.; Ma, J.; Zhang, M.; Li, X.; Sun, Y.; Zhang, M.; Ding, Z. Auxin promotes hypocotyl elongation by enhancing BZR1 nuclear accumulation in Arabidopsis. Sci. Adv. 2023, 9, eade2493. [Google Scholar] [CrossRef] [PubMed]
- Bernardo-Garcia, S.; de Lucas, M.; Martinez, C.; Espinosa-Ruiz, A.; Daviere, J.M.; Prat, S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes. Dev. 2014, 28, 1681–1694. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, X.; Lai, Y.; He, G.; Wen, S.; He, H.; Li, Z.; Zhang, B.; Zhang, D. Transcriptomic analysis reveals the significant effects of fertilization on the biosynthesis of sesquiterpenes in Phoebe bournei. Genomics 2022, 114, 110375. [Google Scholar] [CrossRef]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nolan, T.M.; Ye, H.; Zhang, M.; Tong, H.; Xin, P.; Chu, J.; Chu, C.; Li, Z.; Yin, Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell 2017, 29, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-Interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef]
- Mecchia, M.A.; García-Hourquet, M.; Lozano-Elena, F.; Planas-Riverola, A.; Blasco-Escamez, D.; Marquès-Bueno, M.; Mora-García, S.; Caño-Delgado, A.I. The BES1/BZR1-family transcription factor MpBES1 regulates cell division and differentiation in Marchantia polymorpha. Curr. Biol. 2021, 31, 4860–4869.e8. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, Y.; Li, L.; Li, C. Comprehensive analysis of the BES1 gene family and its expression under abiotic stress and hormone treatment in Populus trichocarpa. Plant Physiol. Biochem. 2022, 173, 1–13. [Google Scholar] [CrossRef]
- Song, X.; Ma, X.; Li, C.; Hu, J.; Yang, Q.; Wang, T.; Wang, L.; Wang, J.; Guo, D.; Ge, W.; et al. Comprehensive analyses of the BES1 gene family in Brassica napus and examination of their evolutionary pattern in representative species. BMC Genom. 2018, 19, 346. [Google Scholar] [CrossRef]
- Liu, Z.; Qanmber, G.; Lu, L.; Qin, W.; Liu, J.; Li, J.; Ma, S.; Yang, Z.; Yang, Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci. China Life Sci. 2018, 61, 1566–1582. [Google Scholar] [CrossRef]
- Yu, H.; Feng, W.; Sun, F.; Zhang, Y.; Qu, J.; Liu, B.; Lu, F.; Yang, L.; Fu, F.; Li, W. Cloning and characterization of BES1/BZR1 transcription factor genes in maize. Plant Growth Regul. 2018, 86, 235–249. [Google Scholar] [CrossRef]
- Gao, Y.; Hu, J.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-wide Identification and Expression Pattern Analysis of BRI1-EMS–suppressor Transcription Factors in Tomato under Abiotic Stresses. J. Am. Soc. Hortic. Sci. 2018, 143, 84–90. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor super-family in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef]
- Xie, L.; Yang, C.; Wang, X. Brassinosteroids can regulate cellulose biosynthesis by con-trolling the expression of CESA genes in Arabidopsis. J. Exp. Bot. 2011, 62, 4495–4506. [Google Scholar] [CrossRef]
- Ye, Q.; Zhu, W.; Li, L.; Zhang, S.; Yin, Y.; Ma, H.; Wang, X. Brassinosteroids control male fertility by regulating the expression of key genes involved in Arabidopsis anther and pollen development. Proc. Natl. Acad. Sci. USA 2010, 107, 6100–6105. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Macho, A.P.; Boutrot, F.; Segonzac, C.; Somssich, I.E.; Zipfel, C. The transcriptional regulator BZR1 mediates trade-off between plant innate immunity and growth. eLife 2013, 2, e00983. [Google Scholar] [CrossRef]
- Li, Q.-F.; Lu, J.; Yu, J.-W.; Zhang, C.-Q.; He, J.-X.; Liu, Q.-Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta (BBA) 2018, 1861, 561–571. [Google Scholar] [CrossRef]
- Wenkai, A.N.; Dan, C.; Fuchun, Z. Expression Characteristics of Transcription Factor BES1/BRI1 of Cotton Seedling in Response to Brassinosteroid under Drought Stress. Acta Bot. Boreal.-Occident. Sin. 2015, 35, 1311–1316. [Google Scholar]
- Li, X.; Liu, L.; Sun, S.; Li, Y.; Jia, L.; Ye, S.; Yu, Y.; Dossa, K.; Luan, Y. Leaf-transcriptome profiles of Phoebe bournei provide insights into temporal drought stress responses. Front. Plant Sci. 2022, 13, 1010314. [Google Scholar] [CrossRef]
- Carnicer, J.; Alegria, A.; Giannakopoulos, C.; Di Giuseppe, F.; Karali, A.; Koutsias, N.; Lionello, P.; Parrington, M.; Vitolo, C. Global warming is shifting the relationships between fire weather and realized fire-induced CO2 emissions in Europe. Sci. Rep. 2022, 12, 10365. [Google Scholar] [CrossRef]
- Dai, A. Drought under global warming: A review. WIREs Clim. Chang. 2010, 2, 45–65. [Google Scholar] [CrossRef]
- Mansoor, S.; Kour, N.; Manhas, S.; Zahid, S.; Wani, O.A.; Sharma, V.; Wijaya, L.; Alyemeni, M.N.; Alsahli, A.A.; El-Serehy, H.A.; et al. Biochar as a tool for effective management of drought and heavy metal toxicity. Chemosphere 2020, 271, 129458. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; He, X.; Wang, J.; Jiang, B.; Ye, R.; Lin, X. Physiological and biochemical responses of Phoebe bournei seedlings to water stress and recovery. Acta Physiol. Plant. 2014, 36, 1241–1250. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef]
- Kolodny, R. Searching protein space for ancient sub-domain segments. Curr. Opin. Struct. Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Nosaki, S.; Miyakawa, T.; Xu, Y.; Nakamura, A.; Hirabayashi, K.; Asami, T.; Nakano, T.; Tanokura, M. Structural basis for brassinosteroid response by BIL1/BZR1. Nat. Plants 2018, 4, 771–776. [Google Scholar] [CrossRef]
- Wang, X.; Niu, Y.; Zheng, Y. Multiple Functions of MYB Transcription Factors in Abiotic Stress Responses. Int. J. Mol. Sci. 2021, 22, 6125. [Google Scholar] [CrossRef]
- Gendron, J.M.; Wang, Z.Y. Multiple mechanisms modulate Brassinosteroid signaling. Curr. Opin. Plant Biol. 2007, 10, 436–441. [Google Scholar] [CrossRef]
- Cheng, M.; Yuan, H.; Wang, R.; Wang, W.; Zhang, L.; Fan, F.; Li, S. Identification and characterization of BES1 genes involved in grain size development of Oryza sativa L. Int. J. Biol. Macromol. 2023, 253, 127327. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Leng, F.; Ma, C.; Zhang, C.; Wang, S. Genome-wide identification, characterization and gene expression of BES1 transcription factor family in grapevine (Vitis vinifera L.). Sci. Rep. 2023, 13, 240. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Xiang, W.; Wen, L.; Lu, W.; Shi, Y.; Liu, Y.; Li, Z. Genome-wide identification, characterization and expression analysis of BES1 gene family in tomato. BMC Plant Biol. 2021, 21, 161. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Cui, Y.; Zhao, Z.; Li, S.; Liang, D.; Wang, C.; Feng, G.; Wang, J.; Liu, Z. Genome-wide identification and characterization of the BES/BZR gene family in wheat and foxtail millet. BMC Genom. 2021, 22, 682. [Google Scholar] [CrossRef]
- Izadi, F.; Zarrini, H.N.; Jelodar, N.B. In silico analysis of BES1 transcription factor in citrus senescence. Res. J. Life Sci. Bioinform. Pharm. Chem. Sci. 2016, 2, 29. [Google Scholar]
- Mahesh, K.; Balaraju, P.; Ramakrishna, B.; Rao, S.S.R. Effect of Brassinosteroids on Germination and Seedling Growth of Radish (Raphanus sativus L.) under PEG-6000 Induced Water Stress. Am. J. Plant Sci. 2013, 4, 2305–2313. [Google Scholar] [CrossRef]
- Anwar, A.; Liu, Y.; Dong, R.; Bai, L.; Yu, X.; Li, Y. The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 2018, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.H.; Li, X.Q.; Wang, G.G.; Zhu, X.T. Brassinosteroids alleviate high-temperature injury in Ficus concinna seedlings via maintaining higher antioxidant defence and glyoxalase systems. AoB Plants 2015, 7, plv009. [Google Scholar] [CrossRef]
- Kagale, S.; Divi, U.K.; Krochko, J.E.; Keller, W.A.; Krishna, P. Brassinosteroid confers tolerance in Arabidopsis thaliana and Brassica napus to a range of abiotic stresses. Planta 2007, 225, 353–364. [Google Scholar] [CrossRef]
- Martins, S.; Montiel-Jorda, A.; Cayrel, A.; Huguet, S.; Roux, C.P.-L.; Ljung, K.; Vert, G. Brassinosteroid signaling-dependent root responses to prolonged elevated ambient temperature. Nat. Commun. 2017, 8, 309. [Google Scholar] [CrossRef]
- Choudhary, S.P.; Yu, J.Q.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Benefits of brassinosteroid crosstalk. Trends Plant Sci. 2012, 17, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, J.; Han, S.; Chong, S.L.; Meng, G.; Song, M.; Wang, Y.; Zhou, S.; Liu, C.; Lou, L.; et al. The chromosome-scale genome of Phoebe bournei reveals contrasting fates of terpene synthase (TPS)-a and TPS-b subfamilies. Plant Commun. 2022, 3, 100410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhu, Y.; Pan, Y.; Huang, H.; Li, C.; Li, G.; Tong, Z. Transcriptomic profiling and identification of candidate genes in two Phoebe bournei ecotypes with contrasting cold stress responses. Trees 2018, 32, 1315–1333. [Google Scholar] [CrossRef]
| Sequence ID | Number of Amino Acid | Molecular Weight (kDa) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) |
|---|---|---|---|---|---|---|
| PbBES1.1 | 341 | 36,251.28 | 8.33 | 58.07 | 55.87 | −0.568 |
| PbBES1.2 | 340 | 36,470.59 | 7.14 | 59.20 | 50.56 | −0.598 |
| PbBES1.3 | 230 | 24,733.89 | 10.21 | 92.65 | 61.22 | −0.632 |
| PbBES1.4 | 318 | 33,920.70 | 9.14 | 75.55 | 56.89 | −0.629 |
| PbBES1.5 | 652 | 73,573.82 | 5.81 | 43.65 | 72.42 | −0.485 |
| PbBES1.6 | 696 | 78,314.37 | 5.65 | 43.90 | 71.47 | −0.454 |
| PbBES1.7 | 229 | 24,521.34 | 9.34 | 53.06 | 57.16 | −0.592 |
| PbBES1.8 | 134 | 15,094.98 | 5.72 | 69.01 | 73.58 | −0.880 |
| PbBES1.9 | 656 | 73,616.07 | 5.59 | 38.65 | 75.66 | −0.450 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Sun, H.; Wang, Y.; Fan, D.; Zhu, Q.; Zhang, J.; Zhong, K.; Yang, H.; Chang, W.; Cao, S. Genome-Wide Identification, Characterization, and Expression Analysis of the BES1 Family Genes under Abiotic Stresses in Phoebe bournei. Int. J. Mol. Sci. 2024, 25, 3072. https://doi.org/10.3390/ijms25053072
Li J, Sun H, Wang Y, Fan D, Zhu Q, Zhang J, Zhong K, Yang H, Chang W, Cao S. Genome-Wide Identification, Characterization, and Expression Analysis of the BES1 Family Genes under Abiotic Stresses in Phoebe bournei. International Journal of Molecular Sciences. 2024; 25(5):3072. https://doi.org/10.3390/ijms25053072
Chicago/Turabian StyleLi, Jingshu, Honggang Sun, Yanhui Wang, Dunjin Fan, Qin Zhu, Jiangyonghao Zhang, Kai Zhong, Hao Yang, Weiyin Chang, and Shijiang Cao. 2024. "Genome-Wide Identification, Characterization, and Expression Analysis of the BES1 Family Genes under Abiotic Stresses in Phoebe bournei" International Journal of Molecular Sciences 25, no. 5: 3072. https://doi.org/10.3390/ijms25053072
APA StyleLi, J., Sun, H., Wang, Y., Fan, D., Zhu, Q., Zhang, J., Zhong, K., Yang, H., Chang, W., & Cao, S. (2024). Genome-Wide Identification, Characterization, and Expression Analysis of the BES1 Family Genes under Abiotic Stresses in Phoebe bournei. International Journal of Molecular Sciences, 25(5), 3072. https://doi.org/10.3390/ijms25053072








