Orphan Nuclear Receptor Family 4A (NR4A) Members NR4A2 and NR4A3 Selectively Modulate Elements of the Monocyte Response to Buffered Hypercapnia
Abstract
1. Introduction
2. Results
2.1. NR4A2 and NR4A3 Are CO2 Responsive in the Presence of LPS
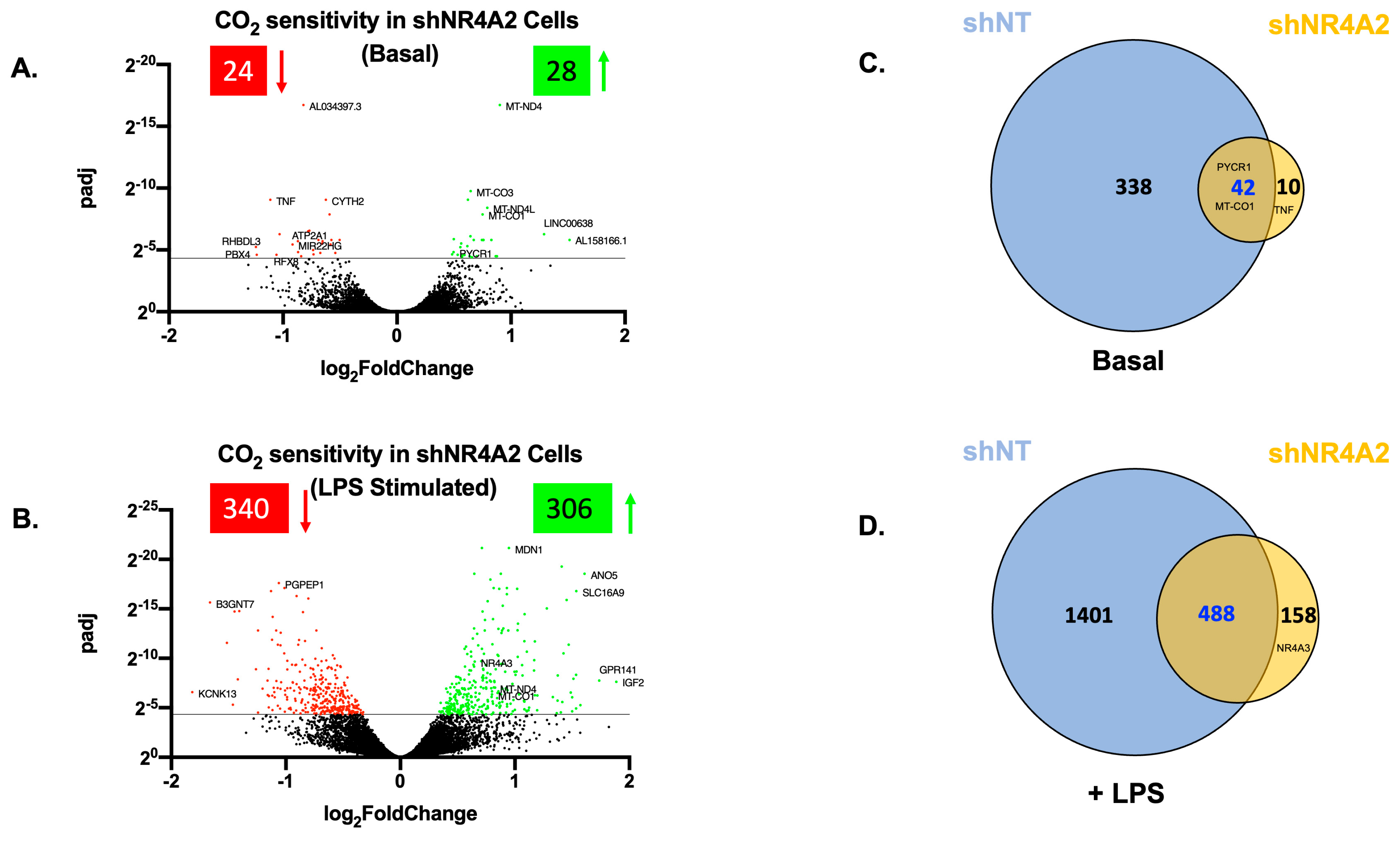
2.2. The Cellular Response to Elevated CO2 Is Modulated by shNR4A2
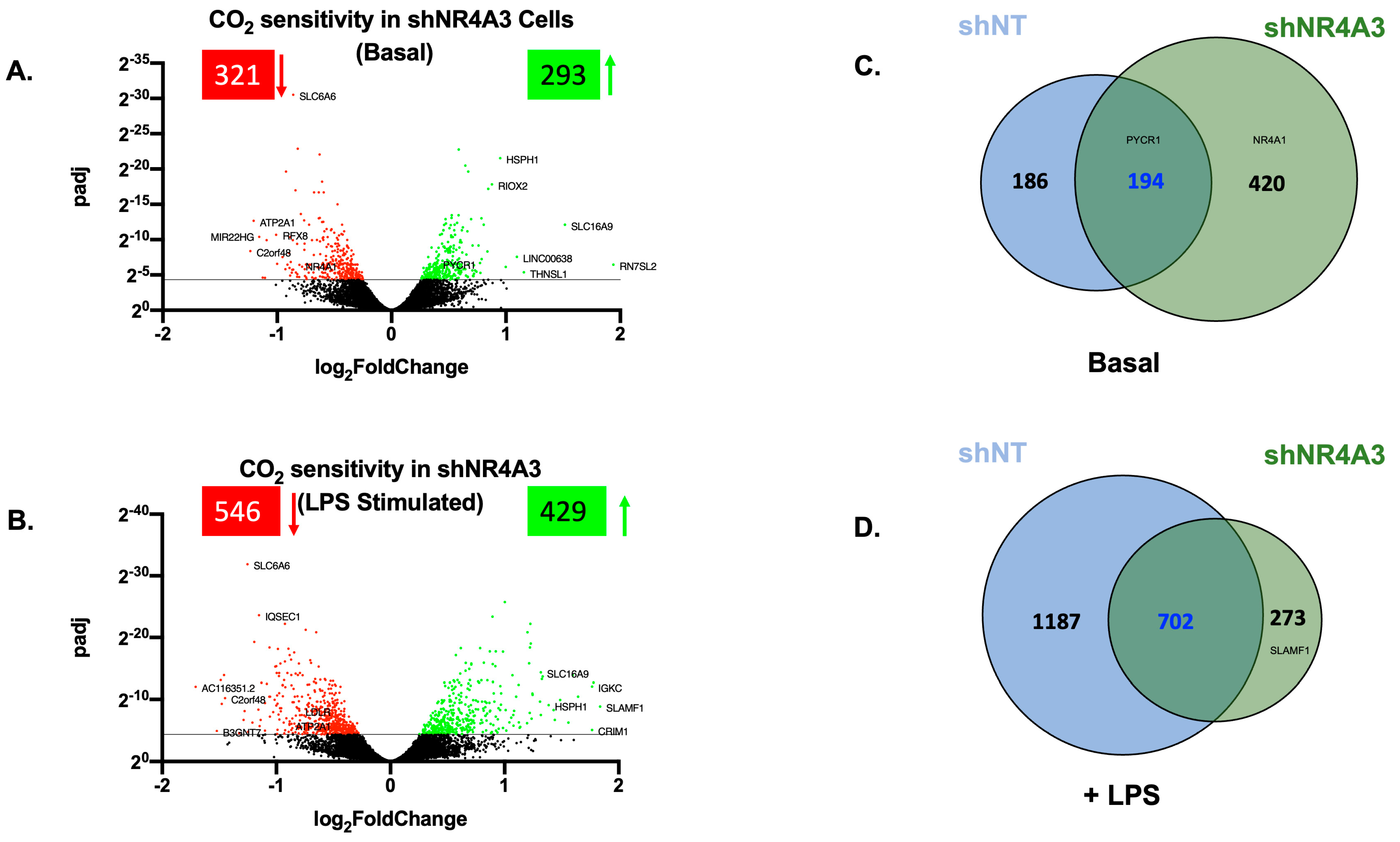
2.3. The Cellular Response to Elevated CO2 Is Modulated by shNR4A3
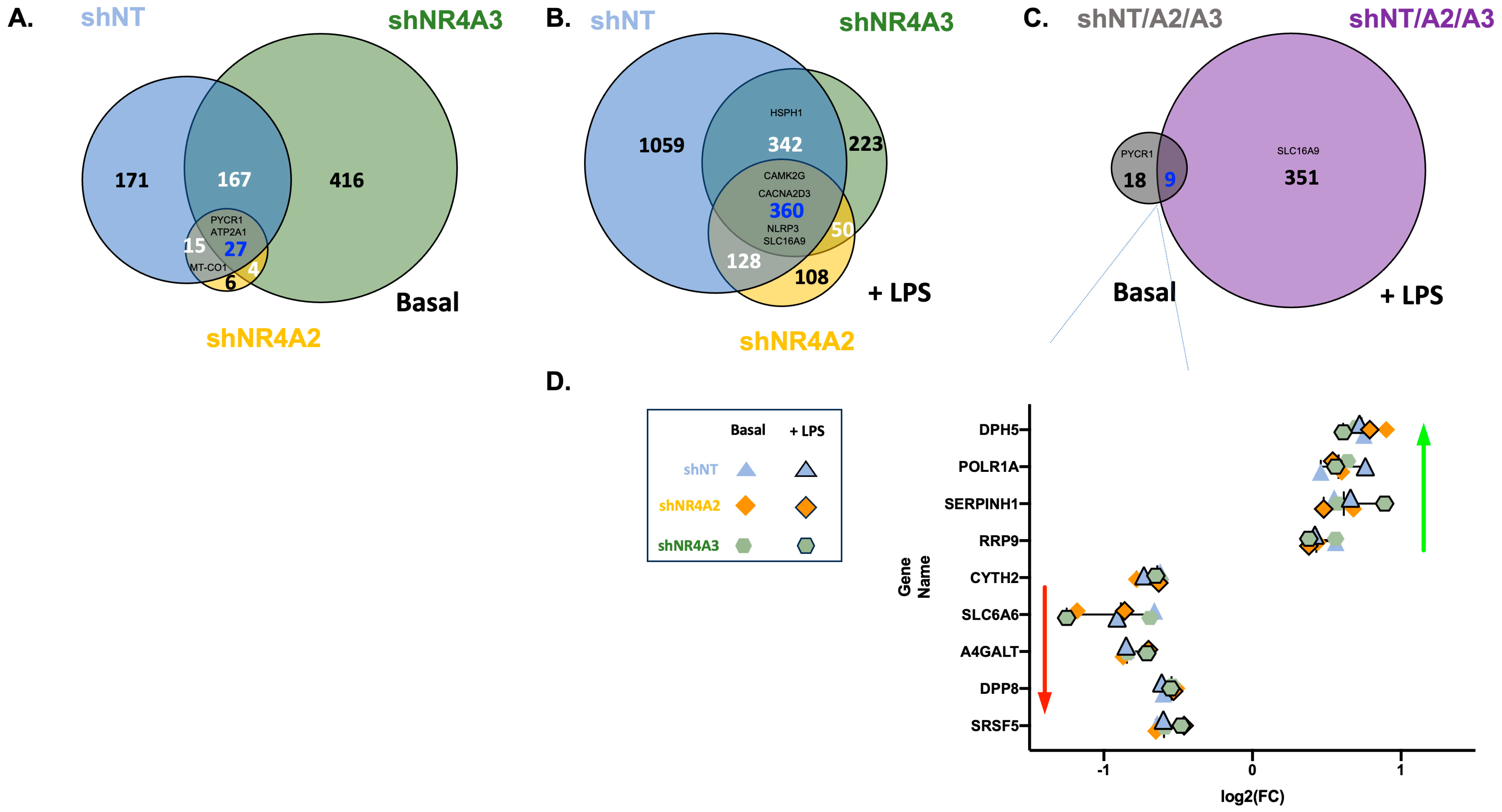
2.4. Comparison of CO2 Responsiveness in Cells Depleted of NR4A2 and NR4A3
Many Genes Are Commonly Regulated by CO2 in shNT, shNR4A2, and shNR4A3 Cells
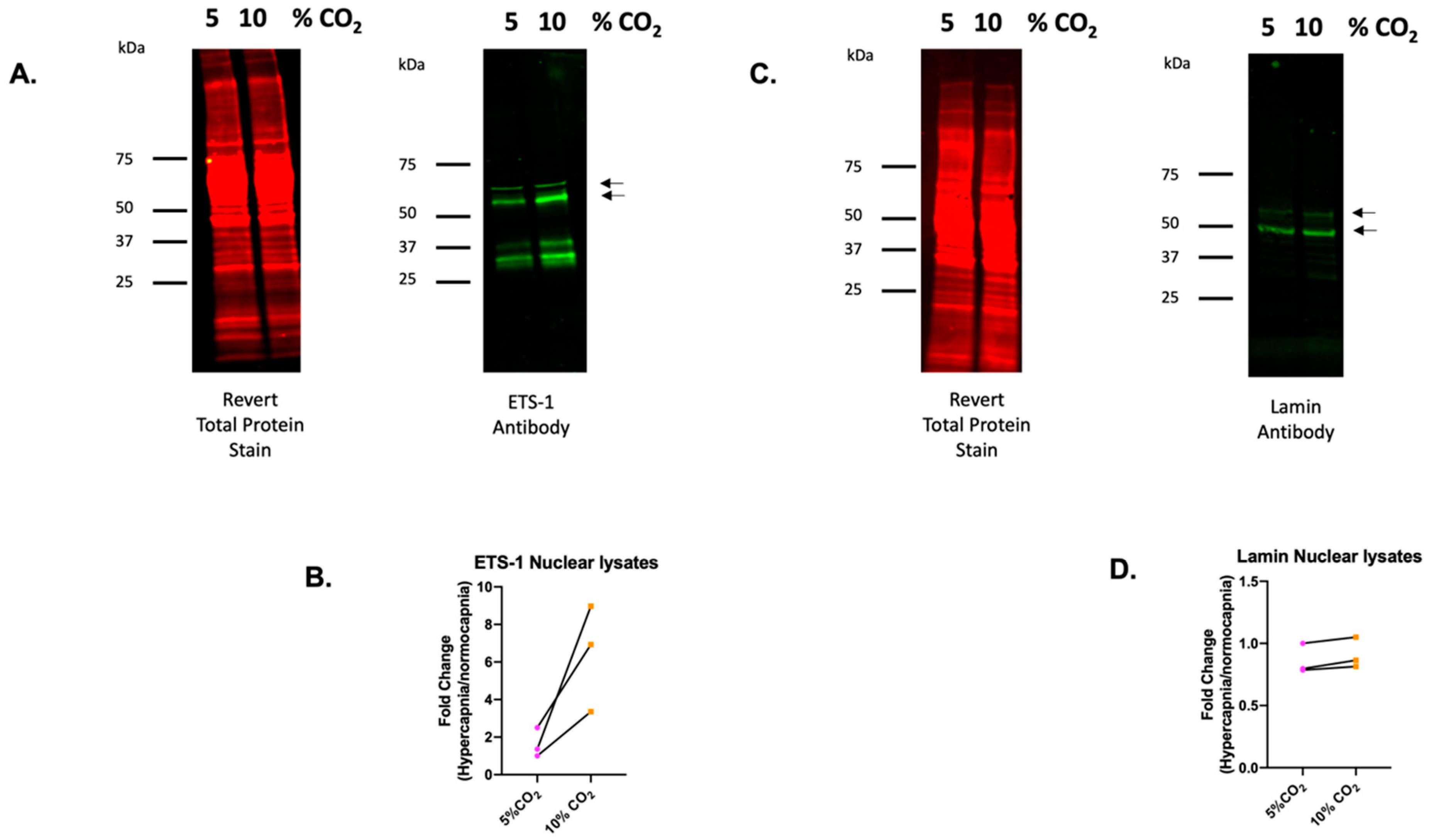
2.5. ETS-1 Is a CO2 Responsive Transcription Factor in Monocytes
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.1.1. THP-1 Monocytes
4.1.2. Hypercapnic Exposures
4.1.3. RNA-Seq
4.2. Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
- NR4A1 F: gttctctggaggtcatccgcaag R: gcagggaccttgagaaggcca
- NR4A3 F: ccaagccttagcctgcctgtc R: agcctgtcccttactctggtgg
- B-actin F: cgacaggatgcagaaggaga R:catctgctggaaggtggaca
4.3. TaqMan Primer Details
- NR4A2: Applied Biosystems Hs01117527_g1 (4331182)
- B-actin: Applied Biosystems (Code:4333762F)
4.4. Western Blot
4.5. Bioinformatic Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nin, N.; Muriel, A.; Penuelas, O.; Brochard, L.; Lorente, J.A.; Ferguson, N.D.; Raymondos, K.; Rios, F.; Violi, D.A.; Thille, A.W.; et al. Severe hypercapnia and outcome of mechanically ventilated patients with moderate or severe acute respiratory distress syndrome. Intensive Care Med. 2017, 43, 200–208. [Google Scholar] [CrossRef]
- Cummins, E.P.; Strowitzki, M.J.; Taylor, C.T. Mechanisms and Consequences of Oxygen and Carbon Dioxide Sensing in Mammals. Physiol. Rev. 2020, 100, 463–488. [Google Scholar] [CrossRef]
- Phelan, D.E.; Mota, C.; Lai, C.; Kierans, S.J.; Cummins, E.P. Carbon dioxide-dependent signal transduction in mammalian systems. Interface Focus. 2021, 11, 20200033. [Google Scholar] [CrossRef]
- Maxwell, M.A.; Muscat, G.E. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal 2006, 4, e002. [Google Scholar] [CrossRef]
- Pearen, M.A.; Muscat, G.E. Minireview: Nuclear hormone receptor 4A signaling: Implications for metabolic disease. Mol. Endocrinol. 2010, 24, 1891–1903. [Google Scholar] [CrossRef]
- Aherne, C.M.; McMorrow, J.; Kane, D.; FitzGerald, O.; Mix, K.S.; Murphy, E.P. Identification of NR4A2 as a transcriptional activator of IL-8 expression in human inflammatory arthritis. Mol. Immunol. 2009, 46, 3345–3357. [Google Scholar] [CrossRef]
- Ralph, J.A.; Ahmed, A.U.; Santos, L.L.; Clark, A.R.; McMorrow, J.; Murphy, E.P.; Morand, E.F. Identification of NURR1 as a mediator of MIF signaling during chronic arthritis: Effects on glucocorticoid-induced MKP1. Am. J. Pathol. 2010, 177, 2366–2378. [Google Scholar] [CrossRef]
- Nus, M.; Basatemur, G.; Galan, M.; Cros-Brunso, L.; Zhao, T.X.; Masters, L.; Harrison, J.; Figg, N.; Tsiantoulas, D.; Geissmann, F.; et al. NR4A1 Deletion in Marginal Zone B Cells Exacerbates Atherosclerosis in Mice-Brief Report. Arter. Thromb. Vasc. Biol. 2020, 40, 2598–2604. [Google Scholar] [CrossRef]
- Wu, H.; Bi, J.; Peng, Y.; Huo, L.; Yu, X.; Yang, Z.; Zhou, Y.; Qin, L.; Xu, Y.; Liao, L.; et al. Nuclear receptor NR4A1 is a tumor suppressor down-regulated in triple-negative breast cancer. Oncotarget 2017, 8, 54364–54377. [Google Scholar] [CrossRef]
- Crean, D.; Murphy, E.P. Targeting NR4A Nuclear Receptors to Control Stromal Cell Inflammation, Metabolism, Angiogenesis, and Tumorigenesis. Front. Cell Dev. Biol. 2021, 9, 589770. [Google Scholar] [CrossRef]
- Safe, S.; Jin, U.H.; Morpurgo, B.; Abudayyeh, A.; Singh, M.; Tjalkens, R.B. Nuclear receptor 4A (NR4A) family—Orphans no more. J. Steroid Biochem. Mol. Biol. 2016, 157, 48–60. [Google Scholar] [CrossRef]
- Mohankumar, K.; Wright, G.; Kumaravel, S.; Shrestha, R.; Zhang, L.; Abdelrahim, M.; Chapkin, R.S.; Safe, S. Bis-indole-derived NR4A1 antagonists inhibit colon tumor and splenic growth and T-cell exhaustion. Cancer Immunol. Immunother. 2023, 72, 3985–3999. [Google Scholar] [CrossRef]
- Zhao, Y.; Bruemmer, D. NR4A orphan nuclear receptors: Transcriptional regulators of gene expression in metabolism and vascular biology. Arter. Thromb. Vasc. Biol. 2010, 30, 1535–1541. [Google Scholar] [CrossRef]
- Rodriguez-Calvo, R.; Tajes, M.; Vazquez-Carrera, M. The NR4A subfamily of nuclear receptors: Potential new therapeutic targets for the treatment of inflammatory diseases. Expert. Opin. Ther. Targets 2017, 21, 291–304. [Google Scholar] [CrossRef]
- Phelan, D.E.; Shigemura, M.; Aldhafiri, S.; Mota, C.; Hall, T.J.; Sznajder, J.I.; Murphy, E.P.; Crean, D.; Cummins, E.P. Transcriptional Profiling of Monocytes Deficient in Nuclear Orphan Receptors NR4A2 and NR4A3 Reveals Distinct Signalling Roles Related to Antigen Presentation and Viral Response. Front. Immunol. 2021, 12, 676644. [Google Scholar] [CrossRef]
- Sharabi, K.; Hurwitz, A.; Simon, A.J.; Beitel, G.J.; Morimoto, R.I.; Rechavi, G.; Sznajder, J.I.; Gruenbaum, Y. Elevated CO2 levels affect development, motility, and fertility and extend life span in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2009, 106, 4024–4029. [Google Scholar] [CrossRef]
- Gwozdzinska, P.; Buchbinder, B.A.; Mayer, K.; Herold, S.; Morty, R.E.; Seeger, W.; Vadasz, I. Hypercapnia Impairs ENaC Cell Surface Stability by Promoting Phosphorylation, Polyubiquitination and Endocytosis of beta-ENaC in a Human Alveolar Epithelial Cell Line. Front. Immunol. 2017, 8, 591. [Google Scholar] [CrossRef] [PubMed]
- Gabrielli, N.M.; Mazzocchi, L.C.; Kryvenko, V.; Tello, K.; Herold, S.; Morty, R.E.; Grimminger, F.; Dada, L.A.; Seeger, W.; Sznajder, J.I.; et al. TRAF2 Is a Novel Ubiquitin E3 Ligase for the Na,K-ATPase beta-Subunit That Drives Alveolar Epithelial Dysfunction in Hypercapnia. Front. Cell Dev. Biol. 2021, 9, 689983. [Google Scholar] [CrossRef] [PubMed]
- Casalino-Matsuda, S.M.; Nair, A.; Beitel, G.J.; Gates, K.L.; Sporn, P.H. Hypercapnia Inhibits Autophagy and Bacterial Killing in Human Macrophages by Increasing Expression of Bcl-2 and Bcl-xL. J. Immunol. 2015, 194, 5388–5396. [Google Scholar] [CrossRef] [PubMed]
- Casalino-Matsuda, S.M.; Berdnikovs, S.; Wang, N.; Nair, A.; Gates, K.L.; Beitel, G.J.; Sporn, P.H.S. Hypercapnia selectively modulates LPS-induced changes in innate immune and DNA replication-related gene transcription in the macrophage. Interface Focus. 2021, 11, 20200039. [Google Scholar] [CrossRef] [PubMed]
- Linthwaite, V.L.; Pawloski, W.; Pegg, H.B.; Townsend, P.D.; Thomas, M.J.; So, V.K.H.; Brown, A.P.; Hodgson, D.R.W.; Lorimer, G.H.; Fushman, D.; et al. Ubiquitin is a carbon dioxide-binding protein. Sci. Adv. 2021, 7, eabi5507. [Google Scholar] [CrossRef]
- Keogh, C.E.; Scholz, C.C.; Rodriguez, J.; Selfridge, A.C.; von Kriegsheim, A.; Cummins, E.P. Carbon dioxide-dependent regulation of NF-kB family members RelB and p100 gives molecular insight into CO2-dependent immune regulation. J. Biol. Chem. 2017, 292, 11561–11571. [Google Scholar] [CrossRef]
- Masterson, C.; O’Toole, D.; Leo, A.; McHale, P.; Horie, S.; Devaney, J.; Laffey, J.G. Effects and Mechanisms by Which Hypercapnic Acidosis Inhibits Sepsis-Induced Canonical Nuclear Factor-kappaB Signaling in the Lung. Crit. Care Med. 2016, 44, e207–e217. [Google Scholar] [CrossRef]
- Saijo, K.; Winner, B.; Carson, C.T.; Collier, J.G.; Boyer, L.; Rosenfeld, M.G.; Gage, F.H.; Glass, C.K. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell 2009, 137, 47–59. [Google Scholar] [CrossRef]
- Crean, D.; Cummins, E.P.; Bahar, B.; Mohan, H.; McMorrow, J.P.; Murphy, E.P. Adenosine Modulates NR4A Orphan Nuclear Receptors To Attenuate Hyperinflammatory Responses in Monocytic Cells. J. Immunol. 2015, 195, 1436–1448. [Google Scholar] [CrossRef]
- Phelan, D.E.; Mota, C.; Strowitzki, M.J.; Shigemura, M.; Sznajder, J.I.; Crowe, L.; Masterson, J.C.; Hayes, S.E.; Reddan, B.; Yin, X.; et al. Hypercapnia alters mitochondrial gene expression and acylcarnitine production in monocytes. Immunol. Cell Biol. 2023, 101, 556–577. [Google Scholar] [CrossRef]
- Pei, L.; Castrillo, A.; Chen, M.; Hoffmann, A.; Tontonoz, P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 2005, 280, 29256–29262. [Google Scholar] [CrossRef]
- McEvoy, C.; de Gaetano, M.; Giffney, H.E.; Bahar, B.; Cummins, E.P.; Brennan, E.P.; Barry, M.; Belton, O.; Godson, C.G.; Murphy, E.P.; et al. NR4A Receptors Differentially Regulate NF-kB Signaling in Myeloid Cells. Front. Immunol. 2017, 8, 7. [Google Scholar] [CrossRef]
- Oliver, K.M.; Lenihan, C.R.; Bruning, U.; Cheong, A.; Laffey, J.G.; McLoughlin, P.; Taylor, C.T.; Cummins, E.P. Hypercapnia induces cleavage and nuclear localization of RelB protein, giving insight into CO2 sensing and signaling. J. Biol. Chem. 2012, 287, 14004–14011. [Google Scholar] [CrossRef]
- Herring, J.A.; Elison, W.S.; Tessem, J.S. Function of Nr4a Orphan Nuclear Receptors in Proliferation, Apoptosis and Fuel Utilization Across Tissues. Cells 2019, 8, 1373. [Google Scholar] [CrossRef]
- Koenis, D.S.; Medzikovic, L.; van Loenen, P.B.; van Weeghel, M.; Huveneers, S.; Vos, M.; Evers-van Gogh, I.J.; Van den Bossche, J.; Speijer, D.; Kim, Y.; et al. Nuclear Receptor Nur77 Limits the Macrophage Inflammatory Response through Transcriptional Reprogramming of Mitochondrial Metabolism. Cell Rep. 2018, 24, 2127–2140.e7. [Google Scholar] [CrossRef] [PubMed]
- Paez, H.G.; Ferrandi, P.J.; Pitzer, C.R.; Mohamed, J.S.; Alway, S.E. Loss of NOR-1 represses muscle metabolism through mTORC1-mediated signaling and mitochondrial gene expression in C2C12 myotubes. FASEB J. 2023, 37, e23050. [Google Scholar] [CrossRef] [PubMed]
- Murshid, A.; Gong, J.; Calderwood, S.K. The role of heat shock proteins in antigen cross presentation. Front. Immunol. 2012, 3, 63. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Song, D.; Li, H.; He, M.L. Stress proteins: The biological functions in virus infection, present and challenges for target-based antiviral drug development. Signal Transduct. Target. Ther. 2020, 5, 125. [Google Scholar] [CrossRef]
- Lu, Z.; Casalino-Matsuda, S.M.; Nair, A.; Buchbinder, A.; Budinger, G.R.S.; Sporn, P.H.S.; Gates, K.L. A role for heat shock factor 1 in hypercapnia-induced inhibition of inflammatory cytokine expression. FASEB J. 2018, 32, 3614–3622. [Google Scholar] [CrossRef] [PubMed]
- Malewicz, M.; Kadkhodaei, B.; Kee, N.; Volakakis, N.; Hellman, U.; Viktorsson, K.; Leung, C.Y.; Chen, B.; Lewensohn, R.; van Gent, D.C.; et al. Essential role for DNA-PK-mediated phosphorylation of NR4A nuclear orphan receptors in DNA double-strand break repair. Genes Dev. 2011, 25, 2031–2040. [Google Scholar] [CrossRef] [PubMed]
- Selfridge, A.C.; Cavadas, M.A.; Scholz, C.C.; Campbell, E.L.; Welch, L.C.; Lecuona, E.; Colgan, S.P.; Barrett, K.E.; Sporn, P.H.; Sznajder, J.I.; et al. Hypercapnia Suppresses the HIF-dependent Adaptive Response to Hypoxia. J. Biol. Chem. 2016, 291, 11800–11808. [Google Scholar] [CrossRef]
- Townsend, P.; Holliday, P.; Fenyk, S.; Hess, K.; Gray, M.; Hodgson, D.; Cann, M. Stimulation of mammalian G-protein-responsive adenylyl cyclases by carbon dioxide. J. Biol. Chem. 2009, 284, 784–791. [Google Scholar] [CrossRef]
- Jaitovich, A.; Angulo, M.; Lecuona, E.; Dada, L.A.; Welch, L.C.; Cheng, Y.; Gusarova, G.; Ceco, E.; Liu, C.; Shigemura, M.; et al. High CO2 levels cause skeletal muscle atrophy via AMP-activated kinase (AMPK), FoxO3a protein, and muscle-specific Ring finger protein 1 (MuRF1). J. Biol. Chem. 2015, 290, 9183–9194. [Google Scholar] [CrossRef]
- Otulakowski, G.; Engelberts, D.; Arima, H.; Hirate, H.; Bayir, H.; Post, M.; Kavanagh, B.P. alpha-Tocopherol transfer protein mediates protective hypercapnia in murine ventilator-induced lung injury. Thorax 2017, 72, 538–549. [Google Scholar] [CrossRef]
- Bolshette, N.; Ezagouri, S.; Dandavate, V.; Karavaeva, I.; Golik, M.; Wang, H.; Espenshade, P.J.; Osborne, T.F.; Han, X.; Asher, G. Carbon dioxide regulates cholesterol levels through SREBP2. PLoS Biol. 2023, 21, e3002367. [Google Scholar] [CrossRef]
- Okuda, T.; Nishimura, M.; Nakao, M.; Fujita, Y. RUNX1/AML1: A central player in hematopoiesis. Int. J. Hematol. 2001, 74, 252–257. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhou, A.; Li, Q. Whole transcriptome analyis of human lung tissue to identify COPD-associated genes. Genomics 2020, 112, 3135–3141. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.S.; Holzner, M.; Weng, M.; Srivastava, Y.; Jauch, R. SOX17 in cellular reprogramming and cancer. Semin. Cancer Biol. 2020, 67 Pt 1, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Sangam, S.; Sun, X.; Schwantes-An, T.H.; Yegambaram, M.; Lu, Q.; Shi, Y.; Cook, T.; Fisher, A.; Frump, A.L.; Coleman, A.; et al. SOX17 Deficiency Mediates Pulmonary Hypertension: At the Crossroads of Sex, Metabolism, and Genetics. Am. J. Respir. Crit. Care Med. 2023, 207, 1055–1069. [Google Scholar] [CrossRef]
- Stanulovic, V.S.; Cauchy, P.; Assi, S.A.; Hoogenkamp, M. LMO2 is required for TAL1 DNA binding activity and initiation of definitive haematopoiesis at the haemangioblast stage. Nucleic Acids Res. 2017, 45, 9874–9888. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.Y.; Tan, Y.; Luo, X.J.; Wu, J.F.; Ni, Y.R. The roles of ETS transcription factors in liver fibrosis. Hum. Cell 2023, 36, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.Y.; Sieweke, M.; Ogawa, E.; Wee, H.J.; Englmeier, U.; Graf, T.; Ito, Y. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 1999, 18, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.R.; Kinston, S.; Knezevic, K.; de Bruijn, M.F.; Wilson, N.; Nottingham, W.T.; Peitz, M.; Edenhofer, F.; Pimanda, J.E.; Ottersbach, K.; et al. Runx genes are direct targets of Scl/Tal1 in the yolk sac and fetal liver. Blood 2008, 111, 3005–3014. [Google Scholar] [CrossRef]
- Laitem, C.; Leprivier, G.; Choul-Li, S.; Begue, A.; Monte, D.; Larsimont, D.; Dumont, P.; Duterque-Coquillaud, M.; Aumercier, M. Ets-1 p27: A novel Ets-1 isoform with dominant-negative effects on the transcriptional properties and the subcellular localization of Ets-1 p51. Oncogene 2009, 28, 2087–2099. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Shigemura, M.; Lecuona, E.; Angulo, M.; Dada, L.A.; Edwards, M.B.; Welch, L.C.; Casalino-Matsuda, S.M.; Sporn, P.H.S.; Vadasz, I.; Helenius, I.T.; et al. Elevated CO2 regulates the Wnt signaling pathway in mammals, Drosophila melanogaster and Caenorhabditis elegans. Sci. Rep. 2019, 9, 18251. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phelan, D.E.; Reddan, B.; Shigemura, M.; Sznajder, J.I.; Crean, D.; Cummins, E.P. Orphan Nuclear Receptor Family 4A (NR4A) Members NR4A2 and NR4A3 Selectively Modulate Elements of the Monocyte Response to Buffered Hypercapnia. Int. J. Mol. Sci. 2024, 25, 2852. https://doi.org/10.3390/ijms25052852
Phelan DE, Reddan B, Shigemura M, Sznajder JI, Crean D, Cummins EP. Orphan Nuclear Receptor Family 4A (NR4A) Members NR4A2 and NR4A3 Selectively Modulate Elements of the Monocyte Response to Buffered Hypercapnia. International Journal of Molecular Sciences. 2024; 25(5):2852. https://doi.org/10.3390/ijms25052852
Chicago/Turabian StylePhelan, David E., Ben Reddan, Masahiko Shigemura, Jacob I. Sznajder, Daniel Crean, and Eoin P. Cummins. 2024. "Orphan Nuclear Receptor Family 4A (NR4A) Members NR4A2 and NR4A3 Selectively Modulate Elements of the Monocyte Response to Buffered Hypercapnia" International Journal of Molecular Sciences 25, no. 5: 2852. https://doi.org/10.3390/ijms25052852
APA StylePhelan, D. E., Reddan, B., Shigemura, M., Sznajder, J. I., Crean, D., & Cummins, E. P. (2024). Orphan Nuclear Receptor Family 4A (NR4A) Members NR4A2 and NR4A3 Selectively Modulate Elements of the Monocyte Response to Buffered Hypercapnia. International Journal of Molecular Sciences, 25(5), 2852. https://doi.org/10.3390/ijms25052852




