Abstract
MADS-box transcription factors have crucial functions in numerous physiological and biochemical processes during plant growth and development. Previous studies have reported that two MADS-box genes, SlMBP21 and SlMADS1, play important regulatory roles in the sepal development of tomato, respectively. However, the functional relationships between these two genes are still unknown. In order to investigate this, we simultaneously studied these two genes in tomato. Phylogenetic analysis showed that they were classified into the same branch of the SEPALLATA (SEP) clade. qRT-PCR displayed that both SlMBP21 and SlMADS1 transcripts are preferentially accumulated in sepals, and are increased with flower development. During sepal development, SlMBP21 is increased but SlMADS1 is decreased. Using the RNAi, tomato plants with reduced SlMBP21 mRNA generated enlarged and fused sepals, while simultaneous inhibition of SlMBP21 and SlMADS1 led to larger (longer and wider) and fused sepals than that in SlMBP21-RNAi lines. qRT-PCR results exhibited that the transcripts of genes relating to sepal development, ethylene, auxin and cell expansion were dramatically changed in SlMBP21-RNAi sepals, especially in SlMBP21-SlMADS1-RNAi sepals. Yeast two-hybrid assay displayed that SlMBP21 can interact with SlMBP21, SlAP2a, TAGL1 and RIN, and SlMADS1 can interact with SlAP2a and RIN, respectively. In conclusion, SlMBP21 and SlMADS1 cooperatively regulate sepal development in tomato by impacting the expression or activities of other related regulators or via interactions with other regulatory proteins.
1. Introduction
MADS-box genes encode an important family of transcription factors that widely distributed in eukaryotes (i.e., plants, animals and fungi) [1]. In plants, this large gene family is divided into two super clades, the type I and the type II, on the basis of their evolutionary origin [2,3]. The MADS-box genes from type I lineage encode the SRF-like domain proteins that can be further classified into three subfamilies (Mα, Mβ, and Mγ) [4]. While the MADS-box genes of type II lineage which cover the most well-known MADS genes encode plant-specific MIKC-type proteins and MEF2-like proteins [5,6,7]. Type I MADS-box proteins only contain the MADS domain, whereas type II MADS-box proteins harbor four recognized domains (MADS (M), intervening (I), keratin-like (K) and C-terminal (C) domains) [5,8,9,10]. In the type II lineage, MIKC-type MADS-box proteins can be further classified into MIKC* and MIKCC groups based on their different encoded I and K domains [11].
In plants, the most well-identified MADS-box transcription factors are the members of the MIKCC group. They play essential roles in numerous critical biological processes of plant growth and development, including abiotic stress response, vegetative growth and reproductive development [12,13,14]. In some plant species, the MADS-box genes have been proved to be the important regulators for plants to adapt challenging environmental conditions. For example, OsMADS26 controls various stress responses by regulating the expression of biosynthesis genes related to JA(jasmonic acid), SA (salicylic acid), ET (ethylene) and ROS (reactive oxygen species) in rice [15]. Overexpression of SlMBP11 in tomato enhanced tolerance to salt stress by reducing the relative electrolyte leakage and MDA content, and increasing the water and chlorophyll content in transgenic plants [16]. SlMBP8 function as a negative stress-responsive transcription factor in the drought and high-salinity stress signaling pathways by regulating multiple stress-related genes [17].
In addition to the abiotic stress response, MADS-box transcription factors also function as the significant regulators in plant vegetative growth. Overexpression of SlFYFL in tomato delayed leaf and sepal senescence and fruit ripening, and affected abscission zone development [18]. Overexpressing the dominant negative form of SlMBP20 resulted in leaf simplification and partly repressed the increase in tomato leaf complexity [19]. Up-regulation of SlMBP11 in tomato led to decreased internode length, leaf size and plant height; increased node and leaf number; and excessively more branches were generated in each leaf axil [20]. In Arabidopsis thaliana, agl21 mutants showed fewer and shorter lateral roots, while AtAGL21 overexpression plants produced more and longer lateral roots; AtAGL21 was found to positively regulate auxin accumulation in lateral root primordia and lateral roots by enhancing local auxin biosynthesis, thus stimulating lateral root initiation and growth [21]. Similarly, OsMADS25 was reported to regulate root system development via auxin signaling in rice [22]. In tomato, four MADS-box genes, MC (MACROCALYX), JOINTLESS, SlFYFL, SlMBP21, are involved in regulating abscission zone development [18,23,24,25].
Nowadays, numerous MADS-box genes have been proved to play crucial roles in regulating inflorescence development, such as PAP2 (PANICLE PHYTOMER2), OsMADS14, OsMADS15, and OsMADS18 in rice [26], MC (MACROCALYX), JOINTLESS, SlMBP21, SlCMB1, SlMADS1 in tomato [27,28,29,30], and four SEPALLATA (SEP) MADS-box genes (SEP1–4) in Arabidopsis [31,32], and so on. These MADS-box genes generally regulate inflorescence development by regulating the expression of genes related to floral meristem development or hormone synthesis. Moreover, MADS-box transcription factors act as the important regulators in plant parthenocarpy, cuticle development, flowering time, fruit development and ripening, embryo and seed development and floral organ identity determination, etc. For instance, down-regulation of the MADS-box gene (TM29) or mutation of SlAGAMOUS-LIKE 6 (SlAGL6) in tomato lead to parthenocarpic fruit development [33,34]; tomato MADS-box gene TAGL1 (TOMATO AGAMOUS-LIKE1) regulates cuticle development by participating in the transcriptional control of cuticle development mediating the biosynthesis of cuticle components [35,36]. Five MADS-box genes, SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1), AGL24 (AGAMOUS-LIKE GENE 24), FLC (FLOWERING LOCUS C), AP1 (APETALA1) and SVP (SHORT VEGETATIVE PHASE), are required for flowering time in Arabidopsis [37,38,39,40,41,42]. AtAGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization [42]. The rice plants with increased OsMADS18 levels or overexpressing the MADS-box gene UNS (UNSHAVEN) in petunia results in an advanced flowering time, respectively [43,44]. MADS-box genes also act as vital regulators in regulating fruit ripening, and these genes generally function at key control sites in the regulatory network of fruit ripening or ethylene synthesis. For example, RIN, a tomato MADS-box transcription factor, is a significant regulator of fruit ripening, in mutant inhibited fruit ripening with a failure to fruit ripening, including the reduced respiratory climacteric, associated ethylene evolution, and carotenoid accumulation, suppressed softening and production of flavor compounds [29]. In addition to RIN, other MADS-box genes are also involved in regulating tomato fruit ripening, such as FUL1(FRUITFULL1), FUL2 (SlMBP7), TAGL1, SlMADS1 and SlFYFL [18,45,46,47].
The regulation of floral organ identity is the most notable role for MADS-box genes in flowering plants. Numerous MADS-box genes are identified as five classes (A, B, C, D and E) following their functions in floral organ identity according to the ABCDE model. As described in this model, A-class genes are required for sepal and petal development, such as APETALA1 (AP1) and AP2 in Arabidopsis [48,49,50], and MC in tomato [29]; B-class genes regulate the development of petal and stamen, such as PISTILLATA (PI) and AP3 in Arabidopsis [51,52], and GLO (GLOBOSA) and DEF (DEFICIENS) in Antirrhinum majus [53]; C-class genes refer to stamen and carpel, containing AGAMOUS (AG) in Arabidopsis [54]; class D genes specify ovule, including FBP7 (FLORAL B IND ING PROTEIN 7) and FBP11 in Petunia [55], and STK (SEEDSTICK), SHATTERPROOF1 (SHP1) and SHP2 in Arabidopsis [56]; and E class genes function in all four whorls of flowers through forming protein complexes with other classes, such as SEP1–4 in Arabidopsis [31,32,57]. Moreover, many other MADS-box transcription factors that are involved in the development of floral organs also have been characterized. For instance, overexpression of SlMBP11 in tomato exhibited shorter style, split ovary, polycarpous fruits and delayed perianth senescence [20], and the suppression of an SEP MADS-box gene, SlCMB1, generates enlarged and fused sepals due to the reduced expression of sepal-related genes (MC, AP2a and GOBLET (GOB)) [30]. Overexpressing TM8 in tomato leads to the anomalous stamens with poorly viable pollen and altered expression of several floral identity genes, among them B-, C- and E-function ones [58]. After the ABCDE model of flower development, the floral quartet model was proposed by Theißen [59]. This model suggests that the identity of four different floral organs was determined by a tetramer of four floral homeotic proteins, respectively. The complex, which is composed of two class A proteins AP1 and two class E proteins SEP (2A + 2SEP), determines sepal identity; a complex harboring one AP1 protein, two class B proteins PI and AP3, and one SEP protein (A + 2B + SEP) specifies petal identity; a complex of one class C protein AG, two class B proteins PI and AP3, and one SEP protein (2B + C+SEP) determines stamen identity; a complex of two AG proteins and two SEP proteins (2C + 2SEP) specifies carpel identity; a complex of one AG protein, one SEP protein and one of each of D class proteins STK and/or SHP (C + 2D + SEP) determines ovule identity [59,60].
SEP MADS-box proteins play fundamental roles in the identity of floral organs and other biological processes. In Arabidopsis, the inner three whorls of floral organs (petal, stamen, and carpel) developed into sepals in sep1sep2sep3 triple mutants, and the indetermination of flower development was observed [32]. While in sep1sep2sep3sep4 quadruple mutants, the four whorls of floral organs were replaced by vegetative leaves, and flower development become more indeterminate [31]. In tomato, six SEP MADS-box transcription factors (TM5, TM29, SlCMB1, SlMBP21, RIN, and SlMADS1) have been identified to regulate different biological processes by protein interaction and/or gene expression regulation [24,29,30,33,47,61,62,63,64]. For instance, SlCMB1 is involved in fruit ripening, inflorescence and sepal development [30,63]; SlMBP21 plays essential roles in the development of an abscission zone, inflorescence, and sepals [24,27,64]; SlMADS1 regulates fruit ripening and inflorescence development [27,47]; and the down-regulation of TM29 results in floral reversion and parthenocarpic fruit development [33]. The sepals is the outermost whorl of floral organs, and protects flower buds and young fruits. Normal sepal development is crucial for successful reproductive development. As the E class genes, the SEP MADS-box transcription factors are involved in the sepal identity in the ABCDE model and the quartet model during floral organ development [60,65]. In Arabidopsis, four SEP MADS-box genes, SEP1–4, cooperatively specify floral organ identity [31,32]. The floral quartet model demonstrates that two class E SEP proteins are involved in specifying sepal identity [60].
In a previous study of our lab, SEP MADS-box gene SlMADS1 (Solyc03g114840) negatively regulates tomato fruit ripening and its transcripts are primary accumulated in sepals [47]. Liu et al. reported that the antisense of SlMADS1 resulted in elongated sepals in transgenic tomato flowers [24], and Soyk et al. reported that the ej2 mutant with the loss of function of EJ2 (i.e., SlMADS1) generates 50% longer sepals than that of WT [27]. Li et al. reported that the other one SEP MADS-box gene, SlMBP21 (Solyc12g038510), regulates sepal size mediated via auxin and ethylene in tomato and it is primary expressed in flowers and sepals [64]. However, the functional connections between SlMBP21 and SlMADS1 are still unknown. So we speculate that SlMBP21 and SlMADS1 may cooperatively regulate sepal development in tomato. In this study, expression pattern analysis showed that the transcripts of SlMBP21 were increased with flower and sepal development, and it was primarily expressed in sepal and pistil. The expression patterns of SlMADS1 was similar to SlMBP21 with the flower development, but SlMADS1 transcripts decreased during sepal development and were primarily accumulated in sepals. This further suggested that both genes may regulate sepal development in tomato. Therefore, it is appealing to study whether or not SlMBP21 and SlMADS1 have cooperative functions in tomato sepal development. Using the RNAi method, tomato plants with reduced SlMBP21 mRNA (SlMBP21-RNAi) generated enlarged and fused sepals, while the simultaneous inhibition of SlMBP21 and SlMADS1 (SlMBP21-SlMADS1-RNAi) led to fused and larger (longer and wider) sepals than that in SlMBP21-RNAi lines. These results suggested that SlMBP21 and SlMADS1 cooperatively play essential roles in sepal development. Then, we performed functional analysis of these two genes through the molecular and phenotypic characterization of single RNAi (SlMBP21-RNAi) and double RNAi (SlMBP21-SlMADS1-RNAi) silencing lines to deeply analyze the molecular mechanisms of the function of SlMBP21 and SlMADS1 in sepal development.
2. Results
2.1. Molecular Characterization of SlMBP21 and SlMADS1
To study the roles of SEP MADS-box proteins played in tomato, we concentrated our work on the two SEP MADS-box genes, SlMBP21 and SlMADS1. Sequence analysis displayed that SlMBP21 harbors a 753 bp ORF encoding a protein with 250 acid residues and this protein has an estimated molecular mass of 28.44 kDa (pI 8.95). SlMADS1 possesses a 741 bp ORF encoding a protein with 246 acid residues, and SlMADS1 has an estimated molecular mass of 28.39 kDa (pI 8.84). Multiple alignment and phylogenetic analysis showed that both SlMBP21 and SlMADS1 proteins are SEP MADS-box proteins with four typical MADS-box domains (i.e., the MADS-box domain, the I domain, and the K-box domain) and their C-terminuses are highly different from other known MADS-box proteins (Figure 1A). Phylogenetic analysis displayed that these two MADS-box proteins belong to the SEP lineage and on the same branch (Figure 1B). SlMBP21 shares the highest similarity (74.2%) with SlMADS1 than other MADS-box proteins, suggesting that they may play similar functions in tomato development [63]. Moreover, SlMBP21 shares 63.7% similarity with SlCMB1 (a regulator of sepal development [30]), and SlMADS1 shares 66.8% similarity with SlCMB1 [30,63].
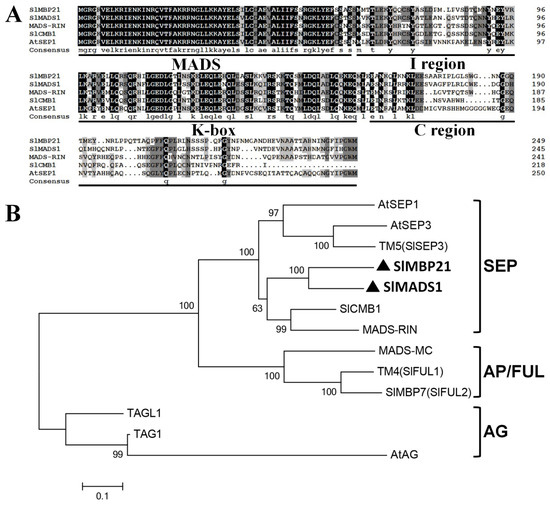
Figure 1.
Sequence and phylogenic analysis of SlMBP21 and SlMADS1. (A) Multiple sequence alignment of SlMBP21 and SlMADS1 with other four known MADS-box proteins. The black part are the identical amino acids, and the gray are the similar amino acids. (The dark grey indicates the homology level between greater than or equal to 75 % and less than 100 %, and the light grey indicates the homology level between greater than or equal to 50 % and less than 75 %). The MADS-box, I region, K-box, and C region are identified. (B) Phylogenetic analysis of SlMBP21 and SlMADS1 with other known MADS-box proteins. SlMBP21 and SlMADS1 are highlighted with black triangles and bold fonts. Accession numbers of these proteins are listed as follows: AtSEP1 (AED92207.1), AtSEP2 (AEE73791.1), AtSEP3 (AEE30503.1), MADS-RIN (AF448522), SlMBP21 (NP_001275579), SlMADS1 (NP_001234380), TM5 (MADS5/LeSEP3) (NP_001234384/AY306153), MC (NP_001234665), LeFUL1 (AY098732.1), LeFUL2 (NM_001307938.1), TAG1 (L26295.1), TAGL1 (AY098735.2), AtAG (NP_001328877).
Moreover, promoter analysis showed that three ethylene-responsive elements (ERE motif, ATTTCAAA), one gibberellin-responsive element (P-Box, CCTTTTG) and one auxin-responsive element (TGA-box, TGACGTAA) were observed in the SlMBP21 promoter region (Figure S1), and three ethylene-responsive elements (ERE motif, ATTTCAAA) and three gibberellin-responsive elements (two P-Box, CCTTTTG and one TATC-box, TATCCCA) were found in the SlMADS1 promoter region (Figure S2). All these results indicate that SlMBP21 and SlMADS1 may play significant roles in tomato growth and development via the response to plant hormones.
2.2. Expression Profiles of SlMBP21 and SlMADS1 in Wild-Type Tomato Tissues
To study the underlying roles of SlMBP21 and SlMADS1 in tomato sepal development, we performed qRT-PCR (quantitative real time PCR) to detect the relative transcript levels of SlMBP21 and SlMADS1 in flowers and sepals at different developmental stages and in floral organs using specific primers (Table S2) of these two genes. The results displayed that SlMBP21 was highly expressed in sepals than other tissues and flowers and the transcripts of SlMBP21 increased with flower and sepal development (Figure 2A–C). In floral organs, SlMBP21 was mainly expressed in the sepal, petal and pistil, and lower expression was observed in the stamen (Figure 2D). SlMADS1 had higher expression levels in sepals and flowers and it had similar expression patterns to SlMBP21 with flower development, but the transcript levels of SlMADS1 declined with sepal development (Figure 2E–G). In floral organs, SlMADS1 transcripts were preferentially accumulated in sepals, and lower accumulation was observed in other floral organs (petal, stamen and pistil) (Figure 2H). These results suggest that both SlMBP21 and SlMADS1 may play significant roles in sepal development.
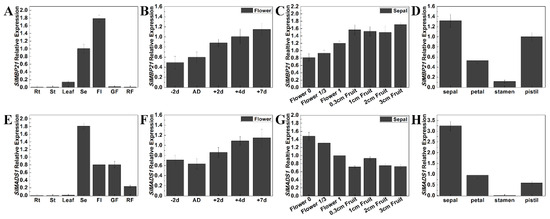
Figure 2.
Expression analysis of SlMBP21 and SlMADS1 in different tissues of AC++ (Mill. cv. Ailsa Craig). (A) Relative expression of SlMBP21 in different tissues of AC. Rt, roots; St, stems; Leaf; Se, sepal; FL, flowers at anthesis; GF, green fruits (25 DPA); RF, red fruits (7 days fruits after breaker). (B) Relative expression of SlMBP21 in the different developmental stages of flowers. -2d, -2day-flowers before anthesis; AD, anthesis day; +2d, +2d-flowers after anthesis; +4d, +4d-flowers after anthesis; +7d, +7day-flowers after anthesis; DPA, days postanthesis. (C) Relative expression of SlMBP21 in the different developmental stages of sepals. Flower 0, sepals of -2d-flowers before anthesis; Flower 1/3, sepals of flowers at anthesis; Flower 1, sepals of 2 days after anthesis; 0.3 cm Fruit, sepals of fruit (the diameter of fruit is 0.3 cm); 1 cm Fruit, sepals of fruit (the diameter of fruit is 1 cm); 2 cm Fruit, sepals of fruit (the diameter of fruit is 2 cm); 3 cm Fruit, sepals of fruit (the diameter of fruit is 3 cm). (D) Relative expression of SlMBP21 in different floral organs of WT. (E) Relative expression of SlMADS1 in different tissues of AC. (F) Relative expression of SlMADS1 in the different developmental stages of flowers. (G) Relative expression of SlMBP21 in the different developmental stages of sepals. (H) Relative expression of SlMADS1 in different floral organs of WT. Error bars indicate SE.
2.3. Generation of SlMBP21-RNAi Lines and SlMBP21-SlMADS1-RNAi Lines
To further investigate the functions of SlMBP21 and SlMADS1 in tomato development, we constructed the single RNAi vector of SlMBP21 (SlMBP21-RNAi) (Figure S3A) and double RNAi vector of SlMBP21 and SlMADS1 (SlMBP21-SlMADS1-RNAi) (Figure S3B). These two RNAi vectors were transferred into wild-type tomato plants to obtain transgenic lines, respectively. In total, we obtained twelve single RNAi lines, in which SlMBP21 was significantly diminished from twelve independent transgenic lines, and three lines (SRNAi02, SRNAi05, and SRNAi08) which showed the lowest transcript levels of SlMBP21 were used for further investigation (Figure 3A). For the double RNAi lines (SlMBP21-SlMADS1-RNAi), we obtained eleven independent transgenic lines, and three lines (DRNAi06, DRNAi07, DRNAi09) which simultaneously showed the lowest transcript levels of SlMBP21 and SlMADS1 were used for further investigation (Figure 3B).
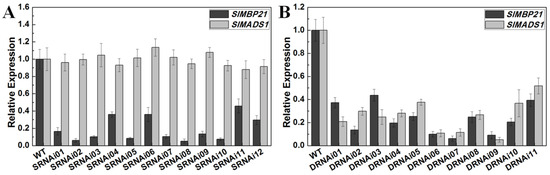
Figure 3.
Relative expression of SlMBP21 and SlMADS1 in sepals of WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines. (A) Relative expression of SlMBP21 and SlMADS1 in sepals of WT and SlMBP21-RNAi lines at 3 DPA (day post-anthesis). (B) Relative expression of SlMBP21 and SlMADS1 in sepals of WT and SlMBP21-SlMADS1-RNAi lines at 3 DPA. Data are the means ± SE of three independent biological replicates.
2.4. Double SlMBP21-SlMADS1-Silenced Lines Generated Larger Sepals Than the Single SlMBP21 Silencing Lines
During the developmental process of tomato plants, the anthesis time was recorded. In addition to abolished development of the tomato flower abscission zone in SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines (Figure 4A,B), we also noticed that the sepals of single SlMBP21 silencing lines grown faster than the wild type. The SlMBP21-RNAi sepals showed larger size than the wild type at the same stage (Figure 3A). Meanwhile, the SlMBP21-SlMADS1-RNAi lines showed faster sepal growth than the single SlMBP21 silencing lines and WT (Figure 3A), and as result, the sepal size of double SlMBP21-SlMADS1 silencing lines was larger than the single SlMBP21 silencing lines at the same stage (Figure 4B,C). Moreover, part of the double RNAi lines, which harbored the strongly simultaneous suppression (more than 90%) of SlMBP21 and SlMADS1, generated larger and leafy sepals than other double RNAi lines which harbored the weak suppression (70–80%) of SlMBP21, and SlMADS1 produced only enlarged but not leafy sepals (Figure 4C). The increased sepal size with the decreased expression levels of these two genes in the double RNAi lines was observed. These results indicated that the sepal size was impacted by different suppression of SlMBP21 and SlMADS1 expression levels.

Figure 4.
Sepal phenotypes in SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines. (A) Sepals of WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines at different developmental stages; −1d, −1-day flowers before anthesis; AD, anthesis day; +3d, +3day-flowers after anthesis; +7d, +7-day flowers after anthesis; DPA, days post-anthesis. (B) Sepals of WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines at 15 DPA; SRNAi, SlMBP21-RNAi lines; DRNAi, SlMBP21-SlMADS1-RNAi lines; DPA, days post-anthesis. (C) Sepals of WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines at 20 DPA; weak, double RNAi lines which harbor weakly simultaneous suppression (70–80%) of SlMBP21 and SlMADS1; strong, double RNAi lines which harbor strongly simultaneous suppression (more than 90%) of SlMBP21 and SlMADS1 generated larger and leafy sepals. Weak and strong representative images of phenotypic classes of sepals found in SlMBP21-SlMADS1-RNAi lines.
In view of the enlarged sepal size in single and double RNAi lines compared with WT, some metrical data of wild-type and transgenic sepals at 3 DPA (day post-anthesis) and 15 DPA after anthesis were obtained. On the third day (3 DPA) after anthesis, the SlMBP21-RNAi sepals were about 10 mm longer than the wild-type, and the double SlMBP21-SlMADS1 silencing sepals were about 12 mm longer than the SlMBP21-RNAi sepals, while the double RNAi sepals were about 22 mm longer than the wild-type ones (Figure 5A). The 3 DPA sepal width of WT, single and double RNAi lines was also measured. The results showed that the SlMBP21-RNAi sepals were about 1.0–1.6 mm wider than WT, and the SlMBP21-SlMADS1 silencing sepals were about 0.8–2.1 mm wider than SlMBP21-RNAi sepals, while the double RNAi sepals were approximate 2.2–3 mm wilder than the wild-type (Figure 5B). Twelve days later (15 DPA), the SlMBP21-RNAi sepals still remained 10 mm longer than the wild-type, and the double SlMBP21-SlMADS1 silencing sepals were approximate 11–18 mm longer than the SlMBP21-RNAi sepals, while the double RNAi sepals were about 20–26 mm longer than WT (Figure 5A). The width of SlMBP21-RNAi sepals was about 1–1.8 mm wider than the wild-type, and the double SlMBP21-SlMADS1 silencing sepals were approximate 1.3–2.1 mm wider than the SlMBP21-RNAi sepals, while the SlMBP21-SlMADS1 silencing sepals were about 2.7–3.1 mm wider than WT (Figure 5B). These results indicate that both SlMBP21 and SlMADS1 synergistically control tomato sepal development.
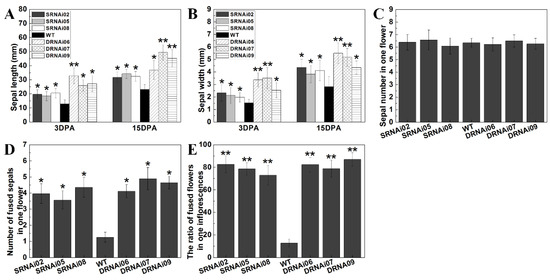
Figure 5.
Statistical analysis of sepal in WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines. (A) The sepal length of WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. (B) The sepal width of WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. (C) The sepal number of one flower in WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. (D) The number of fused sepals in one flower in WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. (E) The ratio of fused flowers in one inflorescence in WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. Data are the means ± SE of three independent biological replicates. The significant differences (p < 0.05, indicated by *) and highly significant difference (p < 0.01, indicated by **) were marked with the asterisks between WT and transgenic lines.
2.5. Both Single SlMBP21 and Double SlMBP21-SlMADS1 Silencing Sepals Fused Together and Generated a Leaf-like Structure
In addition to enlarged sepals, the overwhelming majority of flowers of the single SlMBP21 silencing lines and double SlMBP21-SlMADS1 silencing lines generated fused sepals, in varying degrees, while the WT sepals can split completely with each other (Figure 4A,B). Some sepals of single and double RNAi lines fused together, however, even after the flower opened completely. The transgenic petals failed to unfold normally on account of the constraint of fused sepals. These fused sepals of single SlMBP21 silencing lines and double SlMBP21-SlMADS1 silencing lines could be easily separated by hand. We performed the statistical analysis related to the sepals of WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines, and found that there was no difference in sepal number in one flower among them (Figure 5C), but the number of fused sepals in one flower and the ratio of fused flowers in one inflorescence were markedly higher in the transgenic lines than in WT (Figure 5D,E).
In consideration of the enlarged and fused sepals in single and double RNAi lines, the paraffin section of flowers (3 DPA) and sepals (15 DPA) was analyzed to study the differences among these three lines (WT, single RNAi lines and double RNAi lines). The results showed that the cross-section of the double silencing sepals were the widest, and the sepals of SlMBP21-RNAi lines were wider than WT (Figure 6A). In addition, we also found that the sepals of two kinds of RNAi lines generated a palisade tissue-like structure under the upper and lower epidermis, of which the phenotype was relatively more obvious in the SlMBP21-SlMADS1 silencing sepals than the SlMBP21-RNAi lines (Figure 6A,B). Moreover, based on the cross section of 3 DPA flowers, almost all of the sepals of RNAi lines linked each other with few layers of cells while the wild-type sepals were separated (Figure 6C). These results suggested that SlMBP21 and SlMADS1 are involved in the development of sepal cells.
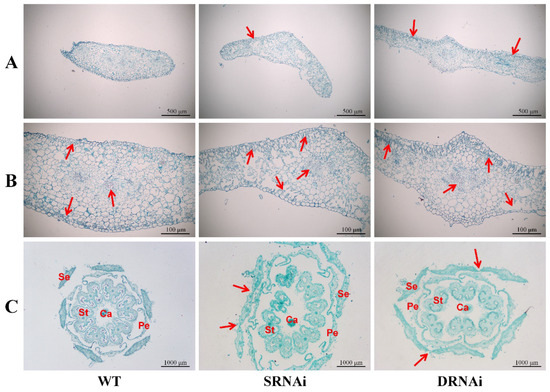
Figure 6.
Morphological study of sepal and flower by paraffin section. (A) Comparison of sepal in WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. Red arrows indicate the palisade tissue in sepals. The palisade tissue was generated in transgenic plants, while it was not found in sepals of WT. (B) Epidermal cell and vascular bundle cell of WT, SlMBP21-RNAi (SRNAi) and SlMBP21-SlMADS1-RNAi (DRNAi) lines. Red arrows indicate the palisade tissue and the vessel cells in sepals of transgenic plants. The palisade tissue was generated in transgenic plants, while it was not found in sepals of WT. The area and number of the vessel cells is larger than that in WT. (C) The flower (3DPA) paraffin section of WT and transgenic lines. The connectives between two sepals were marked by the red arrows. DPA, day post-anthesis; Ca, carpel; St, stamen; Pe, petal; Se, sepal.
2.6. Transcript Analyses of Genes Involved in Sepal Development
Since SlMBP21 and SlMADS1 were primarily expressed in sepals and flowers, and suppression of SlMBP21 and SlMBP21-SlMADS1 resulted in enlarged and fused sepals, the expression of known genes that are involved in sepal development (SlCMB1, MC, SlAP2a and TAGL1) and boundary establishment (GOBLET) were detected in sepals of WT, SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines. Our results displayed that SlCMB1, SlAP2a, TAGL1 and GOBLET were significantly decreased in these two kinds of transgenic lines, but no expression difference of MC was observed in transgenic sepals (Figure 7A–E).
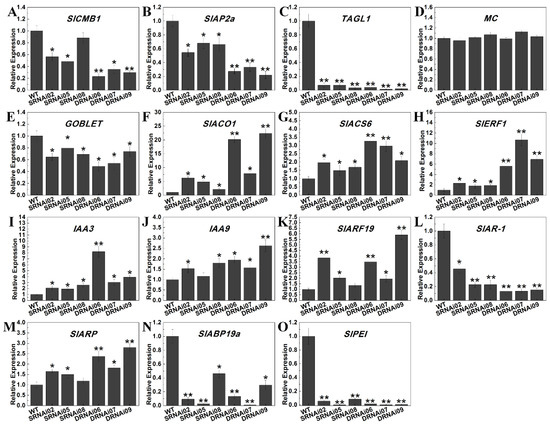
Figure 7.
Relative expression of genes related to different biological processes during sepal development. (A–D). Relative expression levels of genes related to sepal development. (E) Relative expression level of gene related to boundary establishment. (F–H) Relative expression levels of genes related to ethylene. (I–N) Relative expression levels of genes related to auxin. (O) Relative expression level of gene related to cell expansion. The significant differences (p < 0.05, indicated by *) and highly significant differences (p < 0.01, indicated by **) were marked with the asterisks between WT and transgenic lines. Data are the means ± SE of three independent biological replicates.
In 2017, Li et al. reported that SlMBP21 controls sepal size mediated by auxin and ethylene in Micro-Tom, so we also detected some genes related to ethylene (SlACO1, ACS6, ERF1), auxin (SlAR-1, auxin-repressed protein gene; SlARP, auxin-regulated protein gene; SlABP19a, auxin-binding protein gene ABP19a-like and SlARF19, IAA3, and IAA9) and cell expansion (SlPEI, pectinesterase inhibitor), according to their RNA-seq data in our study [64]. Our results displayed that the transcripts of SlACO1, SlACS6, SlERF1, IAA3, IAA9 and SlARF19 were significantly increased in SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi sepals, while the expression levels of SlAR-1, SlARP, SlABP19a and SlPEI were dramatically reduced in these two transgenic lines, compared with WT (Figure 7F–O). It is noteworthy that the transcript levels of some of these fourteen detected genes were markedly changed in SlMBP21-SlMADS1-RNAi lines compared to that in SlMBP21-RNAi lines. Together, suppression of SlMBP21 and SlMADS1 had effect on the transcript levels of some genes related to sepal development.
2.7. SlMBP21 and SlMADS1 Can Interact with Other Sepal Development-Related Proteins
To study the interaction of SlMBP21 and SlMADS1 with other regulators in sepal development, SlAP2a, TAGL1, and the ripening-related protein RIN were selected to carry out the yeast two-hybrid assay [29,30,66,67]. The pGBKT7-SlMBP21 and pGBKT7-SlMADS1 vectors were generated to be used as the bait, respectively (Figure S4A). The pGADT7-SlMBP21, pGADT7-SlAP2a, pGADT7-TAGL1 and pGADT7-RIN vectors were obtained to be used as the prey, respectively (Figure S4B). In addition, self-activation of pGBKT7-SlMBP21 and pGBKT7-SlMADS1 was also performed and the results were negative (Figure S5). Figure 8 showed that the yeast grew on the QDO selective medium and turned blue on the QDO/X medium that contained the X-α-gal indicator, suggesting that SlMBP21 can interact with SlMBP21, SlAP2a, TAGL1, and RIN, while SlMADS1 can only interact with SlAP2a and RIN, and no interaction between SlMBP21 and SlMADS1 was observed. In addition, we performed the yeast two-hybrid assay again to verify the “positive control”, “negative control”, and the interactions of “pGBKT7-SlMBP21 & pGADT7-SlMBP21” and “pGBKT7-SlMBP21 & pGADT7-SlMADS1” (please see Figure S6).
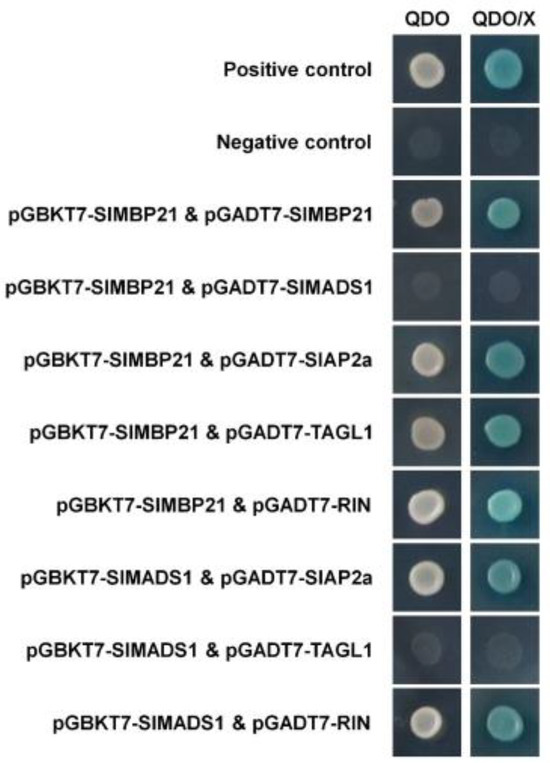
Figure 8.
Yeast two-hybrid assay of SlMBP21 and SlMADS1 with other sepal development-related proteins. SlMBP21 could interact with SlMBP21, SlAP2a, TAGL1 and RIN but not with SlMADS1; SlMADS1 could interact with SlAP2a and RIN, but not with TAGL1, individually. QDO, SD medium lacking Trp, Leu, His, and adenine. QDO/X, SD medium lacking Trp, Leu, His, and adenine with X-α-Gal.
3. Discussion
MADS-box proteins play crucial roles in various plant developmental processes: they are involved in the morphogenesis of almost all plant organs and throughout the whole plant life cycle, contributing to the overall life of plants [14]. Research on MADS-box genes holds potential significances for improving crop cultivation, increasing yield, and adapting plants to challenging environmental conditions. In previous investigations, it was extensively accepted that the SEP subclade is active in specifying floral organ identity, regulating fruit ripening and the development of pedicel abscission zone [31,32,33,62,64,68]. To date, six SEP MADS-box transcription factors, RIN, TM5, TM29, SlCMB1, SlMADS1 and SlMBP21, have been identified and characterized in tomato. RIN is the master regulator that controls fruit ripening of tomato plants, while TM5 regulates the differentiation of the petal, stamen and carpel [29,62]. Suppression of TM29 results in floral reversion and parthenocarpic fruit development [33]. SlCMB1 plays essential roles in fruit ripening, inflorescence architecture and sepal size [30,63]. SlMADS1 is involved in inflorescence and sepal development and functions as a negative regulator during tomato fruit ripening [27,47]. SlMBP21 controls the development of the pedicel abscission zone, inflorescence architecture and sepal size [24,27,64]. It also can be associated with enlarged sepals in SlMBP21 RNAi lines that can better protect inner floral organs and enhance photosynthesis and fruit set ratio [64]. So far, five classes (i.e., A, B, C, D and E) of MADS-box genes have been characterized to determine floral organ identities [69,70]. As E class genes, SEP MADS-box genes play crucial roles in all four whorls of floral organs following the ABCDE model [60,71]. For instance, previous studies have shown that four SEP MADS-box genes, SEP1-SEP4, redundantly regulate the development of floral organs in Arabidopsis [31,32]. Therefore, MADS-box genes, especially those in the SEP class, serve as essential regulators of floral organ identity, which provide crucial insights for plant genetics and genetic engineering.
Previous studies have reported that both SlMBP21 and SlMADS1 shared similar expression profiles as they were preferentially expressed in flowers and sepals [47,64]. As in our study, higher sequence similarity between SlMBP21 and SlMADS1 was observed on the amino acid level, and all these two SEP MADS-box proteins have the four typical MADS-box domains. The conservation of MADS-box domains across different plant species highlights their fundamental biological roles, aiding in phylogenetic and functional analyses. Moreover, the transcripts of SlMBP21 and SlMADS1 increased with flower development, but SlMBP21 expression levels increased with sepal development while SlMADS1 expression levels declined with sepal development. The transcripts of SlMBP21 and SlMADS1 were preferentially accumulated in sepals than the other three whorls of floral organs. The results here suggest that these two genes may be required for the identity of floral organs in tomato, especially for sepal development.
Up to now, it has been reported that some MADS-box genes function redundantly and/or cooperatively during tomato development. For instance, two AGAMOUS clade MADS-box genes, TAG1 and TAGL1, cooperatively control flower development and redundantly regulate sepal and pollen development in tomato [72]. FUL1 (FRUITFULL1) and FUL2 have a redundant function in regulating ethylene-independent aspects of fruit ripening in tomato [73]. Three MADS-box genes (AtAG, AtSHP1, AtSTK) redundantly control carpel and ovule development [56], and the other three closely related and functionally redundant MADS-box genes, AtSEPALLATA1/2/3/4 (AtSEP1/2/3/4), are required for the development of petals, stamens and carpels in Arabidopsis thaliana [31,32]. Liu et al. reported that the antisense of SlMADS1 in tomato results in elongated sepals [24] and Soyk et al. reported that the tomato ej2 (i.e., SlMADS1) mutant generates 50% longer sepals than that of WT [27]. Suppression of SlMBP21 resulted in longer sepals and suppressed pedicel abscission zone development in tomato [24,64]. Similarly, in this study, down-regulation of SlMBP21 also resulted in suppressed pedicel abscission zone development and enlarged and fused sepals in Mill. cv. Ailsa Craig (AC++). The SlMBP21-RNAi sepals were significantly longer and wider than WT, while the SlMBP21-SlMADS1-RNAi plants displayed suppressed pedicel abscission zone development and a larger sepal size than that of WT and the single SlMBP21-RNAi lines. Interestingly, the SlMBP21-SlMADS1-RNAi lines produced different sepals due to the different suppression of SlMBP21 and SlMADS1 expression levels (weak and strong), and the sepal size increased with the decreased expression levels of SlMBP21 and SlMADS1. Simultaneously, the cross-sectional area of 15 DPA sepals of SlMBP21-SlMADS1-RNAi lines was also wider than that of SlMBP21-RNAi lines and WT. Moreover, more of a palisade tissue-like structure in the upper and lower epidermis was observed in the 15 DPA sepals of the SlMBP21-SlMADS1-RNAi lines than that of the single RNAi lines, but not in the WT sepals. Combined with previous investigations and the results obtained in our study, we speculate that SlMBP21 and SlMADS1 may have cooperative functions in controlling sepal size.
Although many functions of MADS-box genes are well-understood in single gene study, their cooperative roles in specific biological processes, such as tissue development or stress responses, require more detailed studies. In the functional study of MADS-box genes, it can be found that silencing or knockout of one gene results in no or few weak phenotypic changes, while silencing or knockout of two or more genes generates the obvious phenotypic changes. And these phenotypic changes are significantly enhanced with the increased number of genes that are silenced or knocked out, such as the TAG1/TAGL1, FUL1/FUL2, AtAG/AtSHP1/AtSTK, and AtSEP1/2/3/4 genes mentioned above [31,32,56,72,73]. In plants, some MADS-box genes can play different roles in various biological processes, while multiple genes can play the same role in the same biological process. However, what the relationship is between these genes that play a common function, and whether these genes have functional cooperation or redundancy are the important part of gene functional investigations. Therefore, simultaneously studying the functional relationship of two or multiple genes in the same biological process will be conducive to further analyzing the molecular mechanism of these genes in regulating this biological process, which will provide new research ideas and enlightenment for the investigation of a complex gene expression regulation network.
In tomato plants, many well-characterized genes have been reported to regulate sepal development, which contribute to the further investigation of sepal formation and development. For instance, the tomato A-class gene MC function as an important regulator in sepal development [29]. Down-regulation of an A-class gene, SlAP2a, led to longer and wider sepals [66]. Suppression of an E-class gene, SlCMB1, results in enlarged and fused sepals [30]. Tomato plants with increased TAGL1 generate fleshy and succulent sepals which can ripen normally with fruit ripening [67]. Our study showed that SlCMB1, SlAP2a and TAGL1 were markedly reduced in SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines, and lower expression levels of these three genes were observed in SlMBP21-SlMADS1-RNAi lines. But another A-class gene, MC, had no significant expression changes both in WT and transgenic sepals. In SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines, we also found that the number of fused sepals in each flower and the ratio of fused flowers in one inflorescence were markedly higher than in WT. The fused sepals linked each other with few layers of cells in cross section of flowers, indicating that the partial boundary morphogenesis of the first whorl was probably impacted. Previous research reported that tomato NAC transcription factor GOBLET is required for the inhibition of congenital fusion with primordia [74]. In our study, the transcripts of GOBLET were significantly reduced in SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi sepals. Based on our results and previous studies, we come to a conclusion that SlMBP21 and SlMADS1 may cooperatively affect sepal development through directly and/or indirectly regulating the expression of A-function genes and control sepals fusion by impacting the transcripts of genes related to boundary establishment.
Previous RNA-seq data showed that the transcripts of genes related to auxin, ethylene and cell expansion were significantly impacted in SlMBP21-RNAi lines of Micro-Tom [64]. In our study, the expression levels of some cell expansion-, auxin- and ethylene-related genes (SlACO1, SlACS6, SlERF1, IAA3, IAA9, SlARF19, SlAR-1, SlARP, SlABP19a, and PEI) that were selected from their RNA-seq data were also dramatically influenced in the SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi lines, and the transcript levels of some of them in the SlMBP21-SlMADS1-RNAi lines were higher/lower than that in SlMBP21-RNAi lines. Furthermore, the SlMBP21 promoter region contains three ethylene-responsive elements (ERE motif, ATTTCAAA), one gibberellin-responsive element (P-Box, CCTTTTG) and one auxin-responsive element (TGA-box, TGACGTAA), and the SlMADS1 promoter region contains three ethylene-responsive elements (ERE motif, ATTTCAAA) and three gibberellin-responsive elements (two P-Box, CCTTTTG and one TATC-box, TATCCCA) indicating the essential roles of SlMBP21 and SlMADS1 in responding to plant hormones. All these results demonstrate that SlMBP21 and SlMADS1 may act cooperatively in sepal formation and development through participating in plant hormone responsive and finally impact the development of sepal cells.
MADS-box transcription factors can form dimers or a multimer with other regulators to control plant growth and development [75,76,77,78]. Although the intricate interactions of MADS-box genes with numerous transcription factors pose challenges for understanding their functions, investigation of these interactions will help to further clarify the function of MADS genes in plant growth and development. For example, the AP1 orthologue MC controls the development of inflorescences, the abscission zone and sepals [23,29]. A recent study showed that SlMBP21 is implicated in regulating sepal size in tomato [64]. Suppression of SlCMB1 results in altered inflorescence architecture and enlarged sepals [30]. Tomato plants with increased TAGL1 transcript levels generate a fleshy sepal, which can ripen normally following fruits [67]. The MADS-box transcription factor, RIN, which is the master regulator of fruit ripening in tomato, regulates ethylene synthesis by increasing the transcript accumulation of ethylene signaling and ethylene biosynthesis genes (e.g., ACS2 and ACS4) [79,80,81]. In addition to the MADS-box proteins mentioned above, AP2a, a transcription factor of the AP2 family, has been proven to regulate tomato sepal development [66]. In this study, the results of yeast two-hybrid assay showed that SlMBP21 can interact with SlMBP21, SlAP2a, TAGL1 and RIN, and SlMADS1 can interact with SlAP2a and RIN but not with TAGL1, but there was no interaction between SlMBP21 and SlMADS1. Moreover, our previous studies have demonstrated that the MADS-box transcription factor SlCMB1 which plays essential roles in the sepal development, can interact with MC, SlAP2a, SlMBP21, TAGL1 and RIN, respectively [30,63]. In the floral quartet model, the sepal identity is determined by a complex of two AP1 proteins and two AG MADS-box proteins (2A + 2SEP) [59,60], while in the ABCDE model, the sepal development is regulated by A and E class genes [60,71]. Leseberg et al. reported that SlMBP21 can interact with MC, but SlMADS1 can not interact with SlMBP21, MC, TAGL1 [75]. Furthermore, they also reported that both SlMBP21 and SlMADS1 can also interact with MADS-box proteins TM3, JOINTLESS, SlMBP24, and RIN, respectively [75]. So it is reasonable to believe that SlMBP21 and SlMADS1 may form tetramers with other functional proteins during tomato development. So we further speculate that SlMBP21 and SlMADS1 may form trimers or tetramers with other functional proteins (e.g., SlAP2a, RIN, SlCMB1, TAGL1, and MC) via protein bridges to control sepal development through the transcriptional regulation of other related genes.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Tomato plants, Solanum lycopersicum (Mill. cv. Ailsa Craig, AC++), were used as the wild type (WT). The WT and transgenic tomato plants were grown in the standard greenhouse (25 °C, 16 h day and 18 °C, 8 h night cycle, 250 μmolm−2 s−1 light intensity and 80% humidity). The tomato flowers were tagged at anthesis. The DPA (day post-anthesis) was used to record the days of flower and sepal development. All samples were frozen with liquid nitrogen immediately and stored at −80 °C.
4.2. Total RNA Extraction and Sequence Analysis of SlMBP21 and SlMADS1
Total RNA was isolated from each sample using the RNAiso Plus (Takara, Dalian, China) following the manufacturer’s protocol. In the reverse transcription, 1 μg total RNA samples were used to synthesize the first-strand cDNA using the M-MLV reverse transcriptase (Promega, Madison, WI, USA) with oligo(dT)20 primer.
To clone the full length of SlMBP21 and SlMADS1, 1–2 μL cDNA was used to employ the PCR reactions with primers SlMBP21-Full-F/-R and SlMADS1-Full-F/-R, respectively (Table S1). After being tailed with a DNA A-Tailing kit (Takara, Dalian, China), the amplified products of SlMBP21 and SlMADS1 were linked into the vector pMD18-T (Takara, Dalian, China) to obtain SlMBP21-pMD18-T and SlMADS1-pMD18-T, respectively. Positive clones of each vector were picked out through Escherichia coli DH5α transformation and were conformed by sequencing.
To analyze putative cis-elements in the SlMBP21 and SlMADS1 promoter region, promoter analysis was carried out using the promoter sequences (2800 bp regions upstream initiation codon ATG of gene-predicted ORF) of each gene on the Plant CARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 3 July 2023).
4.3. Construction of SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi Vectors and Plant Transformation
In order to down-regulate SlMBP21 expression, a RNAi vector was generated using PHANNIBAL and pBIN19 vector. A 381 bp specific DNA fragment of SlMBP21 was amplified using SlMBP21-i-F/R primers (Table S1). After being purified, the amplified fragments were digested with the estriction enzyme Kpn I/Xho I and Hind III/Xba I, respectively. Then, the SlMBP21-RNAi vector (Figure S3A) was generated using the same method following our previous study described [30].
In order to repress the expression of SlMBP21 and SlMADS1 simultaneously in tomato, the double RNAi vector of SlMBP21 and SlMADS1 was constructed using pDH51, PHANNIBAL and pBIN19 vector. Firstly, the 381 bp SlMBP21-specific DNA fragments and the 515 bp SlMADS1 specific DNA fragments were amplified with the primers SlMBP21-1(p)-F/SlMBP21-1(X)-R and SlMADS1-1(X)-F/SlMADS1-1(B)-R (Table S1), respectively. Secondly, after being digested with pst I and Xba I, the purified SlMBP21 fragments were linked into the pDH51 plasmid at the pst I and Xba I restriction site to generate the pDH51-SlMBP21 vector. Thirdly, the digested SlMADS1 fragments using estriction enzyme Xba I and BamH I were cloned into the pDH51-SlMBP21 plasmid at the Xba I/BamH I restriction site to yield pDH51-SlMBP21-SlMADS1. Fourthly, the START DNA polymerase (Takara) was used to amplify the tandem fragment of SlMBP21 and SlMADS1 using the primers SlMBP21-2(K, H)-F/SlMADS1-2(X, B)-R (Table S1). Fifthly, the purified tandem fragments of SlMBP21-SlMADS1 were digested by Kpn I/Xho I and Hind III/BamH I and linked into the pHANNIBAL vector at the Kpn I/Xho I and the Hind III/BamH I restriction site, respectively. At last, the double-stranded RNA expression unit, possessing the cauliflower mosaic virus 35S promoter, the sense-orientated SlMBP21-SlMADS1 tandem fragment, PDK intron, the antisense-orientated SlMBP21-SlMADS1 tandem fragment, and the OCS terminator, was digested with Spe I/Sac I and cloned into pBIN19 vector to generate the Pbin19-SlMBP21-SlMADS1 vector (Figure S3B).
The obtained binary plasmids of SlMBP21 and SlMBP21-SlMADS1 were transferred into Agrobacterium tumefaciens (strain LBA4404), respectively. After that, the transformation of tomato cotyledon explants mediated by Agrobacterium tumefaciens was performed to obtain SlMBP21-RNAi lines and SlMBP21-SlMADS1-RNAi lines [82]. The tissue culture plants were screened by kanamycin and the PCR reactions were carried out to select the positive transgenic plants with the NPTII-F/R primers (Table S1).
4.4. qRT-PCR (Quantitative Real-Time PCR) Analysis
Total RNA extraction of each sample and reverse transcription were carried out following the method as described above. Then, the synthesized cDNAs were diluted to 15 ng/μL using RNase/DNase-free water. The qRT-PCR reactions of gene expression analysis were performed using SYBR Premix Go Taq (Promega, Madison, America) in a 10 μL total reaction volume (5 μL SYBR Premix Go Taq, 3 μL ddH2O, 0.25 μL each primer (10 mM), and 1.5 μL cDNA). No template control (NTC) and no reverse transcription control (NRT) experiments of each gene were performed. Tomato SlCAC gene was used as the internal standard [83]. The 2−∆∆CT method was used to analyze relative transcript levels of each gene [84]. All specific primers of genes used in this study were listed in Table S2. Analyses of gene transcript levels were carried out in three independent biological repeats.
4.5. Anatomical Analyses of Flowers and Sepals
Flowers at 3 DPA and sepals at 20 DPA were fixed in FAA solution (50% absolute ethanol, 5% glacial acetic acid, 5% glycerin and 5% formalin) for 24 h. The paraffin section were performed following the same method of our previous study [30]. A microscope (OLYMPUS IX71, Tokyo, Japan) was used to observe the paraffin section of each sample.
4.6. Statistics of Fused Sepals and Sepal Length and Width
The number of fused sepals in a single flower and flowers with fused sepals of WT and transgenic lines were recorded. Sepal length and width of WT and transgenic (single and double RNAi) flowers were measured at 3 DPA and 20 DPA. In our statistics, the employed flowers were all the first and second flowers of each inflorescences. The 3 DPA flowers were used to observe the fused sepals. At least twenty flowers of each lines were contained in our statistics.
4.7. Yeast Two-Hybrid Assay
The yeast two-hybrid assay was carried out using the MATCHMAKER GAL4 Two-Hybrid System III following the manufacturer’s protocol (Clontech, Los Angeles, America). The ORFs of SlMBP21, SlMADS1, SlAP2a, TAGL1 and RIN were amplified with primers SlMBP21(Y2H)F/R, SlMADS1(Y2H)F/R, SlAP2a(Y2H)F/R and TAGL1(Y2H)F/R, RIN(Y2H)F/R, respectively (Table S1). The amplified products of SlMBP21 and SlMADS1 were linked into the EcoR I/BamH I restriction site of the pGBKT7 vector to generate the pGBKT7-SlMBP21 and pGBKT7-SlMADS1 vector, respectively (Figure S4A). The amplified products of SlMBP21, SlAP2a, TAGL1 and RIN were inserted into the EcoR I/BamH I restriction site of the pGADT7 vector to obtain the pGADT7-SlMBP21, pGADT7-SlAP2a, pGADT7-TAGL1 and pGADT7-RIN (Figure S4B), respectively. The pGADT7-SlMBP21, pGADT7-SlAP2a, pGADT7-TAGL1 and pGADT7-RIN vectors were transferred into the Y187 yeast strain and the pGBKT7-SlMBP21 and pGBKT7-SlMADS1 vectors were transferred into the Y2Hgold yeast strain, respectively. After that, the yeast two-hybrid assay was performed following the same method of our previous study [30].
4.8. Statistical Analysis
All obtained data were presented as mean ± standard deviation (± SD) in this study. Student’s t-test (p < 0.05, p < 0.01) was performed to analyze the significant difference between WT and transgenic lines. The measurement values indicate the mean values of three biological replicates.
5. Conclusions
Taken together, previous studies and our data from SlMBP21-RNAi and SlMBP21-SlMADS1-RNAi sepals prove that SlMBP21 and SlMADS1 cooperatively play significant roles in tomato sepal development through influencing the activities and/or expression of other regulators or via interacting with other regulatory proteins. Although the precise regulatory mechanisms of SlMBP21 and SlMADS1 in tomato sepal development remain to be discovered, these two MADS-box genes should be important targets for investigating the mechanisms underlying sepal development and also will provide a new tool for improving the yield of tomato and other crops. However, higher levels of regulatory cascades of sepal development-associated regulators still need to be discovered, for instance, the targets regulated directly or indirectly by SlMBP21 and SlMADS1 and the interactions of these regulators, which will probably be conducive to further studying the regulatory network of sepal development in tomato. Despite intensive research on some MADS-box family members, the functions of many MADS-box genes remain unexplored, hindering a comprehensive understanding. In summary, MADS-box genes play crucial roles in plant development, but their complexity and diversity present both challenges and opportunities for researchers in the field of plant biology.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25052489/s1.
Author Contributions
J.Z.: conceptualization; data curation; funding acquisition; software; visualization; writing—original draft. Z.H.: project administration; validation; writing—review and editing. Q.X.: visualization; writing—review and editing. T.D.: software. J.L.: visualization. G.C.: conceptualization; funding acquisition; validation; writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (nos. 31572129, 31872121) funded by the Chinese Government, Natural Science Foundation of Shandong Province (no. ZR2021MC079) funded by the Shandong Provincial Government of China.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data and materials supporting the conclusions of this study are included within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Messenguy, F.; Dubois, E. Role of MADS box proteins and their cofactors in combinatorial control of gene expression and cell development. Gene 2003, 316, 1–21. [Google Scholar] [CrossRef]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; De Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef]
- Shore, P.; Sharrocks, A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995, 229, 1–13. [Google Scholar] [CrossRef]
- Smaczniak, C.; Immink, R.G.; Angenent, G.C.; Kaufmann, K. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 2012, 139, 3081–3098. [Google Scholar] [CrossRef]
- Theißen, G.; Kim, J.T.; Saedler, H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 1996, 43, 484–516. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Liljegren, S.S.; Gold, S.E.; Burgeff, C.; Ditta, G.S. MADS-box gene evolution beyond flowers: Expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2010, 24, 457–466. [Google Scholar]
- Stefanie, D.B.; Jeroen, R.; Yves, V.D.P.; Günter, T. And then there were many: MADS goes genomic. Trends Plant Sci. 2003, 8, 475–483. [Google Scholar]
- Norman, C.; Runswick, M.; Pollock, R.; Treisman, R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell 1988, 55, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jang, S.; Chae, S.; Chung, K.M.; Moon, Y.H.; An, G.; Jang, S.K. Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol. Biol. 1999, 40, 419. [Google Scholar] [CrossRef]
- Yang, Y.; Fanning, L.; Jack, T. The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 2003, 33, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Katrin, H.; Rumiko, K.; Mitsuyasu, H.; Heinz, S.; Thomas, M.; Günter, T. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 2002, 19, 801–814. [Google Scholar]
- Causier, B.; Kieffer, M.; Davies, B. MADS-Box Genes Reach Maturity. Science 2002, 296, 275–276. [Google Scholar] [CrossRef]
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar] [CrossRef]
- Schilling, S.; Pan, S.; Kennedy, A.; Melzer, R. MADS-box genes and crop domestication: The jack of all traits. J. Exp. Bot. 2018, 69, 1447–1469. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Woo, Y.-M.; Ryu, S.-I.; Shin, Y.-D.; Kim, W.T.; Park, K.Y.; Lee, I.-J.; An, G. Further characterization of a rice AGL12 group MADS-box gene, OsMADS261[C][W][OA]. Plant Physiol. 2008, 147, 156–168. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chen, G.; Cui, B.; Gao, Q.; Guo, J.E.; Li, A.; Zhang, L.; Hu, Z. Solanum lycopersicum agamous-like MADS-box protein AGL15-like gene, SlMBP11, confers salt stress tolerance. Mol. Breed. 2016, 36, 125. [Google Scholar] [CrossRef]
- Yin, W.; Hu, Z.; Hu, J.; Zhu, Z.; Yu, X.; Cui, B.; Chen, G. Tomato (Solanum lycopersicum) MADS-box transcription factor SlMBP8 regulates drought, salt tolerance and stress-related genes. Plant Growth Regul. 2017, 83, 55–68. [Google Scholar] [CrossRef]
- Xie, Q.; Hu, Z.; Zhu, Z.; Dong, T.; Zhao, Z.; Cui, B.; Chen, G. Overexpression of a novel MADS-box gene SlFYFL delays senescence, fruit ripening and abscission in tomato. Sci. Rep. 2014, 4, 4367–4376. [Google Scholar] [CrossRef] [PubMed]
- Burko, Y.; Shleizer-Burko, S.; Yanai, O.; Shwartz, I.; Zelnik, I.D.; Jacob-Hirsch, J.; Kela, I.; Eshed-Williams, L.; Ori, N. A Role for APETALA1/FRUITFULL Transcription Factors in Tomato Leaf Development. Plant Cell Online 2013, 25, 2070–2083. [Google Scholar] [CrossRef]
- Guo, X.; Chen, G.; Naeem, M.; Yu, X.; Tang, B.; Li, A.; Hu, Z. The MADS-box gene SlMBP11 regulates plant architecture and affects reproductive development in tomato plants. Plant Sci. 2017, 258, 90–101. [Google Scholar] [CrossRef]
- Yu, L.H.; Miao, Z.Q.; Qi, G.F.; Wu, J.; Cai, X.T.; Mao, J.L.; Xiang, C.B. MADS-Box Transcription Factor AGL21 Regulates Lateral Root Development and Responds to Multiple External and Physiological Signals. Mol. Plant 2014, 7, 1653–1669. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, N.; Chen, H.; Wang, G.; Huang, J. OsMADS25 regulates root system development via auxin signaling in rice. Plant J. 2018, 95, 1004–1022. [Google Scholar] [CrossRef]
- Nakano, T.; Kimbara, J.; Fujisawa, M.; Kitagawa, M.; Ihashi, N.; Maeda, H.; Kasumi, T.; Ito, Y. MACROCALYX and JOINTLESS Interact in the Transcriptional Regulation of Tomato Fruit Abscission Zone Development. Plant Physiol. 2012, 158, 439–450. [Google Scholar] [CrossRef]
- Liu, D.; Wang, D.; Qin, Z.; Zhang, D.; Yin, L.; Wu, L.; Colasanti, J.; Li, A.; Mao, L. The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J. 2014, 77, 284–296. [Google Scholar] [CrossRef]
- Mao, L.; Begum, D.; Chuang, H.-w.; Budiman, M.A.; Szymkowiak, E.J.; Irish, E.E.; Wing, R.A. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 2000, 406, 910–913. [Google Scholar] [CrossRef]
- Kobayashi, K.; Yasuno, N.; Sato, Y.; Yoda, M.; Yamazaki, R.; Kimizu, M.; Yoshida, H.; Nagamura, Y.; Kyozuka, J. Inflorescence Meristem Identity in Rice Is Specified by Overlapping Functions of Three AP1/FUL-Like MADS Box Genes and PAP2, a SEPALLATA MADS Box Gene. Plant Cell 2012, 24, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell 2017, 169, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
- Szymkowiak, E.J.; Irish, E.E. JOINTLESS suppresses sympodial identity in inflorescence meristems of tomato. Planta 2006, 223, 646–658. [Google Scholar] [CrossRef]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-Box Gene Necessary for Fruit Ripening at the Tomato Ripening-Inhibitor (Rin) Locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Z.; Wang, Y.; Yu, X.; Liao, C.; Zhu, M.; Chen, G. Suppression of a tomato SEPALLATA MADS-box gene, SlCMB1, generates altered inflorescence architecture and enlarged sepals. Plant Sci. 2018, 272, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 Gene of Arabidopsis thaliana Functions in Floral Organ and Meristem Identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Pelaz, S.; Ditta, G.S.; Baumann, E.; Wisman, E.; Yanofsky, M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 2000, 405, 200–203. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Morris, B.A.; Sutherland, P.; Veit, B.; Yao, J.-L. Down-Regulation of TM29, a Tomato SEPALLATA Homolog, Causes Parthenocarpic Fruit Development and Floral Reversion. Plant Physiol. 2002, 130, 605–617. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2016, 15, 634–647. [Google Scholar] [CrossRef]
- Giménez, E.; Dominguez, E.; Pineda, B.; Heredia, A.; Moreno, V.; Lozano, R.; Angosto, T. Transcriptional Activity of the MADS Box ARLEQUIN/TOMATO AGAMOUS-LIKE1 Gene Is Required for Cuticle Development of Tomato Fruit. Plant Physiol. 2015, 168, 1036–1048. [Google Scholar] [CrossRef]
- Garceau, D.C.; Batson, M.K.; Pan, I.L. Variations on a theme in fruit development: The PLE lineage of MADS-box genes in tomato (TAGL1) and other species. Planta 2017, 246, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Sanda, S.L.; Amasino, R.M. Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol. Gen. Genet. MGG 1996, 251, 69–74. [Google Scholar]
- Hartmann, U.; Höhmann, S.; Nettesheim, K.; Wisman, E.; Saedler, H.; Huijser, P. Molecular cloning of SVP: A negative regulator of the floral transition in Arabidopsis. Plant J. 2000, 21, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Jihyun, M.; Sung-Suk, S.; Horim, L.; Kyu-Ri, C.; Choo Bong, H.; Nam-Chon, P.; Sang-Gu, K.; Ilha, L. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003, 35, 613–623. [Google Scholar]
- Liu, C.; Zhou, J.; Bracha-Drori, K.; Yalovsky, S.; Ito, T.; Yu, H. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 2007, 134, 1901–1910. [Google Scholar] [CrossRef]
- Yu, H.; Xu, Y.; Tan, E.L.; Kumar, P.P. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc. Natl. Acad. Sci. USA 2002, 99, 16336–16341. [Google Scholar] [CrossRef]
- Michaels, S.D.; Ditta, G.; Gustafson-Brown, C.; Pelaz, S.; Yanofsky, M.; Amasino, R.M. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003, 33, 867–874. [Google Scholar] [CrossRef]
- Fornara, F.; Pařenicová, L.; Falasca, G.; Pelucchi, N.; Masiero, S.; Ciannamea, S.; Lopez-Dee, Z.; Altamura, M.M.; Colombo, L.; Kater, M.M. Functional characterization of OsMADS18, a member of the AP1/SQUA subfamily of MADS box genes. Plant Physiol. 2004, 135, 2207–2219. [Google Scholar] [CrossRef]
- Ferrario, S.; Busscher, J.; Franken, J.; Gerats, T.; Vandenbussche, M.; Angenent, G.C.; Immink, R.G. Ectopic Expression of the Petunia MADS Box Gene UNSHAVEN Accelerates Flowering and Confers Leaf-like Characteristics to Floral Organs in a Dominant-Negative Manner. Plant Cell 2004, 16, 1490–1505. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Pan, I.L.; Arroyo, A.J.M.; McQuinn, R.; Chung, M.; Poole, M.; Rose, J.; Seymour, G.; Grandillo, S.; Giovannoni, J. Fleshy Fruit Expansion and Ripening Are Regulated by the Tomato SHATTERPROOF Gene TAGL1. Plant Cell 2009, 21, 3041–3062. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lu, G.; Hou, Z.; Luo, Z.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. Members of the tomato FRUITFULL MADS-box family regulate style abscission and fruit ripening. J. Exp. Bot. 2014, 65, 3005–3014. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Hu, Z.; Deng, L.; Wang, Y.; Zhu, M.; Zhang, J.; Chen, G. A Tomato MADS-box Transcription Factor, SlMADS1, Acts as a Negative Regulator of Fruit Ripening1[C][W]. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.A.; Gustafsonbrown, C.; Savidge, B.; Yanofsky, M.F. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 1992, 360, 273–277. [Google Scholar] [CrossRef]
- Bowman, J.L.; Alvarez, J.; Weigel, D.; Meyerowitz, E.M.; Smyth, D.R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 1993, 119, 721–743. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Denboer, B.G.W.; Vanmontagu, M.; Okamuro, J.K. Control of Arabidopsis Flower and Seed Development by the Homeotic Gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar]
- Goto, K.; Meyerowitz, E.M. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Gene Dev. 1994, 8, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Jack, T. Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 2004, 55, 45–59. [Google Scholar] [CrossRef]
- Egea-Cortines, M.; Saedler, H.; Sommer, H. Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 1999, 18, 5370–5379. [Google Scholar] [CrossRef] [PubMed]
- Mizukami, Y.; Ma, H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 1992, 71, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Angenent, G.C.; Franken, J.; Busscher, M.; van Dijken, A.; van Went, J.L.; Dons, H.; van Tunen, A.J. A Novel Class of MADS Box Genes Is Involved in Ovule Development in Petunia. Plant Cell 1995, 7, 1569–1582. [Google Scholar]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Pelaz, S.; Tapia-López, R.; Alvarez-Buylla, E.R.; Yanofsky, M.F. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 2001, 11, 182–184. [Google Scholar] [CrossRef]
- Daminato, M.; Masiero, S.; Resentini, F.; Lovisetto, A.; Casadoro, G. Characterization of TM8, a MADS-box gene expressed in tomato flowers. BMC Plant Biol. 2014, 14, 319. [Google Scholar] [CrossRef] [PubMed]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Theißen, G.; Melzer, R.; Rümpler, F. MADS-domain transcription factors and the floral quartet model of flower development: Linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Soon Ju, P.; Ke, J.; Schatz, M.C.; Lippman, Z.B. Rate of meristem maturation determines inflorescence architecture in tomato. Proc. Natl. Acad. Sci. USA 2012, 109, 639–644. [Google Scholar]
- Pnueli, L.; Hareven, D.; Broday, L.; Hurwitz, C.; Lifschitz, E. The TM5 MADS Box Gene Mediates Organ Differentiation in the Three Inner Whorls of Tomato Flowers. Plant Cell 1994, 6, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hu, Z.; Yao, Q.; Guo, X.; Nguyen, V.; Li, F.; Chen, G. A tomato MADS-box protein, SlCMB1, regulates ethylene biosynthesis and carotenoid accumulation during fruit ripening. Sci. Rep. 2018, 8, 3413–3427. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Huang, B.; Tang, N.; Jian, W.; Zou, J.; Chen, J.; Cao, H.; Habib, S.; Dong, X.; Wei, W. The MADS-box Gene SlMBP21 Regulates Sepal Size Mediated by Ethylene and Auxin in Tomato. Plant Cell Physiol. 2017, 58, 2241–2256. [Google Scholar] [CrossRef] [PubMed]
- Aceto, S.; Gaudio, L. The MADS and the Beauty: Genes Involved in the Development of Orchid Flowers. Curr. Genom. 2011, 12, 342–356. [Google Scholar] [CrossRef]
- Karlova, R.; Rosin, F.M.; Busscher-Lange, J.; Parapunova, V.; Do, P.T.; Fernie, A.R.; Fraser, P.D.; Baxter, C.; Angenent, G.C.; de Maagd, R.A. Transcriptome and Metabolite Profiling Show That APETALA2a Is a Major Regulator of Tomato Fruit Ripening. Plant Cell 2011, 23, 923–941. [Google Scholar] [CrossRef] [PubMed]
- Giménez, E.; Pineda, B.; Capel, J.; Antón, M.T.; Atarés, A.; Pérez-Martín, F.; García-Sogo, B.; Angosto, T.; Moreno, V.; Lozano, R. Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 2010, 5, e14427. [Google Scholar] [CrossRef]
- Seymour, G.B.; Ryder, C.D.; Cevik, V.; Hammond, J.P.; Popovich, A.; King, G.J.; Vrebalov, J.; Giovannoni, J.J.; Manning, K. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 2011, 62, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31. [Google Scholar] [CrossRef]
- Theissen, G.; Saedler, H. Plant biology: Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Ma, H. The ABCs of Floral Evolution. Cell 2000, 101, 5–8. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, E.; Castañeda, L.; Pineda, B.; Pan, I.L.; Moreno, V.; Angosto, T.; Lozano, R. TOMATO AGAMOUS1 and ARLEQUIN/TOMATO AGAMOUS-LIKE1 MADS-box genes have redundant and divergent functions required for tomato reproductive development. Plant Mol. Biol. 2016, 91, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Bemer, M.; Karlova, R.; Ballester, A.R.; Tikunov, Y.M.; Bovy, A.G.; Woltersarts, M.; Rossetto, P.D.B.; Angenent, G.C.; Maagd, R.A.D. The Tomato FRUITFULL Homologs TDR4/FUL1 and MBP7/FUL2 Regulate Ethylene-Independent Aspects of Fruit Ripening. Plant Cell 2012, 24, 4437–4451. [Google Scholar] [CrossRef] [PubMed]
- Berger, Y.; Harpaz-Saad, S.; Brand, A.; Melnik, H.; Sirding, N.; Alvarez, J.P.; Zinder, M.; Samach, A.; Eshed, Y.; Ori, N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 2009, 136, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Leseberg, C.H.; Eissler, C.L.; Wang, X.; Johns, M.A.; Duvall, M.R.; Mao, L. Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 2008, 59, 2253–2265. [Google Scholar] [CrossRef] [PubMed]
- Favaro, R.; Immink, R.; Ferioli, V.; Bernasconi, B.; Byzova, M.; Angenent, G.; Kater, M.; Colombo, L. Ovule-specific MADS-box proteins have conserved protein-protein interactions in monocot and dicot plants. Mol. Genet. Genom. 2002, 268, 152–159. [Google Scholar] [CrossRef]
- Shchennikova, A.V.; Shulga, O.A.; Immink, R.; Skryabin, K.G.; Angenent, G.C. Identification and Characterization of Four Chrysanthemum MADS-box Genes, Belonging to the APETALA1/FRUITFULL and SEPALLATA3 Subfamilies. Plant Physiol. 2004, 134, 1632–1641. [Google Scholar] [CrossRef]
- De Folter, S.; Shchennikova, A.V.; Franken, J.; Busscher, M.; Baskar, R.; Grossniklaus, U.; Angenent, G.C.; Immink, R.G. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006, 47, 934–946. [Google Scholar] [CrossRef]
- Ito, Y.; Kitagawa, M.; Ihashi, N.; Yabe, K.; Kimbara, J.; Yasuda, J.; Ito, H.; Inakuma, T.; Hiroi, S.; Kasumi, T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008, 55, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, M.; Nakano, T.; Ito, Y. Identification of potential target genes for the tomato fruit-ripening regulator RIN by chromatin immunoprecipitation. BMC Plant Biol. 2011, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The Tomato MADS-box Transcription Factor RIPENING INHIBITOR Interacts with Promoters Involved in Numerous Ripening Processes in a COLORLESS NONRIPENING-Dependent Manner1[W][OA]. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Hackett, R.; Walker, D.; Taylor, A.; Lin, Z.; Grierson, D. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 2004, 136, 2641–2651. [Google Scholar] [CrossRef] [PubMed]
- Expósito-Rodríguez, M.; Borges, A.A.; Borges-Pérez, A.; Pérez, J.A. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).