Emerging Role of GCN1 in Disease and Homeostasis
Abstract
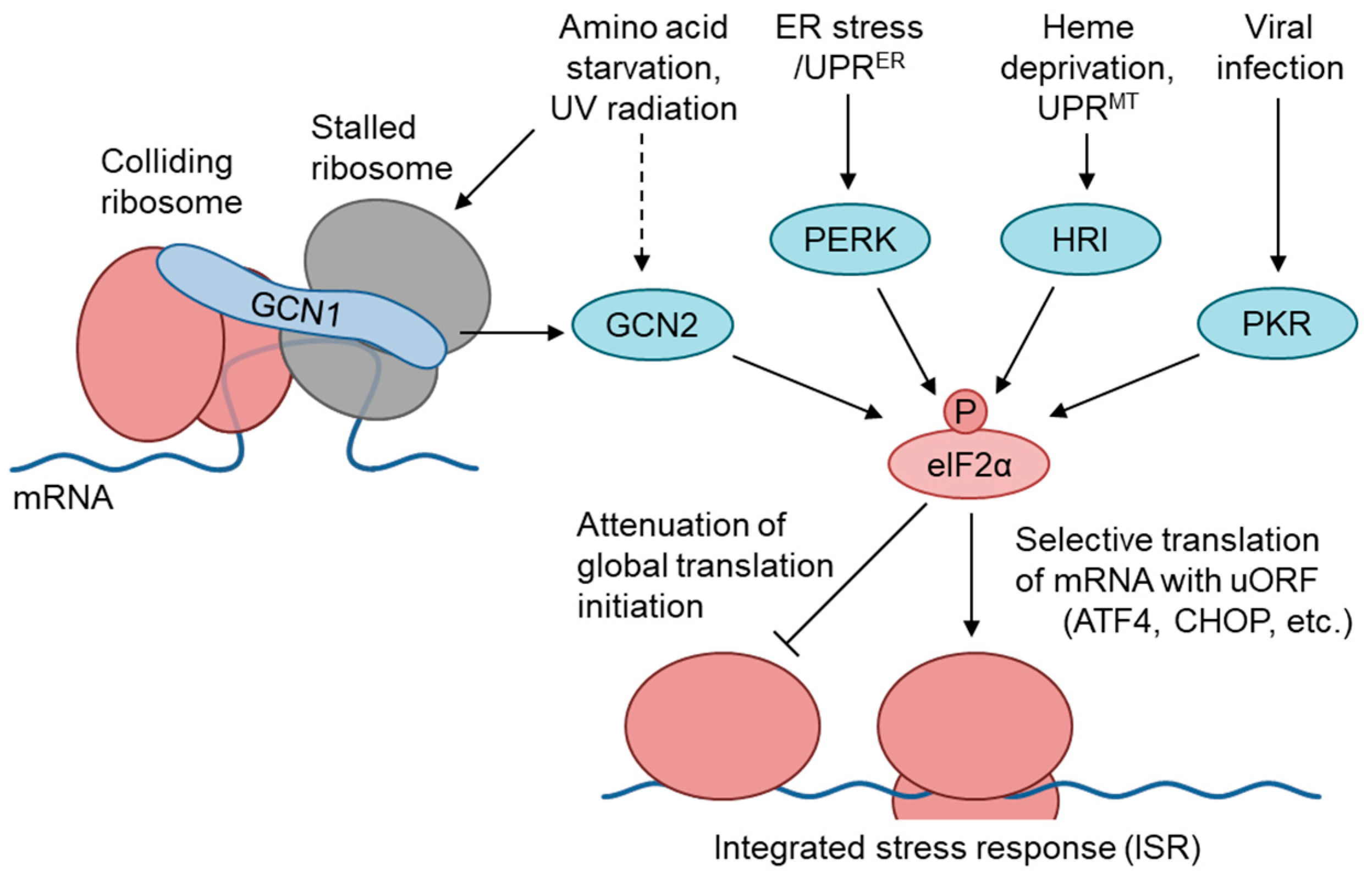
1. The GCN1–GCN2 Pathway Regulates the Amino Acid Starvation Branch of the Integrated Stress Response
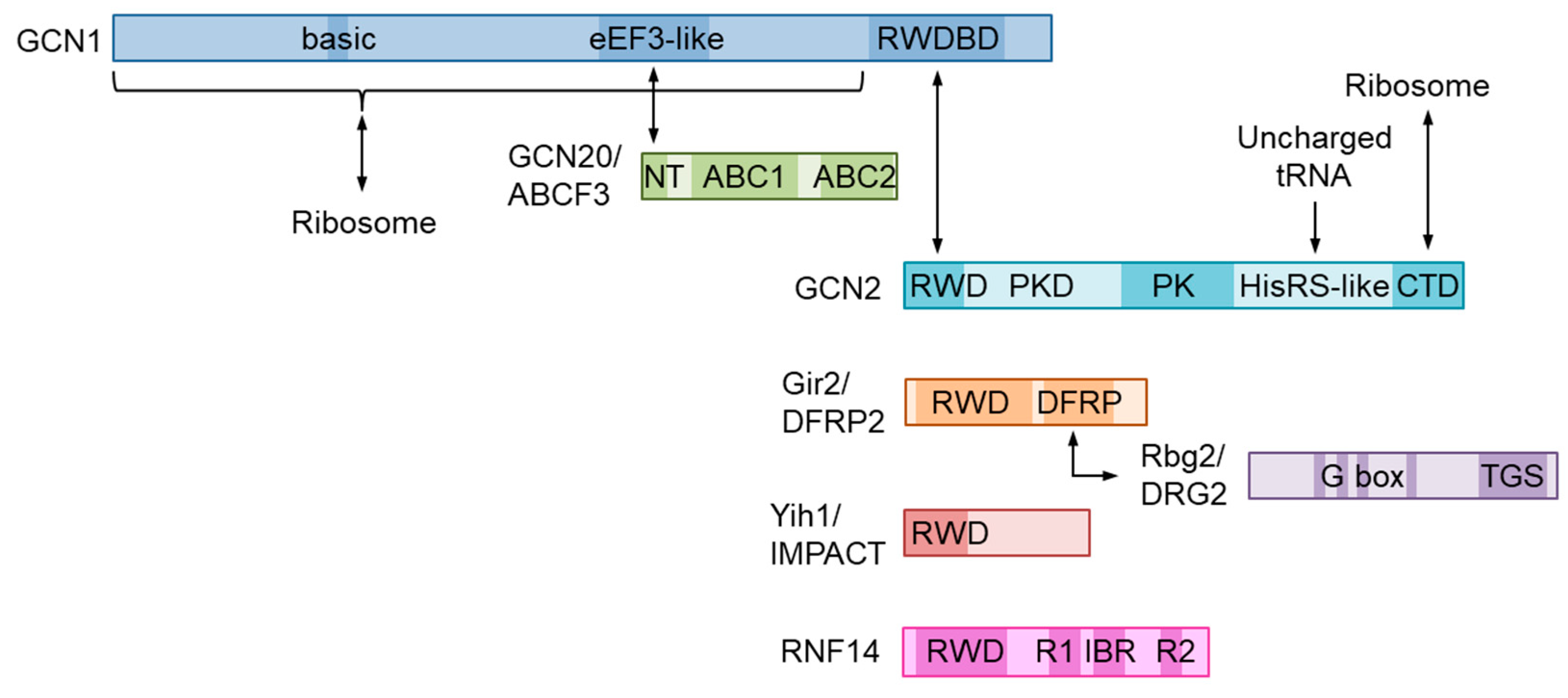
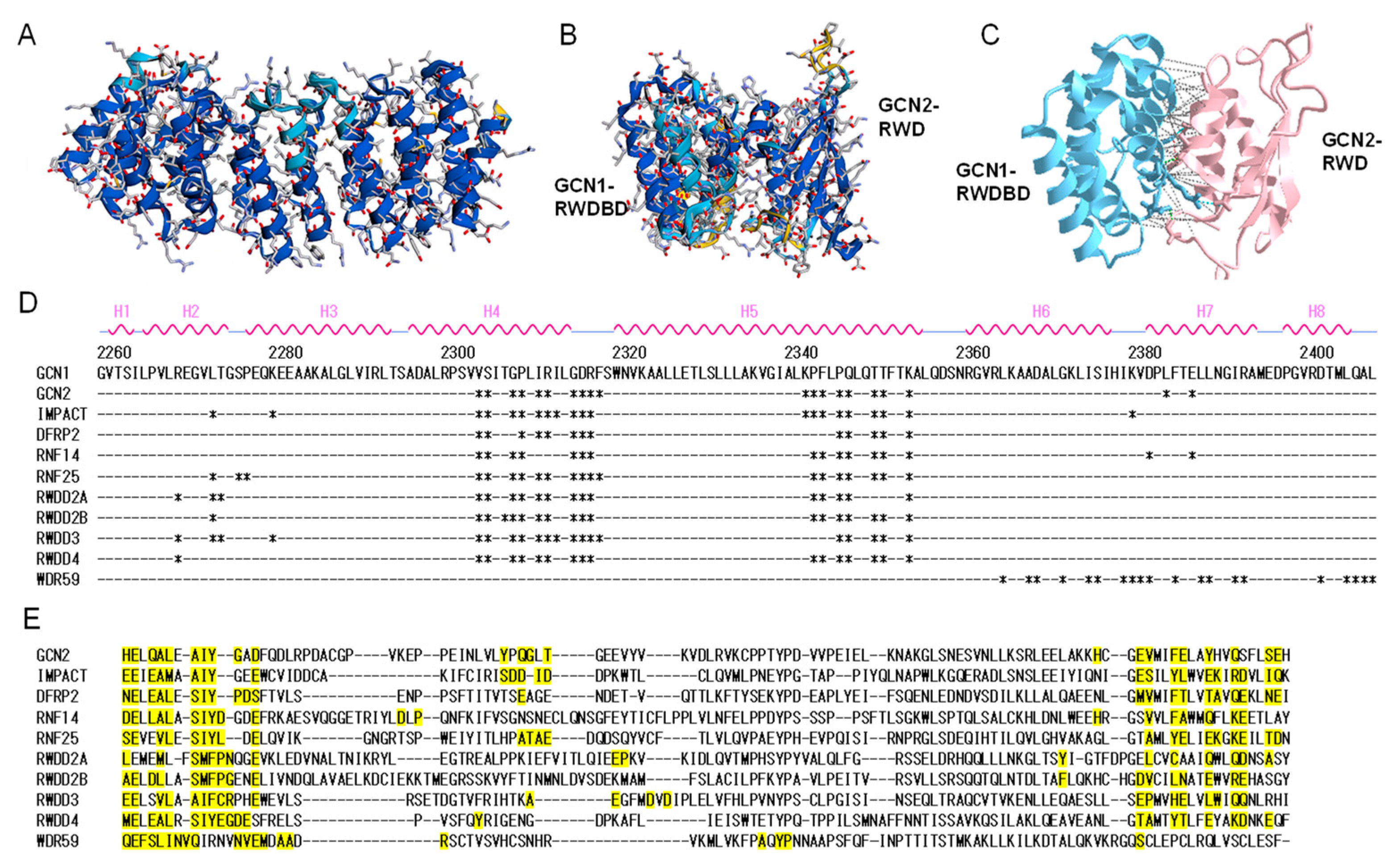
2. Molecular Structure of GCN1 and Its Role in ISR Activation
3. Emerging Role of GCN1 through GCN2-Independent Pathways
3.1. Identification of an Alternative Amino Acid Deprivation Response Distinct from the Canonical GCN2 Pathway in Mammalian Cells
3.2. Role of GCN1-Dependent and GCN2-Independent Mechanisms in Mammalian Development
3.3. GCN2-Independent Pathway in Species Other Than Mammals
4. Function and Structure of the GCN1-Interacting RWD-Domain-Containing Proteins
4.1. GCN2/EIF2AK4
4.2. IMPACT
4.3. DFRP2/RWDD1
4.4. RNF14 and RNF25
4.5. RESUME/RWDD3
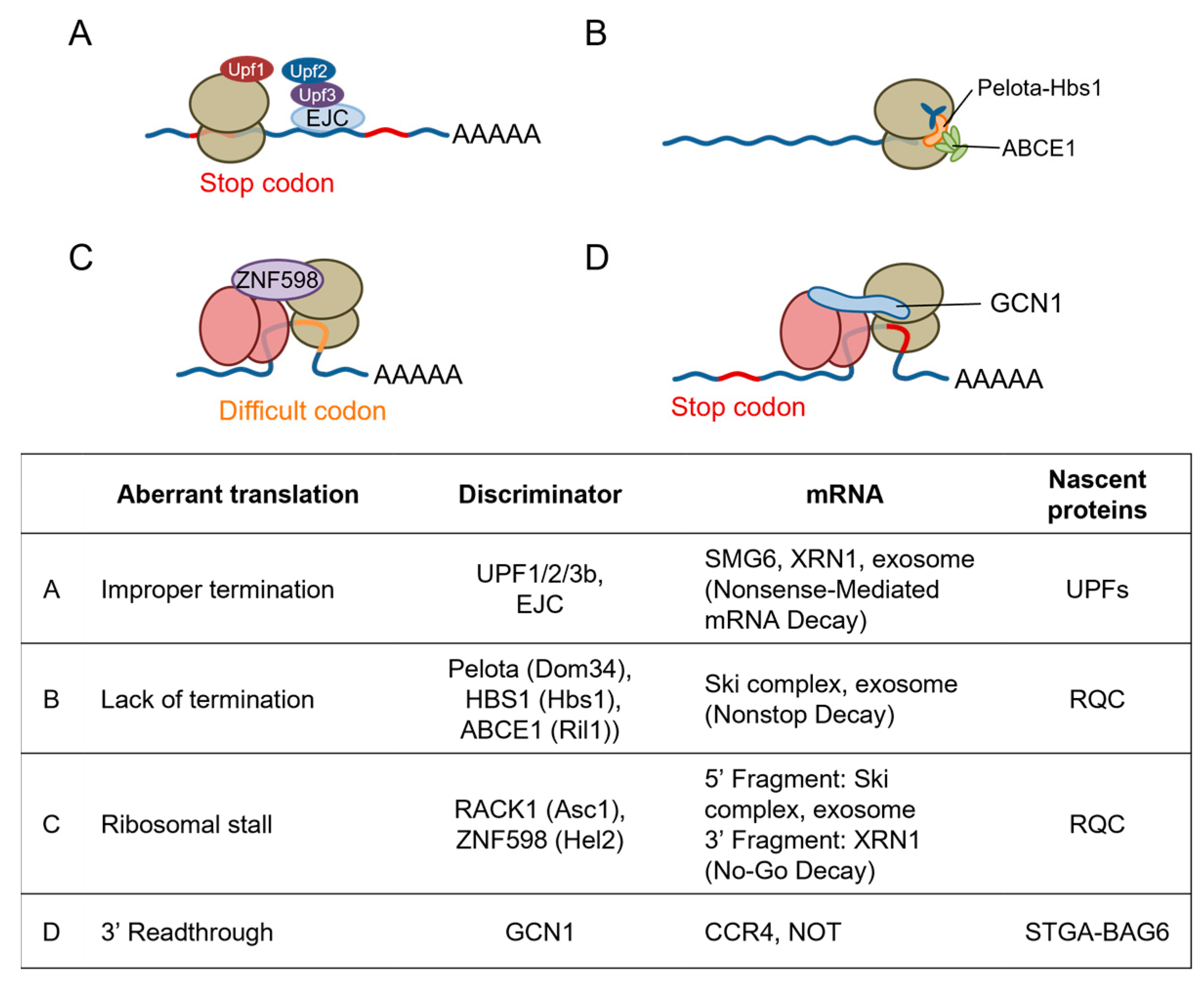
5. Emerging Role of GCN1 in Co-Translational Protein Quality Control
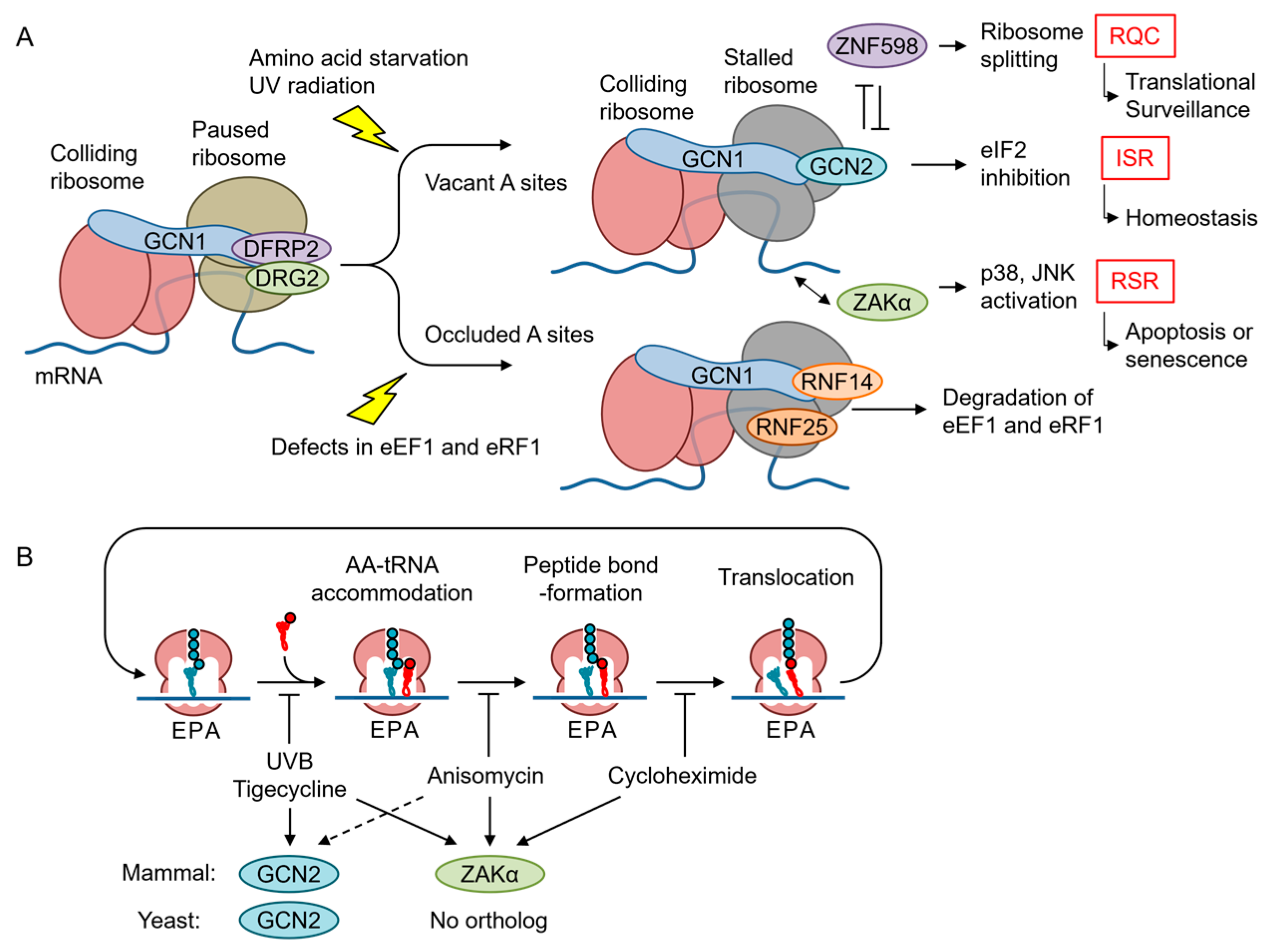
5.1. Quality Control Mechanisms for Stalled Ribosomes
5.2. GCN2 Branch of the ISR
5.3. RSR
6. Function of GCN1 in Basal-State Translational Regulation
7. Role of GCN1-Mediated Responses in Energy Homeostasis
7.1. Role of the GCN1–GCN2 Pathway in the Regulation of Insulin Secretion and Sensitivity
7.2. GCN1 Regulates Energy Storage and Usage
8. Potential Role of GCN1 in Aging and Disease
8.1. Potential Role of GCN1 in Aging
8.2. Potential Role of GCN1 in Neurodegenerative Diseases
9. Future Perspectives
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAS | Amino acid starvation |
| ABC | ATP-binding cassette |
| ABCE1 | ATP-binding cassette sub-family E member 1 |
| ABCF1 | ATP-binding cassette sub-family F member 1 |
| ABCF2 | ATP-binding cassette sub-family F member 2 |
| ATF4 | Cyclic AMP-dependent transcription factor ATF-4 |
| CCR4/NOT | CCR4-Not complex 3’-5’-exoribonuclease subunit Ccr4 |
| Cdk1 | Cyclin-dependent kinase 1 |
| CHOP | CCAAT/Enhancer-binding protein homologous protein |
| DELE1 | DAP3-binding cell death enhancer 1 |
| DFRP2 | RWD domain-containing protein 1 |
| DRG2 | Developmentally regulated GTP-binding protein 2 |
| EDF1 | Endothelial differentiation-related factor 1 |
| eEF1α | Eukaryotic elongation factor-1α |
| eEF3 | Eukaryotic translation elongation factor 3 |
| EIF4E2 | Eukaryotic translation initiation factor 4E type 2 |
| eIF5A | Eukaryotic translation initiation factor 5A-1 |
| elF2α | Eukaryotic initiation factor 2 alpha |
| eRF1 | Eukaryotic release factor 1 |
| eRF3 | Eukaryotic release factor 3 |
| GCN1 | General control nonderepressible 1 |
| GCN2 | General control nonderepressible 2 |
| GIGYF2 | GRB10-interacting GYF protein 2 |
| Hbs1 | Elongation factor 1 alpha-like protein |
| Hel2 | E3 ubiquitin-protein ligase HEL2 |
| HIF1A | Hypoxia-inducible factor 1-alpha |
| HisRS | Histidyl-tRNA synthetase |
| HRI | Eukaryotic translation initiation factor 2-alpha kinase 1 |
| ISR | Integrated stress response |
| LOX | Lipoxygenase |
| Ltn1 | E3 ubiquitin-protein ligase listerin |
| Mbf1 | Multiprotein-bridging factor 1 |
| mTOR | Mechanistic target of rapamycin |
| NEMF | Nuclear export mediator factor |
| NFKBIA | NF-kappa-B inhibitor alpha |
| NGD | No-go decay |
| NMD | Nonsense-mediated decay |
| NPC | Nascent polypeptide chain |
| NR3C1 | Glucocorticoid receptor |
| NRLP1 | Nod-like receptor 1 |
| NSD | Nonstop decay |
| OMA1 | Metalloendopeptidase OMA1, mitochondrial |
| PERK | Eukaryotic translation initiation factor 2-alpha kinase 3 |
| PIAS | Protein inhibitor of activated STAT 1 |
| PKR | Interferon-induced, double-stranded RNA-activated protein kinase |
| Rbg2 | Ribosome-interacting GTPase 2 |
| Rli1 | RNase L inhibitor 1 |
| RNF14 | Ring finger protein 14 |
| RNF25 | Ring finger protein 25 |
| RPS10 | Small ribosomal subunit protein eS10 |
| RQC | Ribosome-associated quality control |
| RWDBD | RWD-binding domain |
| RWDD2A | RWD domain-containing protein 2A |
| RWDD2B | RWD domain-containing protein 2B |
| RWDD3 | RWD domain-containing protein 3 |
| RWDD4 | RWD domain-containing protein 4 |
| SUMO | Small ubiquitin-related modifier |
| TOP1 | DNA topoisomerase 1 |
| UBC9 | Ubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) |
| uS10 | Small ribosomal subunit protein uS10 |
| WDR59 | WD repeat-containing protein 59 |
| ZAKα | Mitogen-activated protein kinase kinase kinase AZK long isoform |
| ZNF598 | Zinc finger protein 598 |
References
- Castilho, B.A.; Shanmugam, R.; Silva, R.C.; Ramesh, R.; Himme, B.M.; Sattlegger, E. Keeping the eIF2 alpha kinase Gcn2 in check. Biochim. Biophys. Acta 2014, 1843, 1948–1968. [Google Scholar] [CrossRef] [PubMed]
- Pakos-Zebrucka, K.; Koryga, I.; Mnich, K.; Ljujic, M.; Samali, A.; Gorman, A.M. The integrated stress response. EMBO Rep. 2016, 17, 1374–1395. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, S.; Miyake, M.; Tsugawa, K.; Oyadomari, M.; Oyadomari, S. Integrated stress response of vertebrates is regulated by four eIF2alpha kinases. Sci. Rep. 2016, 6, 32886. [Google Scholar] [CrossRef]
- Baird, T.D.; Wek, R.C. Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 2012, 3, 307–321. [Google Scholar] [CrossRef] [PubMed]
- B’Chir, W.; Maurin, A.C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef] [PubMed]
- Iurlaro, R.; Munoz-Pinedo, C. Cell death induced by endoplasmic reticulum stress. FEBS J. 2016, 283, 2640–2652. [Google Scholar] [CrossRef]
- Pitale, P.M.; Gorbatyuk, O.; Gorbatyuk, M. Neurodegeneration: Keeping ATF4 on a Tight Leash. Front. Cell Neurosci. 2017, 11, 410. [Google Scholar] [CrossRef]
- Lee, H.C.; Fu, C.Y.; Lin, C.Y.; Hu, J.R.; Huang, T.Y.; Lo, K.Y.; Tsai, H.Y.; Sheu, J.C.; Tsai, H.J. Poly(U)-specific endoribonuclease ENDOU promotes translation of human CHOP mRNA by releasing uORF element-mediated inhibition. EMBO J. 2021, 40, e104123. [Google Scholar] [CrossRef]
- Anda, S.; Zach, R.; Grallert, B. Activation of Gcn2 in response to different stresses. PLoS ONE 2017, 12, e0182143. [Google Scholar] [CrossRef]
- Yamazaki, H.; Kasai, S.; Mimura, J.; Ye, P.; Inose-Maruyama, A.; Tanji, K.; Wakabayashi, K.; Mizuno, S.; Sugiyama, F.; Takahashi, S.; et al. Ribosome binding protein GCN1 regulates the cell cycle and cell proliferation and is essential for the embryonic development of mice. PLoS Genet. 2020, 16, e1008693. [Google Scholar] [CrossRef]
- Ye, J.; Kumanova, M.; Hart, L.S.; Sloane, K.; Zhang, H.; De Panis, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Ron, D.; Koumenis, C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010, 29, 2082–2096. [Google Scholar] [CrossRef]
- Deng, J.; Harding, H.P.; Raught, B.; Gingras, A.C.; Berlanga, J.J.; Scheuner, D.; Kaufman, R.J.; Ron, D.; Sonenberg, N. Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 2002, 12, 1279–1286. [Google Scholar] [CrossRef]
- Robert, F.; Williams, C.; Yan, Y.; Donohue, E.; Cencic, R.; Burley, S.K.; Pelletier, J. Blocking UV-induced eIF2alpha phosphorylation with small molecule inhibitors of GCN2. Chem. Biol. Drug Des. 2009, 74, 57–67. [Google Scholar] [CrossRef]
- Andrade, M.A.; Petosa, C.; O’Donoghue, S.I.; Muller, C.W.; Bork, P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001, 309, 1–18. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Pochopien, A.A.; Beckert, B.; Kasvandik, S.; Berninghausen, O.; Beckmann, R.; Tenson, T.; Wilson, D.N. Structure of Gcn1 bound to stalled and colliding 80S ribosomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2022756118. [Google Scholar] [CrossRef]
- Nameki, N.; Yoneyama, M.; Koshiba, S.; Tochio, N.; Inoue, M.; Seki, E.; Matsuda, T.; Tomo, Y.; Harada, T.; Saito, K.; et al. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 2004, 13, 2089–2100. [Google Scholar] [CrossRef]
- Rakesh, R.; Krishnan, R.; Sattlegger, E.; Srinivasan, N. Recognition of a structural domain (RWDBD) in Gcn1 proteins that interacts with the RWD domain containing proteins. Biol. Direct 2017, 12, 12. [Google Scholar] [CrossRef]
- Sattlegger, E.; Hinnebusch, A.G. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2alpha kinase GCN2 during amino acid starvation. J. Biol. Chem. 2005, 280, 16514–16521. [Google Scholar] [CrossRef]
- Vazquez de Aldana, C.R.; Marton, M.J.; Hinnebusch, A.G. GCN20, a novel ATP binding cassette protein, and GCN1 reside in a complex that mediates activation of the eIF-2 alpha kinase GCN2 in amino acid-starved cells. EMBO J. 1995, 14, 3184–3199. [Google Scholar] [CrossRef]
- Marton, M.J.; Vazquez de Aldana, C.R.; Qiu, H.; Chakraburtty, K.; Hinnebusch, A.G. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol. Cell Biol. 1997, 17, 4474–4489. [Google Scholar] [CrossRef] [PubMed]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef]
- Tyzack, J.K.; Wang, X.; Belsham, G.J.; Proud, C.G. ABC50 interacts with eukaryotic initiation factor 2 and associates with the ribosome in an ATP-dependent manner. J. Biol. Chem. 2000, 275, 34131–34139. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Horvitz, H.R. The translational regulators GCN-1 and ABCF-3 act together to promote apoptosis in C. elegans. PLoS Genet. 2014, 10, e1004512. [Google Scholar] [CrossRef] [PubMed]
- Murina, V.; Kasari, M.; Takada, H.; Hinnu, M.; Saha, C.K.; Grimshaw, J.W.; Seki, T.; Reith, M.; Putrins, M.; Tenson, T.; et al. ABCF ATPases Involved in Protein Synthesis, Ribosome Assembly and Antibiotic Resistance: Structural and Functional Diversification across the Tree of Life. J. Mol. Biol. 2019, 431, 3568–3590. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Fernandez, I.S.; Gordiyenko, Y.; Ramakrishnan, V. Ribosome-dependent activation of stringent control. Nature 2016, 534, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Sundrud, M.S.; Zhou, C.; Edenius, M.; Zocco, D.; Powers, K.; Zhang, M.; Mazitschek, R.; Rao, A.; Yeo, C.Y.; et al. Aminoacyl-tRNA synthetase inhibition activates a pathway that branches from the canonical amino acid response in mammalian cells. Proc. Natl. Acad. Sci. USA 2020, 117, 8900–8911. [Google Scholar] [CrossRef]
- Sundrud, M.S.; Koralov, S.B.; Feuerer, M.; Calado, D.P.; Kozhaya, A.E.; Rhule-Smith, A.; Lefebvre, R.E.; Unutmaz, D.; Mazitschek, R.; Waldner, H.; et al. Halofuginone inhibits TH17 cell differentiation by activating the amino acid starvation response. Science 2009, 324, 1334–1338. [Google Scholar] [CrossRef]
- De Vito, A.; Lazzaro, M.; Palmisano, I.; Cittaro, D.; Riba, M.; Lazarevic, D.; Bannai, M.; Gabellini, D.; Schiaffino, M.V. Amino acid deprivation triggers a novel GCN2-independent response leading to the transcriptional reactivation of non-native DNA sequences. PLoS ONE 2018, 13, e0200783. [Google Scholar] [CrossRef]
- Palmisano, I.; Della Chiara, G.; D’Ambrosio, R.L.; Huichalaf, C.; Brambilla, P.; Corbetta, S.; Riba, M.; Piccirillo, R.; Valente, S.; Casari, G.; et al. Amino acid starvation induces reactivation of silenced transgenes and latent HIV-1 provirus via down-regulation of histone deacetylase 4 (HDAC4). Proc. Natl. Acad. Sci. USA 2012, 109, E2284–E2293. [Google Scholar] [CrossRef]
- Izquierdo, Y.; Kulasekaran, S.; Benito, P.; Lopez, B.; Marcos, R.; Cascon, T.; Hamberg, M.; Castresana, C. Arabidopsis nonresponding to oxylipins locus NOXY7 encodes a yeast GCN1 homolog that mediates noncanonical translation regulation and stress adaptation. Plant Cell Environ. 2018, 41, 1438–1452. [Google Scholar] [CrossRef]
- Doerks, T.; Copley, R.R.; Schultz, J.; Ponting, C.P.; Bork, P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002, 12, 47–56. [Google Scholar] [CrossRef]
- Pereira, C.M.; Sattlegger, E.; Jiang, H.Y.; Longo, B.M.; Jaqueta, C.B.; Hinnebusch, A.G.; Wek, R.C.; Mello, L.E.; Castilho, B.A. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J. Biol. Chem. 2005, 280, 28316–28323. [Google Scholar] [CrossRef]
- Waller, T.; Lee, S.J.; Sattlegger, E. Evidence that Yih1 resides in a complex with ribosomes. FEBS J. 2012, 279, 1761–1776. [Google Scholar] [CrossRef]
- Wout, P.K.; Sattlegger, E.; Sullivan, S.M.; Maddock, J.R. Saccharomyces cerevisiae Rbg1 protein and its binding partner Gir2 interact on Polyribosomes with Gcn1. Eukaryot. Cell 2009, 8, 1061–1071. [Google Scholar] [CrossRef]
- Ishikawa, K.; Ito, K.; Inoue, J.; Semba, K. Cell growth control by stable Rbg2/Gir2 complex formation under amino acid starvation. Genes. Cells 2013, 18, 859–872. [Google Scholar] [CrossRef]
- Oltion, K.; Carelli, J.D.; Yang, T.; See, S.K.; Wang, H.Y.; Kampmann, M.; Taunton, J. An E3 ligase network engages GCN1 to promote the degradation of translation factors on stalled ribosomes. Cell 2023, 186, 346–362.e17. [Google Scholar] [CrossRef]
- Wu, C.C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Lageix, S.; Zhang, J.; Rothenburg, S.; Hinnebusch, A.G. Interaction between the tRNA-binding and C-terminal domains of Yeast Gcn2 regulates kinase activity in vivo. PLoS Genet. 2015, 11, e1004991. [Google Scholar] [CrossRef]
- Wek, S.A.; Zhu, S.; Wek, R.C. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell Biol. 1995, 15, 4497–4506. [Google Scholar] [CrossRef]
- Zaborske, J.M.; Narasimhan, J.; Jiang, L.; Wek, S.A.; Dittmar, K.A.; Freimoser, F.; Pan, T.; Wek, R.C. Genome-wide analysis of tRNA charging and activation of the eIF2 kinase Gcn2p. J. Biol. Chem. 2009, 284, 25254–25267. [Google Scholar] [CrossRef]
- Costa-Mattioli, M.; Gobert, D.; Harding, H.; Herdy, B.; Azzi, M.; Bruno, M.; Bidinosti, M.; Ben Mamou, C.; Marcinkiewicz, E.; Yoshida, M.; et al. Translational control of hippocampal synaptic plasticity and memory by the eIF2alpha kinase GCN2. Nature 2005, 436, 1166–1173. [Google Scholar] [CrossRef]
- Maurin, A.C.; Jousse, C.; Averous, J.; Parry, L.; Bruhat, A.; Cherasse, Y.; Zeng, H.; Zhang, Y.; Harding, H.P.; Ron, D.; et al. The GCN2 kinase biases feeding behavior to maintain amino acid homeostasis in omnivores. Cell Metab. 2005, 1, 273–277. [Google Scholar] [CrossRef]
- Eyries, M.; Montani, D.; Girerd, B.; Perret, C.; Leroy, A.; Lonjou, C.; Chelghoum, N.; Coulet, F.; Bonnet, D.; Dorfmüller, P.; et al. EIF2AK4 mutations cause pulmonary veno-occlusive disease, a recessive form of pulmonary hypertension. Nat. Genet. 2014, 46, 65–69. [Google Scholar] [CrossRef]
- Roffé, M.; Hajj, G.N.; Azevedo, H.F.; Alves, V.S.; Castilho, B.A. IMPACT is a developmentally regulated protein in neurons that opposes the eukaryotic initiation factor 2α kinase GCN2 in the modulation of neurite outgrowth. J. Biol. Chem. 2013, 288, 10860–10869. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.; Morquette, B.; Kroner, A.; Leong, S.; Madwar, C.; Sanz, R.; Banerjee, S.L.; Antel, J.; Bisson, N.; David, S.; et al. Small-Molecule Stabilization of 14-3-3 Protein-Protein Interactions Stimulates Axon Regeneration. Neuron 2017, 93, 1082–1093.e1085. [Google Scholar] [CrossRef]
- Sattlegger, E.; Swanson, M.J.; Ashcraft, E.A.; Jennings, J.L.; Fekete, R.A.; Link, A.J.; Hinnebusch, A.G. YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J. Biol. Chem. 2004, 279, 29952–29962. [Google Scholar] [CrossRef]
- Cambiaghi, T.D.; Pereira, C.M.; Shanmugam, R.; Bolech, M.; Wek, R.C.; Sattlegger, E.; Castilho, B.A. Evolutionarily conserved IMPACT impairs various stress responses that require GCN1 for activating the eIF2 kinase GCN2. Biochem. Biophys. Res. Commun. 2014, 443, 592–597. [Google Scholar] [CrossRef]
- Ferraz, R.C.; Camara, H.; De-Souza, E.A.; Pinto, S.; Pinca, A.P.; Silva, R.C.; Sato, V.N.; Castilho, B.A.; Mori, M.A. IMPACT is a GCN2 inhibitor that limits lifespan in Caenorhabditis elegans. BMC Biol. 2016, 14, 87. [Google Scholar] [CrossRef]
- Daugeron, M.C.; Prouteau, M.; Lacroute, F.; Seraphin, B. The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1. Nucleic. Acids Res. 2011, 39, 2221–2233. [Google Scholar] [CrossRef]
- Ishikawa, K.; Azuma, S.; Ikawa, S.; Semba, K.; Inoue, J. Identification of DRG family regulatory proteins (DFRPs): Specific regulation of DRG1 and DRG2. Genes Cells 2005, 10, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Westrip, C.A.E.; Zhuang, Q.; Hall, C.; Eaton, C.D.; Coleman, M.L. Developmentally regulated GTPases: Structure, function and roles in disease. Cell Mol. Life Sci. 2021, 78, 7219–7235. [Google Scholar] [CrossRef]
- Müller, M.B.D.; Kasturi, P.; Jayaraj, G.G.; Hartl, F.U. Mechanisms of readthrough mitigation reveal principles of GCN1-mediated translational quality control. Cell 2023, 186, 3227–3244.e3220. [Google Scholar] [CrossRef]
- Xu, K.; Shimelis, H.; Linn, D.E.; Jiang, R.; Yang, X.; Sun, F.; Guo, Z.; Chen, H.; Li, W.; Chen, H.; et al. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 2009, 15, 270–282. [Google Scholar] [CrossRef]
- Alontaga, A.Y.; Ambaye, N.D.; Li, Y.J.; Vega, R.; Chen, C.H.; Bzymek, K.P.; Williams, J.C.; Hu, W.; Chen, Y. RWD Domain as an E2 (Ubc9)-Interaction Module. J. Biol. Chem. 2015, 290, 16550–16559. [Google Scholar] [CrossRef]
- Carbia-Nagashima, A.; Gerez, J.; Perez-Castro, C.; Paez-Pereda, M.; Silberstein, S.; Stalla, G.K.; Holsboer, F.; Arzt, E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell 2007, 131, 309–323. [Google Scholar] [CrossRef]
- Shan, B.; Gerez, J.; Haedo, M.; Fuertes, M.; Theodoropoulou, M.; Buchfelder, M.; Losa, M.; Stalla, G.K.; Arzt, E.; Renner, U. RSUME is implicated in HIF-1-induced VEGF-A production in pituitary tumour cells. Endocr. Relat. Cancer 2012, 19, 13–27. [Google Scholar] [CrossRef]
- Gerez, J.; Fuertes, M.; Tedesco, L.; Silberstein, S.; Sevlever, G.; Paez-Pereda, M.; Holsboer, F.; Turjanski, A.G.; Arzt, E. In silico structural and functional characterization of the RSUME splice variants. PLoS ONE 2013, 8, e57795. [Google Scholar] [CrossRef]
- Druker, J.; Liberman, A.C.; Antunica-Noguerol, M.; Gerez, J.; Paez-Pereda, M.; Rein, T.; Iniguez-Lluhi, J.A.; Holsboer, F.; Arzt, E. RSUME enhances glucocorticoid receptor SUMOylation and transcriptional activity. Mol. Cell Biol. 2013, 33, 2116–2127. [Google Scholar] [CrossRef]
- Kim, K.Q.; Zaher, H.S. Canary in a coal mine: Collided ribosomes as sensors of cellular conditions. Trends Biochem. Sci. 2022, 47, 82–97. [Google Scholar] [CrossRef]
- De, S.; Muhlemann, O. A comprehensive coverage insurance for cells: Revealing links between ribosome collisions, stress responses and mRNA surveillance. RNA Biol. 2022, 19, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.C.; Aloy, P.; Grandi, P.; Krause, R.; Boesche, M.; Marzioch, M.; Rau, C.; Jensen, L.J.; Bastuck, S.; Dumpelfeld, B.; et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 2006, 440, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Hermjakob, H.; Montecchi-Palazzi, L.; Lewington, C.; Mudali, S.; Kerrien, S.; Orchard, S.; Vingron, M.; Roechert, B.; Roepstorff, P.; Valencia, A.; et al. IntAct: An open source molecular interaction database. Nucleic Acids Res. 2004, 32, D452–D455. [Google Scholar] [CrossRef] [PubMed]
- Gurzeler, L.A.; Link, M.; Ibig, Y.; Schmidt, I.; Galuba, O.; Schoenbett, J.; Gasser-Didierlaurant, C.; Parker, C.N.; Mao, X.; Bitsch, F.; et al. Drug-induced eRF1 degradation promotes readthrough and reveals a new branch of ribosome quality control. Cell Rep. 2023, 42, 113056. [Google Scholar] [CrossRef] [PubMed]
- Meydan, S.; Guydosh, N.R. A cellular handbook for collided ribosomes: Surveillance pathways and collision types. Curr. Genet. 2021, 67, 19–26. [Google Scholar] [CrossRef]
- Garzia, A.; Jafarnejad, S.M.; Meyer, C.; Chapat, C.; Gogakos, T.; Morozov, P.; Amiri, M.; Shapiro, M.; Molina, H.; Tuschl, T.; et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat. Commun. 2017, 8, 16056. [Google Scholar] [CrossRef]
- Matsuo, Y.; Ikeuchi, K.; Saeki, Y.; Iwasaki, S.; Schmidt, C.; Udagawa, T.; Sato, F.; Tsuchiya, H.; Becker, T.; Tanaka, K.; et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. Commun. 2017, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Juszkiewicz, S.; Speldewinde, S.H.; Wan, L.; Svejstrup, J.Q.; Hegde, R.S. The ASC-1 Complex Disassembles Collided Ribosomes. Mol. Cell 2020, 79, 603–614 e608. [Google Scholar] [CrossRef]
- Filbeck, S.; Cerullo, F.; Pfeffer, S.; Joazeiro, C.A.P. Ribosome-associated quality-control mechanisms from bacteria to humans. Mol. Cell 2022, 82, 1451–1466. [Google Scholar] [CrossRef]
- Hickey, K.L.; Dickson, K.; Cogan, J.Z.; Replogle, J.M.; Schoof, M.; D’Orazio, K.N.; Sinha, N.K.; Hussmann, J.A.; Jost, M.; Frost, A.; et al. GIGYF2 and 4EHP Inhibit Translation Initiation of Defective Messenger RNAs to Assist Ribosome-Associated Quality Control. Mol. Cell 2020, 79, 950–962.e6. [Google Scholar] [CrossRef]
- Sinha, N.K.; Ordureau, A.; Best, K.; Saba, J.A.; Zinshteyn, B.; Sundaramoorthy, E.; Fulzele, A.; Garshott, D.M.; Denk, T.; Thoms, M.; et al. EDF1 coordinates cellular responses to ribosome collisions. Elife 2020, 9, e58828. [Google Scholar] [CrossRef]
- Juszkiewicz, S.; Slodkowicz, G.; Lin, Z.; Freire-Pritchett, P.; Peak-Chew, S.Y.; Hegde, R.S. Ribosome collisions trigger cis-acting feedback inhibition of translation initiation. Elife 2020, 9, e60038. [Google Scholar] [CrossRef]
- Yan, L.L.; Zaher, H.S. Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes. Mol. Cell 2021, 81, 614–628.e614. [Google Scholar] [CrossRef]
- Fedry, J.; Silva, J.; Vanevic, M.; Fronik, S.; Mechulam, Y.; Schmitt, E.; des Georges, A.; Faller, W.; Forster, F. Visualization of translation reorganization upon persistent ribosome collision stress in mammalian cells. Mol. Cell 2024, 84, 1–12. [Google Scholar] [CrossRef]
- Masson, G.R. Towards a model of GCN2 activation. Biochem. Soc. Trans. 2019, 47, 1481–1488. [Google Scholar] [CrossRef]
- Ishimura, R.; Nagy, G.; Dotu, I.; Chuang, J.H.; Ackerman, S.L. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 2016, 5, e14295. [Google Scholar] [CrossRef]
- Darnell, A.M.; Subramaniam, A.R.; O’Shea, E.K. Translational Control through Differential Ribosome Pausing during Amino Acid Limitation in Mammalian Cells. Mol. Cell 2018, 71, 229–243.e11. [Google Scholar] [CrossRef]
- Stoneley, M.; Harvey, R.F.; Mulroney, T.E.; Mordue, R.; Jukes-Jones, R.; Cain, K.; Lilley, K.S.; Sawarkar, R.; Willis, A.E. Unresolved stalled ribosome complexes restrict cell-cycle progression after genotoxic stress. Mol. Cell 2022, 82, 1557–1572.e7. [Google Scholar] [CrossRef]
- Wu, C.C.; Zinshteyn, B.; Wehner, K.A.; Green, R. High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol. Cell 2019, 73, 959–970.e955. [Google Scholar] [CrossRef]
- Lee, S.J.; Swanson, M.J.; Sattlegger, E. Gcn1 contacts the small ribosomal protein Rps10, which is required for full activation of the protein kinase Gcn2. Biochem J. 2015, 466, 547–559. [Google Scholar] [CrossRef]
- Snieckute, G.; Genzor, A.V.; Vind, A.C.; Ryder, L.; Stoneley, M.; Chamois, S.; Dreos, R.; Nordgaard, C.; Sass, F.; Blasius, M.; et al. Ribosome stalling is a signal for metabolic regulation by the ribotoxic stress response. Cell Metab. 2022, 34, 2036–2046.e2038. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, L.; Chen, J.; Zhang, W.W.; Zhang, X.Y.; Wang, B.; Zhang, C.; Wang, Y.; Huang, Y.C.; Wang, H.; et al. Mitochondrial ROS-mediated ribosome stalling and GCN2 activation are partially involved in 1-nitropyrene-induced steroidogenic inhibition in testes. Environ. Int. 2022, 167, 107393. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Cordes, J.; Caban, K.M.; Götz, M.J.; Mackens-Kiani, T.; Veltri, A.J.; Sinha, N.K.; Weickert, P.; Kaya, S.; Hewitt, G.; et al. RNF14-dependent atypical ubiquitylation promotes translation-coupled resolution of RNA-protein crosslinks. Mol. Cell 2023, 83, 4290–4303.e4299. [Google Scholar] [CrossRef]
- Ryder, L.; Arendrup, F.S.; Martínez, J.F.; Snieckute, G.; Pecorari, C.; Shah, R.A.; Lund, A.H.; Blasius, M.; Bekker-Jensen, S. Nitric oxide-induced ribosome collision activates ribosomal surveillance mechanisms. Cell Death Dis. 2023, 14, 467. [Google Scholar] [CrossRef]
- Inglis, A.J.; Masson, G.R.; Shao, S.; Perisic, O.; McLaughlin, S.H.; Hegde, R.S.; Williams, R.L. Activation of GCN2 by the ribosomal P-stalk. Proc. Natl. Acad. Sci. USA 2019, 116, 4946–4954. [Google Scholar] [CrossRef]
- Farooq, Z.; Kusuma, F.; Burke, P.; Dufour, C.R.; Lee, D.; Tabatabaei, N.; Toboz, P.; Radovani, E.; Greenblatt, J.F.; Rehman, J.; et al. The amino acid sensor GCN2 suppresses terminal oligopyrimidine (TOP) mRNA translation via La-related protein 1 (LARP1). J. Biol. Chem. 2022, 298, 102277. [Google Scholar] [CrossRef]
- Iordanov, M.S.; Pribnow, D.; Magun, J.L.; Dinh, T.H.; Pearson, J.A.; Chen, S.L.; Magun, B.E. Ribotoxic stress response: Activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol. Cell Biol. 1997, 17, 3373–3381. [Google Scholar] [CrossRef]
- Vind, A.C.; Genzor, A.V.; Bekker-Jensen, S. Ribosomal stress-surveillance: Three pathways is a magic number. Nucleic Acids Res. 2020, 48, 10648–10661. [Google Scholar] [CrossRef]
- Wang, X.; Mader, M.M.; Toth, J.E.; Yu, X.; Jin, N.; Campbell, R.M.; Smallwood, J.K.; Christe, M.E.; Chatterjee, A.; Goodson, T., Jr.; et al. Complete inhibition of anisomycin and UV radiation but not cytokine induced JNK and p38 activation by an aryl-substituted dihydropyrrolopyrazole quinoline and mixed lineage kinase 7 small interfering RNA. J. Biol. Chem. 2005, 280, 19298–19305. [Google Scholar] [CrossRef]
- Jandhyala, D.M.; Ahluwalia, A.; Obrig, T.; Thorpe, C.M. ZAK: A MAP3Kinase that transduces Shiga toxin- and ricin-induced proinflammatory cytokine expression. Cell Microbiol. 2008, 10, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Jandhyala, D.M.; Wong, J.; Mantis, N.J.; Magun, B.E.; Leong, J.M.; Thorpe, C.M. A Novel Zak Knockout Mouse with a Defective Ribotoxic Stress Response. Toxins 2016, 8, 259. [Google Scholar] [CrossRef]
- Sauter, K.A.; Magun, E.A.; Iordanov, M.S.; Magun, B.E. ZAK is required for doxorubicin, a novel ribotoxic stressor, to induce SAPK activation and apoptosis in HaCaT cells. Cancer Biol. Ther. 2010, 10, 258–266. [Google Scholar] [CrossRef]
- Vind, A.C.; Snieckute, G.; Blasius, M.; Tiedje, C.; Krogh, N.; Bekker-Jensen, D.B.; Andersen, K.L.; Nordgaard, C.; Tollenaere, M.A.X.; Lund, A.H.; et al. ZAKalpha Recognizes Stalled Ribosomes through Partially Redundant Sensor Domains. Mol. Cell 2020, 78, 700–713.e707. [Google Scholar] [CrossRef] [PubMed]
- Robinson, K.S.; Toh, G.A.; Rozario, P.; Chua, R.; Bauernfried, S.; Sun, Z.; Firdaus, M.J.; Bayat, S.; Nadkarni, R.; Poh, Z.S.; et al. ZAKalpha-driven ribotoxic stress response activates the human NLRP1 inflammasome. Science 2022, 377, 328–335. [Google Scholar] [CrossRef]
- Muela-Zarzuela, I.; Suarez-Rivero, J.M.; Gallardo-Orihuela, A.; Wang, C.; Izawa, K.; de Gregorio-Procopio, M.; Couillin, I.; Ryffel, B.; Kitaura, J.; Sanz, A.; et al. NLRP1 inflammasome modulates senescence and senescence-associated secretory phenotype. bioRxiv 2023. [Google Scholar] [CrossRef]
- Snieckute, G.; Ryder, L.; Vind, A.C.; Wu, Z.; Arendrup, F.S.; Stoneley, M.; Chamois, S.; Martinez-Val, A.; Leleu, M.; Dreos, R.; et al. ROS-induced ribosome impairment underlies ZAKalpha-mediated metabolic decline in obesity and aging. Science 2023, 382, eadf3208. [Google Scholar] [CrossRef]
- Doma, M.K.; Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006, 440, 561–564. [Google Scholar] [CrossRef]
- Dimitrova, L.N.; Kuroha, K.; Tatematsu, T.; Inada, T. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 2009, 284, 10343–10352. [Google Scholar] [CrossRef]
- Letzring, D.P.; Wolf, A.S.; Brule, C.E.; Grayhack, E.J. Translation of CGA codon repeats in yeast involves quality control components and ribosomal protein L1. RNA 2013, 19, 1208–1217. [Google Scholar] [CrossRef]
- Simms, C.L.; Hudson, B.H.; Mosior, J.W.; Rangwala, A.S.; Zaher, H.S. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014, 9, 1256–1264. [Google Scholar] [CrossRef]
- Brandman, O.; Hegde, R.S. Ribosome-associated protein quality control. Nat. Struct. Mol. Biol. 2016, 23, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Gamble, C.E.; Brule, C.E.; Dean, K.M.; Fields, S.; Grayhack, E.J. Adjacent Codons Act in Concert to Modulate Translation Efficiency in Yeast. Cell 2016, 166, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Joazeiro, C.A.P. Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu. Rev. Cell Dev. Biol. 2017, 33, 343–368. [Google Scholar] [CrossRef] [PubMed]
- Collart, M.A.; Weiss, B. Ribosome pausing, a dangerous necessity for co-translational events. Nucleic Acids Res. 2020, 48, 1043–1055. [Google Scholar] [CrossRef]
- Lyu, X.; Yang, Q.; Zhao, F.; Liu, Y. Codon usage and protein length-dependent feedback from translation elongation regulates translation initiation and elongation speed. Nucleic Acids Res. 2021, 49, 9404–9423. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.R.; Vo, M.T.; Kim, D.J.; Lee, U.H.; Yoon, J.H.; Kim, H.J.; Kim, J.; Kim, S.R.; Lee, J.Y.; Yang, C.H.; et al. DRG2 Deficient Mice Exhibit Impaired Motor Behaviors with Reduced Striatal Dopamine Release. Int. J. Mol. Sci. 2019, 21, 60. [Google Scholar] [CrossRef] [PubMed]
- Houston, L.; Platten, E.M.; Connelly, S.M.; Wang, J.; Grayhack, E.J. Frameshifting at collided ribosomes is modulated by elongation factor eEF3 and by integrated stress response regulators Gcn1 and Gcn20. RNA 2022, 28, 320–339. [Google Scholar] [CrossRef]
- Schuller, A.P.; Wu, C.C.; Dever, T.E.; Buskirk, A.R.; Green, R. eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205.e195. [Google Scholar] [CrossRef]
- Tesina, P.; Ebine, S.; Buschauer, R.; Thoms, M.; Matsuo, Y.; Inada, T.; Beckmann, R. Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control. Mol. Cell 2023, 83, 607–621.e604. [Google Scholar] [CrossRef]
- Zeng, F.; Li, X.; Pires-Alves, M.; Chen, X.; Hawk, C.W.; Jin, H. Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells. Cell Rep. 2021, 37, 109877. [Google Scholar] [CrossRef]
- Kriachkov, V.; Ormsby, A.R.; Kusnadi, E.P.; McWilliam, H.E.G.; Mintern, J.D.; Amarasinghe, S.L.; Ritchie, M.E.; Furic, L.; Hatters, D.M. Arginine-rich C9ORF72 ALS proteins stall ribosomes in a manner distinct from a canonical ribosome-associated quality control substrate. J. Biol. Chem. 2023, 299, 102774. [Google Scholar] [CrossRef]
- Seo, J.; Fortuno, E.S., 3rd; Suh, J.M.; Stenesen, D.; Tang, W.; Parks, E.J.; Adams, C.M.; Townes, T.; Graff, J.M. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes 2009, 58, 2565–2573. [Google Scholar] [CrossRef]
- Kanno, A.; Asahara, S.I.; Furubayashi, A.; Masuda, K.; Yoshitomi, R.; Suzuki, E.; Takai, T.; Kimura-Koyanagi, M.; Matsuda, T.; Bartolome, A.; et al. GCN2 regulates pancreatic β cell mass by sensing intracellular amino acid levels. JCI Insight 2020, 5, e128820. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yuan, J.; Yue, W.; Bi, Y.; Shen, X.; Gao, J.; Xu, X.; Lu, Z. GCN2 deficiency protects against high fat diet induced hepatic steatosis and insulin resistance in mice. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 3257–3267. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, S.H.; Marti, A.; Jena, J.; Garcia Pena, L.M.; Weatherford, E.T.; Kato, K.; Koneru, J.; Chen, J.; Sood, A.; Potthoff, M.J.; et al. ATF4 Expression in Thermogenic Adipocytes is Required for Cold-Induced Thermogenesis in Mice via FGF21-Independent Mechanisms. Sci. Rep. 2024, 14, 1563. [Google Scholar] [CrossRef]
- Miyake, M.; Zhang, J.; Yasue, A.; Hisanaga, S.; Tsugawa, K.; Sakaue, H.; Oyadomari, M.; Kiyonari, H.; Oyadomari, S. Integrated stress response regulates GDF15 secretion from adipocytes, preferentially suppresses appetite for a high-fat diet and improves obesity. iScience 2021, 24, 103448. [Google Scholar] [CrossRef]
- Yuan, J.; Li, F.; Shen, X.; Gao, J.; Yu, Z.; Luo, K.; Cui, B.; Lu, Z. Genetic and Pharmacological Inhibition of GCN2 Ameliorates Hyperglycemia and Insulin Resistance in Type 2 Diabetic Mice. Antioxidants 2022, 11, 1584. [Google Scholar] [CrossRef]
- Yuan, J.; Yu, Z.; Gao, J.; Luo, K.; Shen, X.; Cui, B.; Lu, Z. Inhibition of GCN2 alleviates hepatic steatosis and oxidative stress in obese mice: Involvement of NRF2 regulation. Redox Biol. 2022, 49, 102224. [Google Scholar] [CrossRef]
- Dehwah, M.A.; Wang, M.; Huang, Q.Y. CDKAL1 and type 2 diabetes: A global meta-analysis. Genet. Mol. Res. 2010, 9, 1109–1120. [Google Scholar] [CrossRef]
- Wei, F.Y.; Suzuki, T.; Watanabe, S.; Kimura, S.; Kaitsuka, T.; Fujimura, A.; Matsui, H.; Atta, M.; Michiue, H.; Fontecave, M.; et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Invest. 2011, 121, 3598–3608. [Google Scholar] [CrossRef]
- Takahashi, N.; Wei, F.Y.; Watanabe, S.; Hirayama, M.; Ohuchi, Y.; Fujimura, A.; Kaitsuka, T.; Ishii, I.; Sawa, T.; Nakayama, H.; et al. Reactive sulfur species regulate tRNA methylthiolation and contribute to insulin secretion. Nucleic Acids Res. 2017, 45, 435–445. [Google Scholar] [CrossRef]
- Miyake, K.; Yang, W.; Hara, K.; Yasuda, K.; Horikawa, Y.; Osawa, H.; Furuta, H.; Ng, M.C.; Hirota, Y.; Mori, H.; et al. Construction of a prediction model for type 2 diabetes mellitus in the Japanese population based on 11 genes with strong evidence of the association. J. Hum. Genet. 2009, 54, 236–241. [Google Scholar] [CrossRef]
- Kanno, A.; Asahara, S.I.; Masuda, K.; Matsuda, T.; Kimura-Koyanagi, M.; Seino, S.; Ogawa, W.; Kido, Y. Compensatory hyperinsulinemia in high-fat diet-induced obese mice is associated with enhanced insulin translation in islets. Biochem. Biophys. Res. Commun. 2015, 458, 681–686. [Google Scholar] [CrossRef]
- Kitakaze, K.; Oyadomari, M.; Zhang, J.; Hamada, Y.; Takenouchi, Y.; Tsuboi, K.; Inagaki, M.; Tachikawa, M.; Fujitani, Y.; Okamoto, Y.; et al. ATF4-mediated transcriptional regulation protects against β-cell loss during endoplasmic reticulum stress in a mouse model. Mol. Metab. 2021, 54, 101338. [Google Scholar] [CrossRef]
- Szabat, M.; Page, M.M.; Panzhinskiy, E.; Skovsø, S.; Mojibian, M.; Fernandez-Tajes, J.; Bruin, J.E.; Bround, M.J.; Lee, J.T.; Xu, E.E.; et al. Reduced Insulin Production Relieves Endoplasmic Reticulum Stress and Induces β Cell Proliferation. Cell Metab. 2016, 23, 179–193. [Google Scholar] [CrossRef]
- Liu, J.; Kasai, S.; Tatara, Y.; Yamazaki, H.; Mimura, J.; Mizuno, S.; Sugiyama, F.; Takahashi, S.; Sato, T.; Ozaki, T.; et al. Inducible Systemic Gcn1 Deletion in Mice Leads to Transient Body Weight Loss upon Tamoxifen Treatment Associated with Decrease of Fat and Liver Glycogen Storage. Int. J. Mol. Sci. 2022, 23, 3201. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Stein, K.C.; Morales-Polanco, F.; van der Lienden, J.; Rainbolt, T.K.; Frydman, J. Ageing exacerbates ribosome pausing to disrupt cotranslational proteostasis. Nature 2022, 601, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, T.; Bi, Z.; Ni, J.Q.; Pastor-Pareja, J.C.; Javid, B. Premature termination codon readthrough in Drosophila varies in a developmental and tissue-specific manner. Sci. Rep. 2020, 10, 8485. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, H.; Mendell, J.T. Ribosome Recycling by ABCE1 Links Lysosomal Function and Iron Homeostasis to 3′ UTR-Directed Regulation and Nonsense-Mediated Decay. Cell Rep. 2020, 32, 107895. [Google Scholar] [CrossRef]
- Shen, P.S.; Park, J.; Qin, Y.; Li, X.; Parsawar, K.; Larson, M.H.; Cox, J.; Cheng, Y.; Lambowitz, A.M.; Weissman, J.S.; et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 2015, 347, 75–78. [Google Scholar] [CrossRef]
- Wu, Z.; Tantray, I.; Lim, J.; Chen, S.; Li, Y.; Davis, Z.; Sitron, C.; Dong, J.; Gispert, S.; Auburger, G.; et al. MISTERMINATE Mechanistically Links Mitochondrial Dysfunction with Proteostasis Failure. Mol. Cell 2019, 75, 835–848.e838. [Google Scholar] [CrossRef]
- Gehrke, S.; Wu, Z.; Klinkenberg, M.; Sun, Y.; Auburger, G.; Guo, S.; Lu, B. PINK1 and Parkin control localized translation of respiratory chain component mRNAs on mitochondria outer membrane. Cell Metab. 2015, 21, 95–108. [Google Scholar] [CrossRef]
- Martin, P.B.; Kigoshi-Tansho, Y.; Sher, R.B.; Ravenscroft, G.; Stauffer, J.E.; Kumar, R.; Yonashiro, R.; Müller, T.; Griffith, C.; Allen, W.; et al. NEMF mutations that impair ribosome-associated quality control are associated with neuromuscular disease. Nat. Commun. 2020, 11, 4625. [Google Scholar] [CrossRef] [PubMed]
- Ishimura, R.; Nagy, G.; Dotu, I.; Zhou, H.; Yang, X.L.; Schimmel, P.; Senju, S.; Nishimura, Y.; Chuang, J.H.; Ackerman, S.L. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 2014, 345, 455–459. [Google Scholar] [CrossRef] [PubMed]
- Sudmant, P.H.; Lee, H.; Dominguez, D.; Heiman, M.; Burge, C.B. Widespread Accumulation of Ribosome-Associated Isolated 3′ UTRs in Neuronal Cell Populations of the Aging Brain. Cell Rep. 2018, 25, 2447–2456.e2444. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, Z.; Tantray, I.; Li, Y.; Chen, S.; Dong, J.; Glynn, S.; Vogel, H.; Snyder, M.; Lu, B. Quality-control mechanisms targeting translationally stalled and C-terminally extended poly(GR) associated with ALS/FTD. Proc. Natl. Acad. Sci. USA 2020, 117, 25104–25115. [Google Scholar] [CrossRef] [PubMed]
- Rimal, S.; Li, Y.; Vartak, R.; Geng, J.; Tantray, I.; Li, S.; Huh, S.; Vogel, H.; Glabe, C.; Grinberg, L.T.; et al. Inefficient quality control of ribosome stalling during APP synthesis generates CAT-tailed species that precipitate hallmarks of Alzheimer’s disease. Acta Neuropathol. Commun. 2021, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Tatara, Y.; Yamazaki, H.; Katsuoka, F.; Chiba, M.; Saigusa, D.; Kasai, S.; Nakamura, T.; Inoue, J.; Aoki, Y.; Shoji, M.; et al. Multiomics and artificial intelligence enabled peripheral blood-based prediction of amnestic mild cognitive impairment. Curr. Res. Transl. Med. 2023, 71, 103367. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, V.; Romani, M.; Mouchiroud, L.; Beck, J.S.; Zhang, H.; D’Amico, D.; Moullan, N.; Potenza, F.; Schmid, A.W.; Rietsch, S.; et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 2017, 552, 187–193. [Google Scholar] [CrossRef]
- Fessler, E.; Eckl, E.M.; Schmitt, S.; Mancilla, I.A.; Meyer-Bender, M.F.; Hanf, M.; Philippou-Massier, J.; Krebs, S.; Zischka, H.; Jae, L.T. A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 2020, 579, 433–437. [Google Scholar] [CrossRef]
- Guo, X.; Aviles, G.; Liu, Y.; Tian, R.; Unger, B.A.; Lin, Y.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 2020, 579, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Fessler, E.; Krumwiede, L.; Jae, L.T. DELE1 tracks perturbed protein import and processing in human mitochondria. Nat. Commun. 2022, 13, 1853. [Google Scholar] [CrossRef] [PubMed]
- Sekine, Y.; Houston, R.; Eckl, E.M.; Fessler, E.; Jae, L.T.; Narendra, D.P.; Sekine, S. A mitochondrial iron-responsive pathway regulated by DELE1. Mol. Cell 2023, 83, 2059–2076. [Google Scholar] [CrossRef]
- Wang, S.F.; Chen, M.S.; Chou, Y.C.; Ueng, Y.F.; Yin, P.H.; Yeh, T.S.; Lee, H.C. Mitochondrial dysfunction enhances cisplatin resistance in human gastric cancer cells via the ROS-activated GCN2-eIF2alpha-ATF4-xCT pathway. Oncotarget 2016, 7, 74132–74151. [Google Scholar] [CrossRef]
- Baker, B.M.; Nargund, A.M.; Sun, T.; Haynes, C.M. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 2012, 8, e1002760. [Google Scholar] [CrossRef]
- Topf, U.; Suppanz, I.; Samluk, L.; Wrobel, L.; Boser, A.; Sakowska, P.; Knapp, B.; Pietrzyk, M.K.; Chacinska, A.; Warscheid, B. Quantitative proteomics identifies redox switches for global translation modulation by mitochondrially produced reactive oxygen species. Nat. Commun. 2018, 9, 324. [Google Scholar] [CrossRef]
| RWD-Domain-Containing Proteins | Gene Symbol | Amino Acids | RWD Domain Region |
|---|---|---|---|
| eIF-2-alpha kinase GCN2 | EIF2AK4 (GCN2) | 1649 | 25–137 |
| Protein IMPACT | IMPACT | 320 | 14–116 |
| Ring finger protein 14 | RNF14 | 474 | 11–137 |
| Ring finger protein 25 | RNF25 | 459 | 18–128 |
| RWD domain-containing protein 1 | RWDD1 (DFRP2) | 243 | 10–114 |
| RWD domain-containing protein 2A | RWDD2A | 292 | 14–134 |
| RWD domain-containing protein 2B | RWDD2B | 319 | 41–165 |
| RWD domain-containing protein 3 | RWDD3 (RESUME) | 267 | 7–114 |
| RWD domain-containing protein 4 | RWDD4 | 188 | 9–111 |
| WD repeat-containing protein 59 | WDR59 | 974 | 393–494 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatara, Y.; Kasai, S.; Kokubu, D.; Tsujita, T.; Mimura, J.; Itoh, K. Emerging Role of GCN1 in Disease and Homeostasis. Int. J. Mol. Sci. 2024, 25, 2998. https://doi.org/10.3390/ijms25052998
Tatara Y, Kasai S, Kokubu D, Tsujita T, Mimura J, Itoh K. Emerging Role of GCN1 in Disease and Homeostasis. International Journal of Molecular Sciences. 2024; 25(5):2998. https://doi.org/10.3390/ijms25052998
Chicago/Turabian StyleTatara, Yota, Shuya Kasai, Daichi Kokubu, Tadayuki Tsujita, Junsei Mimura, and Ken Itoh. 2024. "Emerging Role of GCN1 in Disease and Homeostasis" International Journal of Molecular Sciences 25, no. 5: 2998. https://doi.org/10.3390/ijms25052998
APA StyleTatara, Y., Kasai, S., Kokubu, D., Tsujita, T., Mimura, J., & Itoh, K. (2024). Emerging Role of GCN1 in Disease and Homeostasis. International Journal of Molecular Sciences, 25(5), 2998. https://doi.org/10.3390/ijms25052998







