Differential Expression of Stress Adaptation Genes in a Diatom Ulnaria acus under Different Culture Conditions
Abstract
1. Introduction
2. Results
2.1. MC and MCP Molecular Structure
2.2. Molecular Structure of UaDSP
2.3. Molecular Structure of UaALDH12
2.4. Molecular Structure of UaGSHS
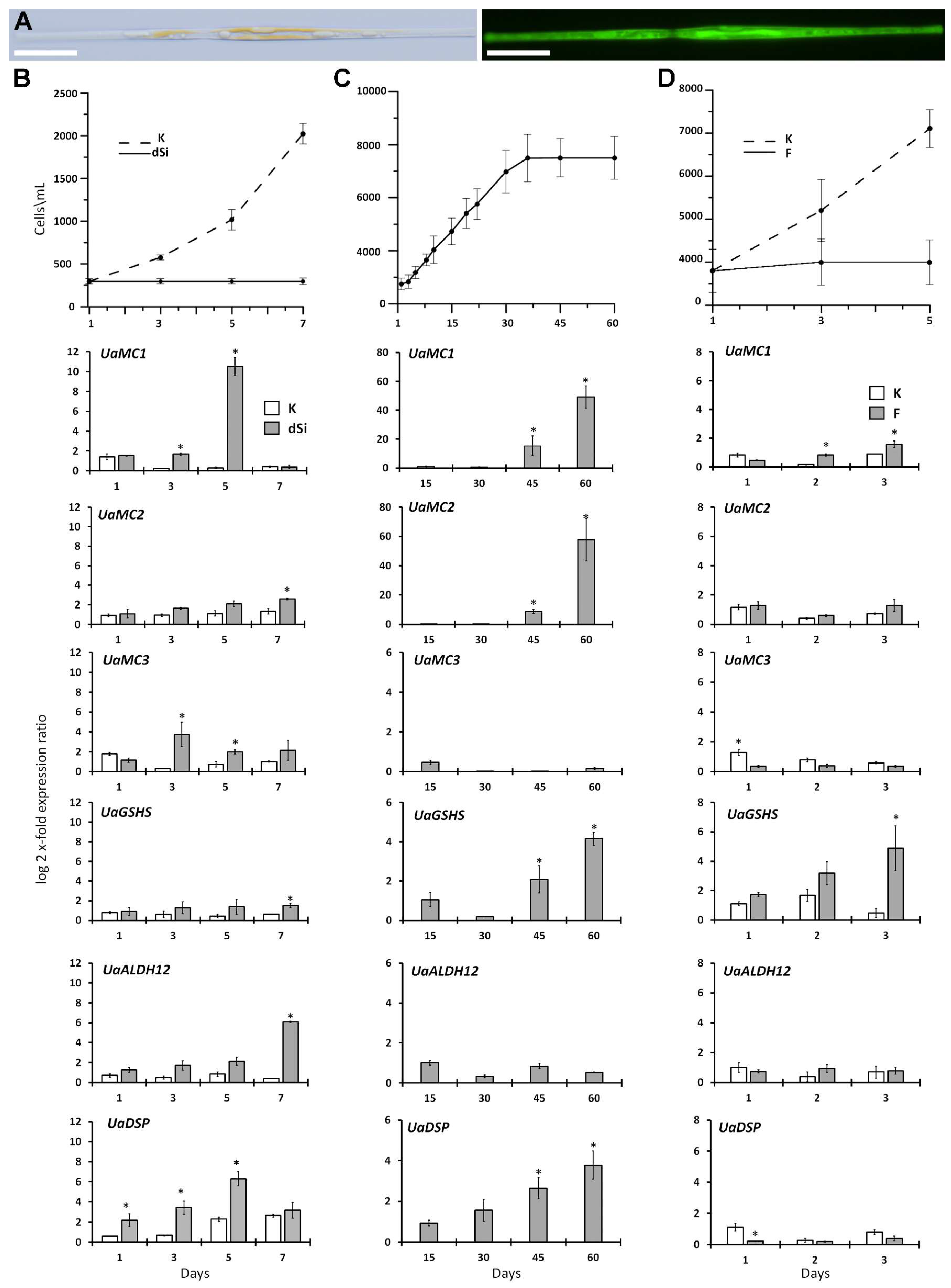
2.5. Differential Expression of Six Studied Genes as a Response to Different Stresses
3. Discussion
3.1. Comparison of the Molecular Structure of the Predicted U. acus Proteins with Their Homologues in Other Organisms
3.2. Changes in MC, ALDH12, GSHS, and DSP Expression
4. Materials and Methods
4.1. Culture Conditions
4.2. Target Gene Identification
4.3. Sanger Sequencing and Sequence Analysis
4.4. Analysis of the Predicted Amino Acid Sequences
4.5. Algicidal Effect
4.6. Silicon Starvation
4.7. Long-Term Cultivation
4.8. Fluorescence Microscopy
4.9. RNA Isolation and cDNA Synthesis0
4.10. Real-Time PCR
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smetacek, V. Diatoms and the Ocean Carbon Cycle. Protist 1999, 150, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.M.; Tréguer, P.; Brzezinski, M.A.; Leynaert, A.; Quéguiner, B. Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 1995, 9, 359–372. [Google Scholar] [CrossRef]
- Spungin, D.; Bidle, K.D.; Berman-Frank, I. Metacaspase involvement in programmed cell death of the marine cyanobacterium Trichodesmium. Environ. Microbiol. 2019, 21, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Vardi, A.; Berman-Frank, I.; Rozenberg, T.; Hadas, O.; Kaplan, A.; Levine, A. Programmed cell death of the dinoflagellate Peridinium gatunense is mediated by CO2 limitation and oxidative stress. Curr. Biol. 1999, 9, 1061–1064. [Google Scholar] [CrossRef] [PubMed]
- Bidle, K.D. Programmed Cell Death in Unicellular Phytoplankton. Curr. Biol. 2016, 26, R594–R607. [Google Scholar] [CrossRef] [PubMed]
- Ianora, A.; Miralto, A. Toxigenic effects of diatoms on grazers, phytoplankton and other microbes: A review. Ecotoxicology 2010, 19, 493–511. [Google Scholar] [CrossRef] [PubMed]
- Coyne, K.J.; Wang, Y.; Johnson, G. Algicidal Bacteria: A Review of Current Knowledge and Applications to Control Harmful Algal Blooms. Front. Microbiol. 2022, 13, 871177. [Google Scholar] [CrossRef]
- Brussaard, C.P. Viral control of phytoplankton populations—A review. J. Eukaryot. Microbiol. 2004, 51, 125–138. [Google Scholar] [CrossRef]
- Alexander, H.; Jenkins, B.D.; Rynearson, T.A.; Dyhrman, S.T. Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proc. Natl. Acad. Sci. USA 2015, 112, E2182–E2190. [Google Scholar] [CrossRef]
- Burson, A.; Stomp, M.; Greenwell, E.; Grosse, J.; Huisman, J. Competition for nutrients and light: Testing advances in resource competition with a natural phytoplankton community. Ecology 2018, 99, 1108–1118. [Google Scholar] [CrossRef]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular Stress Responses: Cell Survival and Cell Death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef]
- Pandey, R.; Vengavasi, K.; Hawkesford, M.J. Plant adaptation to nutrient stress. Plant Physiol. Rep. 2021, 26, 583–586. [Google Scholar] [CrossRef]
- Trachootham, D.; Lu, W.; Ogasawara, M.A.; Rivera-dell Valle, N.; Huang, P. Redox Regulation of Cell Survival. Antioxid. Redox Signal. 2008, 10, 1343–1374. [Google Scholar] [CrossRef]
- Hetz, C.; Papa, F.R. The Unfolded Protein Response and Cell Fate Control. Mol. Cell 2018, 69, 169–181. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006, 12, 440–450. [Google Scholar] [CrossRef]
- Nair, A.; Chauhan, P.; Saha, B.; Kubatzky, K.F. Conceptual Evolution of Cell Signaling. Int. J. Mol. Sci. 2019, 20, 3292. [Google Scholar] [CrossRef]
- Zhang, M.; Lin, Y.; Chen, R.; Yu, H.; Li, Y.; Chen, M.; Dou, C.; Yin, P.; Zhang, L.; Tang, P. Ghost messages: Cell death signals spread. Cell Commun. Signal. 2023, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Rezayian, M.; Niknam, V.; Ebrahimzadeh, H. Oxidative damage and antioxidative system in algae. Toxicol. Rep. 2019, 6, 1309–1313. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.A.; de la Lastra, J.M.P.; Plou, F.J.; Pérez-Lebeña, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- Kari, S.; Subramanian, K.; Altomonte, I.A.; Murugesan, A.; Yli-Harja, O.; Kandhavelu, M. Programmed cell death detection methods: A systematic review and a categorical comparison. Apoptosis 2022, 27, 482–508. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Caspase function in programmed cell death. Cell Death Differ. 2007, 14, 32–43. [Google Scholar] [CrossRef]
- Fujita, J.; Crane, A.M.; Souza, M.K.; Dejosez, M.; Kyba, M.; Flavell, R.A.; Thomson, J.A.; Zwaka, T.P. Caspase Activity Mediates the Differentiation of Embryonic Stem Cells. Cell Stem Cell 2008, 2, 595–601. [Google Scholar] [CrossRef]
- Nakajima, Y.-I.; Kuranaga, E. Caspase-dependent non-apoptotic processes in development. Cell Death Differ. 2017, 24, 1422–1430. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.; Ozmadenci, D.; Ichim, G.; Stupack, D. Caspase-8 function, and phosphorylation, in cell migration. Semin. Cell Dev. Biol. 2018, 82, 105–117. [Google Scholar] [CrossRef]
- Uren, A.G.; O’Rourke, K.; Aravind, L.A.; Pisabarro, M.T.; Seshagiri, S.; Koonin, E.V.; Dixit, V.M. Identification of Paracaspases and Metacaspases: Two Ancient Families of Caspase-like Proteins, One of which Plays a Key Role in MALT Lymphoma. Mol. Cell 2000, 6, 961–967. [Google Scholar] [CrossRef]
- Vercammen, D.; van de Cotte, B.; De Jaeger, G.; Eeckhout, D.; Casteels, P.; Vandepoele, K.; Vandenberghe, I.; Van Beeumen, J.; Inzé, D.; Van Breusegem, F. Type II Metacaspases Atmc4 and Atmc9 of Arabidopsis thaliana Cleave Substrates after Arginine and Lysine. J. Biol. Chem. 2004, 279, 45329–45336. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; Van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis Type I Metacaspases Control Cell Death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.; Kalicharan, R.E.; Kinch, L.; Fernandez, J. Regulating Death and Disease: Exploring the Roles of Metacaspases in Plants and Fungi. Int. J. Mol. Sci. 2022, 24, 312. [Google Scholar] [CrossRef]
- Madeo, F.; Herker, E.; Maldener, C.; Wissing, S.; Lächelt, S.; Herlan, M.; Fehr, M.; Lauber, K.; Sigrist, S.J.; Wesselborg, S.; et al. A Caspase-Related Protease Regulates Apoptosis in Yeast. Mol. Cell 2002, 9, 911–917. [Google Scholar] [CrossRef]
- Zalila, H.; González, I.J.; El-Fadili, A.K.; Delgado, M.B.; Desponds, C.; Schaff, C.; Fasel, N. Processing of metacaspase into a cytoplasmic catalytic domain mediating cell death in Leishmania major. Mol. Microbiol. 2011, 79, 222–239. [Google Scholar] [CrossRef]
- Choi, C.J.; Berges, A.J. New types of metacaspases in phytoplankton reveal diverse origins of cell death proteases. Cell Death Dis. 2013, 4, e490. [Google Scholar] [CrossRef]
- Klemenčič, M.; Funk, C. Structural and functional diversity of caspase homologues in non-metazoan organisms. Protoplasma 2018, 255, 387–397. [Google Scholar] [CrossRef]
- Klemenčič, M.; Funk, C. Type III metacaspases: Calcium-dependent activity proposes new function for the p10 domain. New Phytol. 2018, 218, 1179–1191. [Google Scholar] [CrossRef]
- Acosta-Maspons, A.; Sepúlveda-García, E.; Sánchez-Baldoquín, L.; Marrero-Gutiérrez, J.; Pons, T.; Rocha-Sosa, M.; González, L. Two aspartate residues at the putative p10 subunit of a type II metacaspase from Nicotiana tabacum L. may contribute to the substrate-binding pocket. Planta 2014, 239, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Bidle, K.D.; Bender, S.J. Iron Starvation and Culture Age Activate Metacaspases and Programmed Cell Death in the Marine Diatom Thalassiosira pseudonana. Eukaryot. Cell 2008, 7, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mi, T.; Zhen, Y.; Jing, X.; Liu, Q.; Yu, Z. Metacaspases and programmed cell death in Skeletonema marinoi in response to silicate limitation. J. Plankton Res. 2017, 39, 729–743. [Google Scholar] [CrossRef]
- Wang, H.; Chen, F.; Mi, T.; Liu, Q.; Yu, Z.; Zhen, Y. Responses of Marine Diatom Skeletonema marinoi to Nutrient Deficiency: Programmed Cell Death. Appl. Environ. Microbiol. 2020, 86, e02460-19. [Google Scholar] [CrossRef] [PubMed]
- Ribalet, F.; Wichard, T.; Pohnert, G.; Ianora, A.; Miralto, A.; Casotti, R. Age and nutrient limitation enhance polyunsaturated aldehyde production in marine diatoms. Phytochemistry 2007, 68, 2059–2067. [Google Scholar] [CrossRef] [PubMed]
- Vidoudez, C.; Pohnert, G. Growth phase-specific release of polyunsaturated aldehydes by the diatom Skeletonema marinoi. J. Plankton Res. 2008, 30, 1305–1313. [Google Scholar] [CrossRef]
- Vardi, A.; Bidle, K.D.; Kwityn, C.; Hirsh, D.J.; Thompson, S.M.; Callow, J.A.; Falkowski, P.; Bowler, C. A Diatom Gene Regulating Nitric-Oxide Signaling and Susceptibility to Diatom-Derived Aldehydes. Curr. Biol. 2008, 18, 895–899. [Google Scholar] [CrossRef]
- van Creveld, S.G.; Ben-Dor, S.; Mizrachi, A.; Alcolombri, U.; Hopes, A.; Mock, T.; Rosenwasser, S.; Vardi, A. Biochemical Characterization of a Novel Redox-Regulated Metacaspase in a Marine Diatom. Front. Microbiol. 2021, 12, 688199. [Google Scholar] [CrossRef]
- Chung, C.-C.; Hwang, S.-P.L.; Chang, J. Cooccurrence of ScDSP Gene Expression, Cell Death, and DNA Fragmentation in a Marine Diatom, Skeletonema costatum. Appl. Environ. Microbiol. 2005, 71, 8744–8751. [Google Scholar] [CrossRef]
- Orefice, I.; Musella, M.; Smerilli, A.; Sansone, C.; Chandrasekaran, R.; Corato, F.; Brunet, C. Role of nutrient concentrations and water movement on diatom’s productivity in culture. Sci. Rep. 2019, 9, 1479. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.-B.; Balamurugan, S.; Yang, W.-D.; Wang, X.; Li, H.-Y. Regulatory role of death specific protein in response to nutrient limitation in a marine diatom. Algal Res. 2021, 58, 102392. [Google Scholar] [CrossRef]
- Thamatrakoln, K.; Bailleul, B.; Brown, C.M.; Gorbunov, M.Y.; Kustka, A.B.; Frada, M.; Joliot, P.A.; Falkowski, P.G.; Bidle, K.D. Death-specific protein in a marine diatom regulates photosynthetic responses to iron and light availability. Proc. Natl. Acad. Sci. USA 2013, 110, 20123–20128. [Google Scholar] [CrossRef]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 Is Involved in Cyclic Electron Flow around Photosystem I and Is Essential for Photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Gallina, A.A.; Chung, C.-C.; Casotti, R.; Comunale, V. Expression of death-related genes and reactive oxygen species production in Skeletonema tropicum upon exposure to the polyunsaturated aldehyde octadienal. Adv. Oceanogr. Limnol. 2015, 6, 13–20. [Google Scholar] [CrossRef][Green Version]
- Chung, C.-C.; Hwang, S.-P.L.; Chang, J. Nitric Oxide as a Signaling Factor To Upregulate the Death-Specific Protein in a Marine Diatom, Skeletonema costatum, during Blockage of Electron Flow in Photosynthesis. Appl. Environ. Microbiol. 2008, 74, 6521–6527. [Google Scholar] [CrossRef] [PubMed]
- Rai, G.K.; Kumar, P.; Choudhary, S.M.; Singh, H.; Adab, K.; Kosser, R.; Magotra, I.; Kumar, R.R.; Singh, M.; Sharma, R.; et al. Antioxidant Potential of Glutathione and Crosstalk with Phytohormones in Enhancing Abiotic Stress Tolerance in Crop Plants. Plants 2023, 12, 1133. [Google Scholar] [CrossRef] [PubMed]
- Lauritano, C.; Orefice, I.; Procaccini, G.; Romano, G.; Ianora, A. Key genes as stress indicators in the ubiquitous diatom Skeletonema marinoi. BMC Genom. 2015, 16, 411. [Google Scholar] [CrossRef]
- Hippmann, A.A.; Schuback, N.; Moon, K.M.; McCrow, J.P.; Allen, A.E.; Foster, L.F.; Green, B.R.; Maldonado, M.T. Proteomic analysis of metabolic pathways supports chloroplast-mitochondria cross-talk in a Cu-limited diatom. Plant Direct 2022, 6, e376. [Google Scholar] [CrossRef] [PubMed]
- Szabados, L.; Savouré, A. Proline: A multifunctional amino acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Končitíková, R.; Kopečná, M.; Hájková, E.; Vigouroux, A.; Moréra, S.; Becker, D.F.; Šebela, M.; Tanner, J.J.; Kopečný, D. Structural and biochemical characterization of Aldehyde Dehydrogenase 12, the last enzyme of proline catabolism in plants. J. Mol. Biol. 2019, 431, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jiang, W.; Liu, J.; Li, Y.; Gai, J.; Li, Y. Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genom. 2017, 18, 518. [Google Scholar] [CrossRef]
- Hildebrand, M.; Brzezinski, M.A.; Martin-Jézéquel, V. Silicon Metabolism in Diatoms: Implications for Growth. J. Phycol. 2003, 36, 821–840. [Google Scholar] [CrossRef]
- Bucciarelli, E.; Sunda, W.G. Influence of CO2, nitrate, phosphate, and silicate limitation on intracellular dimethylsulfoniopropionate in batch cultures of the coastal diatom Thalassiosira pseudonana. Limnol. Oceanogr. 2003, 48, 2256–2265. [Google Scholar] [CrossRef]
- Thangaraj, S.; Palanisamy, S.K.; Zhang, G.; Sun, J. Quantitative Proteomic Profiling of Marine Diatom Skeletonema dohrnii in Response to Temperature and Silicate Induced Environmental Stress. Front. Microbiol. 2021, 11, 554832. [Google Scholar] [CrossRef]
- Vaulot, D.; Chisholm, S.W. A simple model of the growth of phytoplankton populations in light/dark cycles. J. Plankton Res. 1987, 9, 345–366. [Google Scholar] [CrossRef]
- Brzezinski, M.A.; Olson, R.J.; Chisholm, S.W. Silicon availability and cell-cycle progression in marine diatoms. Mar. Ecol. Prog. Ser. 1990, 67, 83–96. [Google Scholar] [CrossRef]
- Llamas, A.; Leon-Miranda, E.; Tejada-Jimenez, M. Microalgal and Nitrogen-Fixing Bacterial Consortia: From Interaction to Biotechnological Potential. Plants 2023, 12, 2476. [Google Scholar] [CrossRef]
- Tong, C.Y.; Honda, K.; Derek, C.J.C. A review on microalgal-bacterial co-culture: The multifaceted role of beneficial bacteria towards enhancement of microalgal metabolite production. Environ. Res. 2023, 228, 115872. [Google Scholar] [CrossRef]
- Paul, C.; Pohnert, G. Interactions of the Algicidal Bacterium Kordia algicida with Diatoms: Regulated Protease Excretion for Specific Algal Lysis. PLoS ONE 2011, 6, e21032. [Google Scholar] [CrossRef]
- Havskum, H.; Thingstad, T.F.; Scharek, R.; Peters, F.; Berdalet, E.; Sala, M.M.; Alcaraz, M.; Bangsholt, J.C.; Zweifel, U.L.; Hagström, Å.; et al. Silicate and labile DOC interfere in structuring the microbial food web via algal—Bacterial competition for mineral nutrients: Results of a mesocosm experiment. Limnol. Oceanogr. 2003, 48, 129–140. [Google Scholar] [CrossRef]
- Desbois, A.P.; Lebl, T.; Yan, L.; Smith, V.J. Isolation and structural characterisation of two antibacterial free fatty acids from the marine diatom, Phaeodactylum tricornutum. Appl. Microbiol. Biotechnol. 2008, 81, 755–764. [Google Scholar] [CrossRef]
- Li, Y.; Lei, X.; Zhu, H.; Zhang, H.; Guan, C.; Chen, Z.; Zheng, W.; Fu, L.; Zheng, T. Chitinase producing bacteria with direct algicidal activity on marine diatoms. Sci. Rep. 2016, 6, 21984. [Google Scholar] [CrossRef]
- Wang, H.; Butt, L.; Rooks, P.; Khan, F.; Allen, M.J.; Ali, S.T. Characterisation of algicidal bacterial exometabolites against the lipid-accumulating diatom Skeletonema sp. Algal Res. 2016, 13, 1–6. [Google Scholar] [CrossRef]
- Bedoshvili, Y.; Bayramova, E.; Sudakov, N.; Klimenkov, I.; Kurilkina, M.; Likhoshway, Y.; Zakharova, Y. Impact of Algicidal Bacillus mycoides on Diatom Ulnaria acus from Lake Baikal. Diversity 2021, 13, 469. [Google Scholar] [CrossRef]
- Bayramova, E.M.; Bedoshvili, Y.D.; Likhoshway, Y.V. Molecular and cellular mechanisms of diatom response to environmental changes. Limnol. Freshw. Biol. 2023, 1, 20–30. [Google Scholar] [CrossRef]
- Galachyants, Y.P.; Zakharova, Y.R.; Petrova, D.P.; Morozov, A.A.; Sidorov, I.A.; Marchenkov, A.M.; Logacheva, M.D.; Markelov, M.L.; Khabudaev, K.V.; Likhoshway, Y.V.; et al. Sequencing of the complete genome of an araphid pennate diatom Synedra acus subsp. radians from Lake Baikal. Dokl. Biochem. Biophys. 2015, 461, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Galachyants, Y.P.; Zakharova, Y.R.; Volokitina, N.A.; Morozov, A.A.; Likhoshway, Y.V.; Grachev, M.A. De novo transcriptome assembly and analysis of the freshwater araphid diatom Fragilaria radians, Lake Baikal. Sci. Data 2019, 6, 183. [Google Scholar] [CrossRef]
- Morozov, A.A. Duplications and subdomain shuffling in diatom metacaspases. Limnol. Freshw. Biol. 2019, 3, 240–244. [Google Scholar] [CrossRef]
- Popovskaya, G.I.; Genkal, S.I. On the ecology and morphology of Baikal Synedra acus subsp. radians (Kutz.) Skabitsch. (Bacillariophyta). Inland Water Biol. 1998, 1, 92–95. [Google Scholar]
- Zakharova, Y.; Marchenkov, A.; Petrova, D.; Bukin, Y.; Morozov, A.; Bedoshvili, Y.; Podunay, Y.; Davidovich, O.; Davidovich, N.; Bondar, A.; et al. Delimitation of some taxa of Ulnaria and Fragilaria (Bacillariophyceae) based on genetic, morphological data and mating compatibility. Diversity 2023, 15, 271. [Google Scholar] [CrossRef]
- Usoltseva, M.V.; Titova, L.A.; Firsova, A.D.; Zakharova, Y.R. Long-term dynamics of dominant diatom species abundance of spring phytoplankton in three basins of the pelagic zone of Lake Baikal in 1964-1984 and 2007-2016. Limnol. Freshw. Biol. 2023, 6, 55–62. [Google Scholar] [CrossRef]
- Thumuluri, V.; Armenteros, J.J.A.; Johansen, A.R.; Nielsen, H.; Winther, O. DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 2022, 50, W228–W234. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Seewald, M.; Dadi, T.; Friese, K.; Mi, C.; Boehrer, B.; Schultze, M.; Rinke, K.; Shatwell, T. Unravelling winter diatom blooms in temperate lakes using high frequency data and ecological modeling. Water Res. 2021, 190, 116681. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [PubMed]
- Rigden, D.J.; Galperin, M.Y. The DxDxDG Motif for Calcium Binding: Multiple Structural Contexts and Implications for Evolution. J. Mol. Biol. 2004, 343, 971–984. [Google Scholar] [CrossRef]
- Krell, A.; Funck, D.; Plettner, I.; John, U.; Dieckmann, G. Regulation of Proline Metabolism under Salt Stress in the Psychrophilic Diatom Fragilariopsis cylindrus (Bacillariophyceae)1. J. Phycol. 2007, 43, 753–762. [Google Scholar] [CrossRef]
- Zhang, L.; Becker, D.F. Connecting proline metabolism and signaling pathways in plant senescence. Front. Plant Sci. 2015, 6, 552. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Wang, P.; Wu, X.; Zhou, X.; Ji, G.; Shen, Y.; Gao, Y.; Li, Q.Q.; Liang, J. Distinct genome-wide alternative polyadenylation during the response to silicon availability in the marine diatom Thalassiosira pseudonana. Plant J. 2019, 99, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Dinescu, A.; Anderson, M.E.; Cundari, T.R. Catalytic loop motion in human glutathione synthetase: A molecular modeling approach. Biochem. Biophys. Res. Commun. 2007, 353, 450–456. [Google Scholar] [CrossRef]
- Herrera, K.; Cahoon, R.E.; Kumaran, S.; Jez, J. Reaction mechanism of glutathione synthetase from Arabidopsis thaliana: Site-directed mutagenesis of active site residues. J. Biol. Chem. 2007, 282, 17157–17165. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef]
- Smith, S.R.; Gillard, J.T.F.; Kustka, A.B.; McCrow, J.P.; Badger, J.H.; Zheng, H.; New, A.M.; Dupont, C.L.; Obata, T.; Fernie, A.R.; et al. Transcriptional Orchestration of the Global Cellular Response of a Model Pennate Diatom to Diel Light Cycling under Iron Limitation. PLOS Genet. 2016, 12, e1006490. [Google Scholar] [CrossRef]
- Matthijs, M.; Fabris, M.; Obata, T.; Foubert, I.; Franco-Zorrilla, J.M.; Solano, R.; Fernie, A.R.; Vyverman, W.; Goossens, A. The transcription factor bZIP14 regulates the TCA cycle in the diatom Phaeodactylum tricornutum. EMBO J. 2017, 36, 1559–1576. [Google Scholar] [CrossRef]
- Venekamp, J.H. Regulation of cytosol acidity in plants under conditions of drought. Physiol. Plant. 1989, 76, 112–117. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant, Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Shishlyannikov, S.M.; Zakharova, Y.R.; Volokitina, N.A.; Mikhailov, I.S.; Petrova, D.P.; Likhoshway, Y.V. A procedure for establishing an axenic culture of the diatom Synedra acus subsp. radians (Kütz.) Skabibitsch. from Lake Baikal. Limnol. Oceanogr. Methods 2011, 9, 478–484. [Google Scholar] [CrossRef]
- Thompson, A.S.; Rhodes, J.C.; Pettman, I. Culture Collection of Algae and Protozoa: Catalogue of Strains; Freshwater Biological Association: Ambleside, UK, 1988. [Google Scholar]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009, 23, 205–211. [Google Scholar] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families data base: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, 279–285. [Google Scholar] [CrossRef]
- Marchenkov, A.M.; Petrova, D.P.; Morozov, A.A.; Zakharova, Y.R.; Grachev, M.A.; Bondar, A.A. A family of silicon transporter structural genes in a pennate diatom Synedra ulna subsp. danica (Kütz.) Skabitsch. PLoS ONE 2018, 13, e0203161. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger data sets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, 10.1–10.12. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef]
- Möller, S.; Croning, M.D.R.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef] [PubMed]
- McLuskey, K.; Mottram, J.C. Comparative structural analysis of the caspase family with other clan CD cysteine peptidases. Biochem. J. 2015, 466, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Blom, N.; Sicheritz-Pontén, T.; Gupta, R.; Gammeltoft, S.; Brunak, S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004, 4, 1633–1649. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Owczarzy, R.; Tataurov, A.V.; Wu, Y.; Manthey, J.A.; McQuisten, K.A.; Almabrazi, H.G.; Pedersen, K.F.; Lin, Y.; Garretson, J.; McEntaggart, N.O.; et al. IDT SciTools: A suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res. 2008, 36, W163–W169. [Google Scholar] [CrossRef] [PubMed]
- Alexander, H.; Jenkins, B.D.; Rynearson, T.A.; Saito, M.A.; Mercier, M.L.; Dyhrman, S.T. Identifying reference genes with stable expression from high throughput sequence data. Front. Microbiol. 2012, 3, 385. [Google Scholar] [CrossRef]
- Liu, Q.; Xing, Y.; Li, Y.; Wang, H.; Mi, T.; Zhen, Y.; Yu, Z. Carbon fixation gene expression in Skeletonema marinoi in nitrogen-, phosphate-, silicate-starvation, and low-temperature stress exposure. J. Phycol. 2020, 56, 310–323. [Google Scholar] [CrossRef]
- Xie, F.; Wang, J.; Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef]
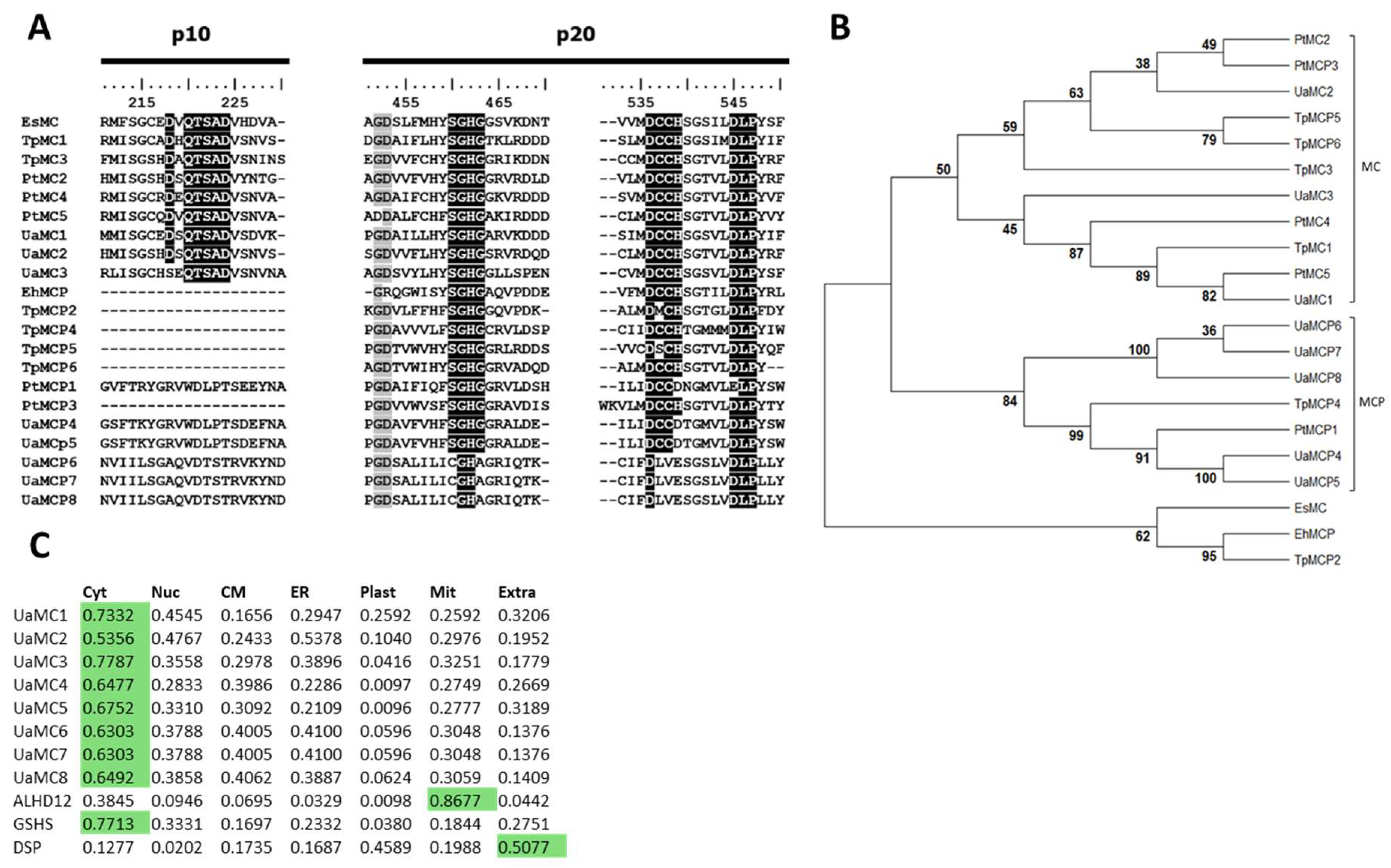

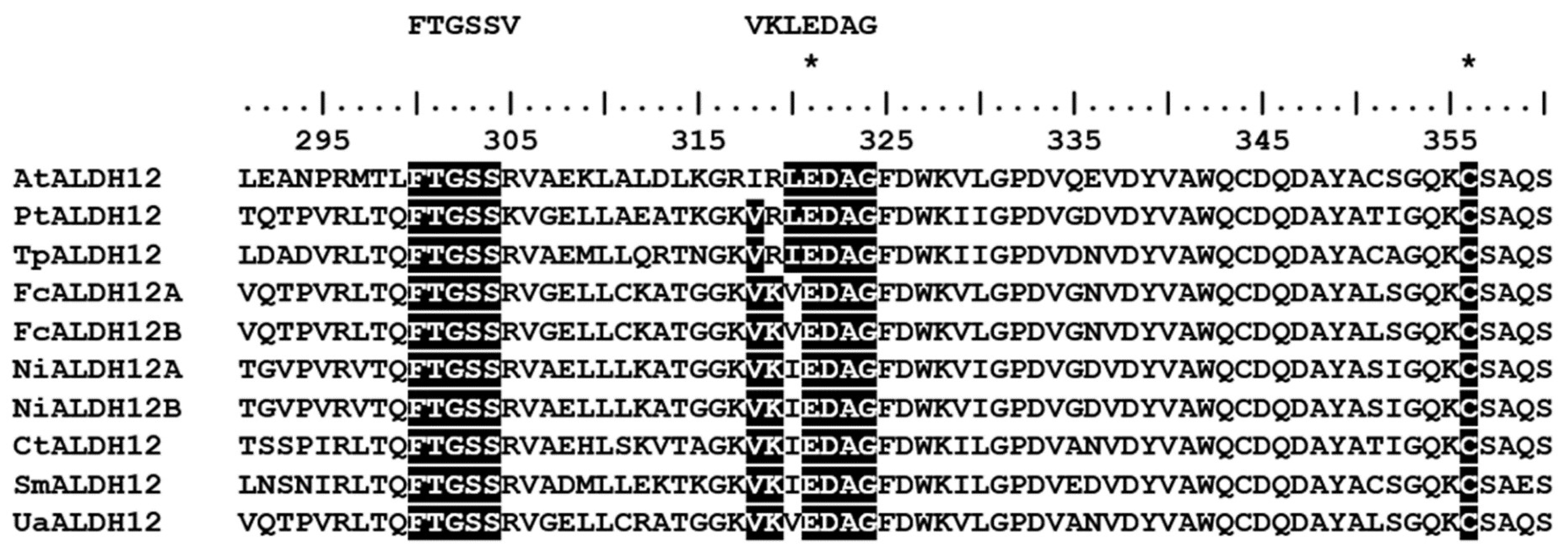
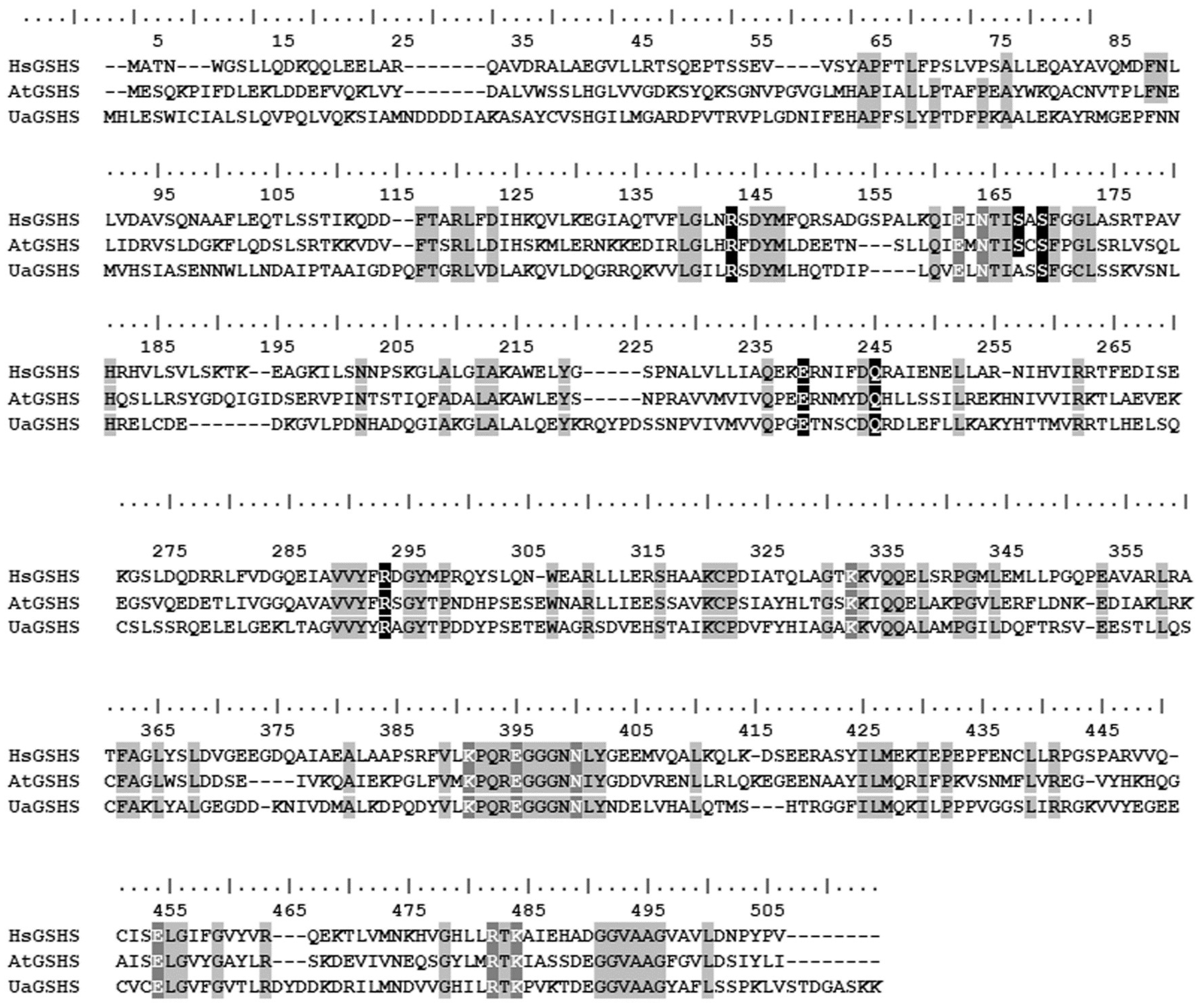
| Species | NCBI Accession Number | Type of Metacaspase | Length, a.a. | Abbreviation | p10 Domain | p20 Domain |
| Phaeodactylum tricornutum | XP_002178108.1 | Metacaspase-like protease | 633 | PtMCP1 | 285–588 | |
| XP_002182518.1 | Metacaspase type III | 369 | PtMC2 | 16–161 | 174–358 | |
| XP_002180075.1 | Metacaspase-like protease | 292 | PtMCP3 | 31–257 | ||
| XP_002182552.1 | Metacaspase type III | 322 | PtMC4 | 7–102 | 116–319 | |
| B7G6D3.2 | Metacaspase type III | 337 | PtMC5 | 6–111 | 124–325 | |
| Thalassiosira pseudonana | XP_002294738.1 | Metacaspase type III | 318 | TpMC1 | 2–112 | 126–315 |
| XP_002292692.1 | Metacaspase-like protease | 208 | TpMCP2 | 20–208 | ||
| XP_002287857.1 | Metacaspase type III | 500 | TpMC3 | 38–273 | 290–484 | |
| XP_002290139.1 | Metacaspase-like protease | 347 | TpMCP4 | 55–304 | ||
| XP_002297230.1 | Metacaspase-like protease | 235 | TpMCP5 | 1–204 | ||
| XP_002295354.1 | Metacaspase-like protease | 159 | TpMCP6 | 11–159 | ||
| Ulnaria acus | OR677812 | Metacaspase type III | 318 | UaMC1 | 7–114 | 127–304 |
| OR677813 | Metacaspase type III | 306 | UaMC2 | 3–103 | 117–291 | |
| OR677814 | Metacaspase type III | 426 | UaMC3 | 7–127 | 141–377 | |
| OR677815 | Metacaspase-like protease | 701 | UaMCP4 | 372–669 | ||
| OR677816 | Metacaspase-like protease | 701 | UaMCP5 | 372–669 | ||
| OR677817 | Metacaspase-like protease | 320 | UaMCP6 | 113–292 | ||
| OR677818 | Metacaspase-like protease | 320 | UaMCP7 | 113–292 | ||
| OR677819 | Metacaspase-like protease | 320 | UaMCP8 | 113–292 | ||
| Out group | ||||||
| Ectocarpus siliculosus | CBN76943.1 | Metacaspase type III | 354 | ESMC | 42–150 | 166–353 |
| Emiliania huxleyi | XP_005790400.1 | Metacaspase-like protease | 388 | EhMCP | 25–263 |
| Gene | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
| 18S rRNA | CTTCTTAGAGGGACGTGCGTTC | TCTCGGCCAAGGTACACTCG |
| E2 | AGGATCAGTGGAGTCCTGCT | TTTCGACGTCCACTCTCGTG |
| MC1 | TGAGCCAGGTGATGCAATCC | CTTCAGCGGCTTGATGATCGT |
| MC2 | CTGGACATGGAAGTCGTGTACG | CTTCGCAAAGGCTTCACAAGC |
| MC3 | ACCAATTTCCCGAGCAGGAT | CCATGACCGGAATAGTGCAGA |
| DSP | TGATGGAAGTGGTACGGTGG | GGAGCTTCGGTCAACCACTC |
| ALDH12 | GTCGGTCATAGATGGCCTTACG | GAAGATCGACATCGGCAGGA |
| GSHS | ACCCGTAGGTGGCTCATTG | CGTCTTCACTGGCTTTGTGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayramova, E.; Petrova, D.; Marchenkov, A.; Morozov, A.; Galachyants, Y.; Zakharova, Y.; Bedoshvili, Y.; Likhoshway, Y. Differential Expression of Stress Adaptation Genes in a Diatom Ulnaria acus under Different Culture Conditions. Int. J. Mol. Sci. 2024, 25, 2314. https://doi.org/10.3390/ijms25042314
Bayramova E, Petrova D, Marchenkov A, Morozov A, Galachyants Y, Zakharova Y, Bedoshvili Y, Likhoshway Y. Differential Expression of Stress Adaptation Genes in a Diatom Ulnaria acus under Different Culture Conditions. International Journal of Molecular Sciences. 2024; 25(4):2314. https://doi.org/10.3390/ijms25042314
Chicago/Turabian StyleBayramova, Elvira, Darya Petrova, Artyom Marchenkov, Alexey Morozov, Yuri Galachyants, Yulia Zakharova, Yekaterina Bedoshvili, and Yelena Likhoshway. 2024. "Differential Expression of Stress Adaptation Genes in a Diatom Ulnaria acus under Different Culture Conditions" International Journal of Molecular Sciences 25, no. 4: 2314. https://doi.org/10.3390/ijms25042314
APA StyleBayramova, E., Petrova, D., Marchenkov, A., Morozov, A., Galachyants, Y., Zakharova, Y., Bedoshvili, Y., & Likhoshway, Y. (2024). Differential Expression of Stress Adaptation Genes in a Diatom Ulnaria acus under Different Culture Conditions. International Journal of Molecular Sciences, 25(4), 2314. https://doi.org/10.3390/ijms25042314







