Association of MDM2 Overexpression in Ameloblastomas with MDM2 Amplification and BRAFV600E Expression
Abstract
1. Introduction
2. Results
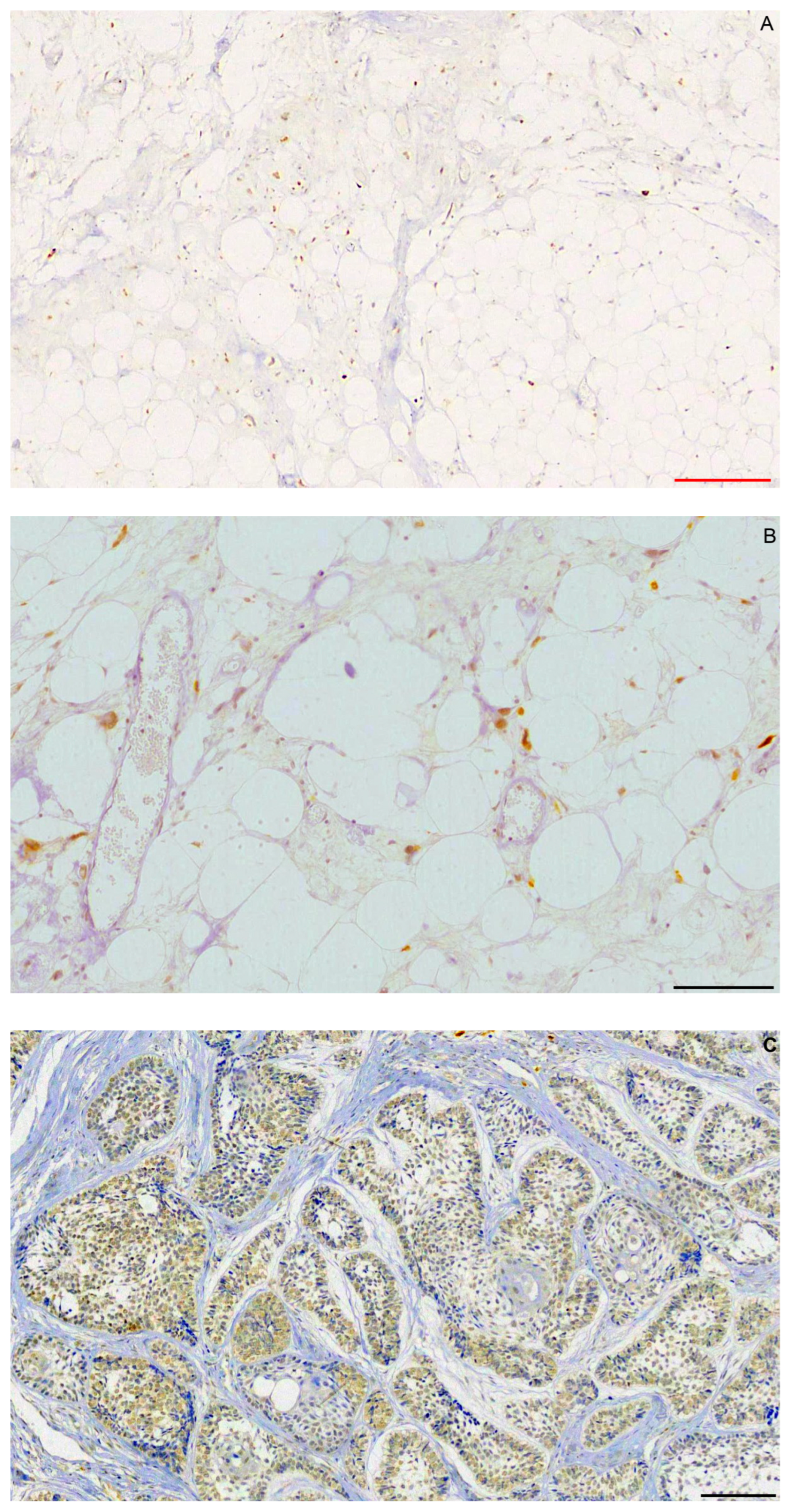
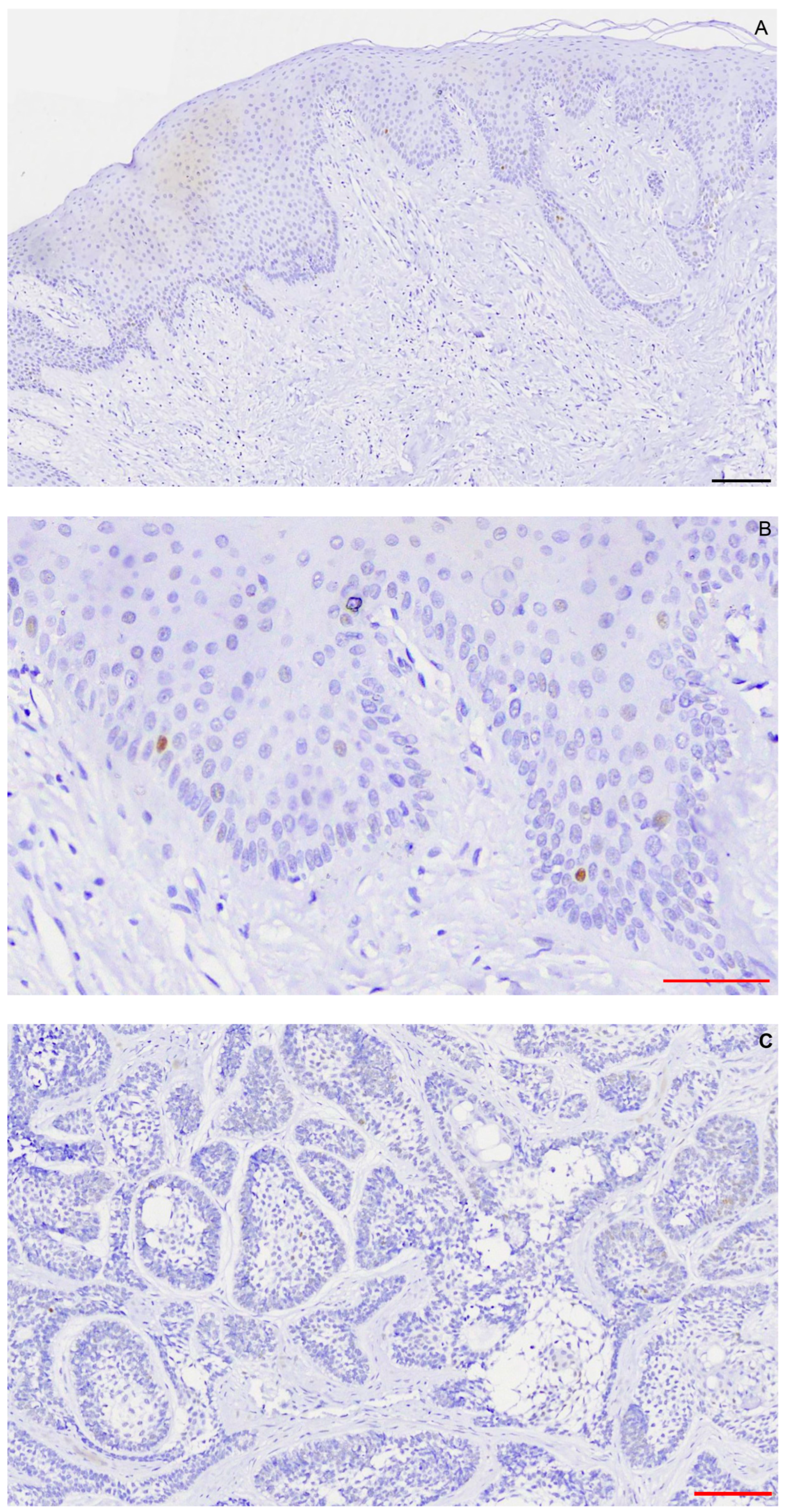
2.1. Immunohistochemistry
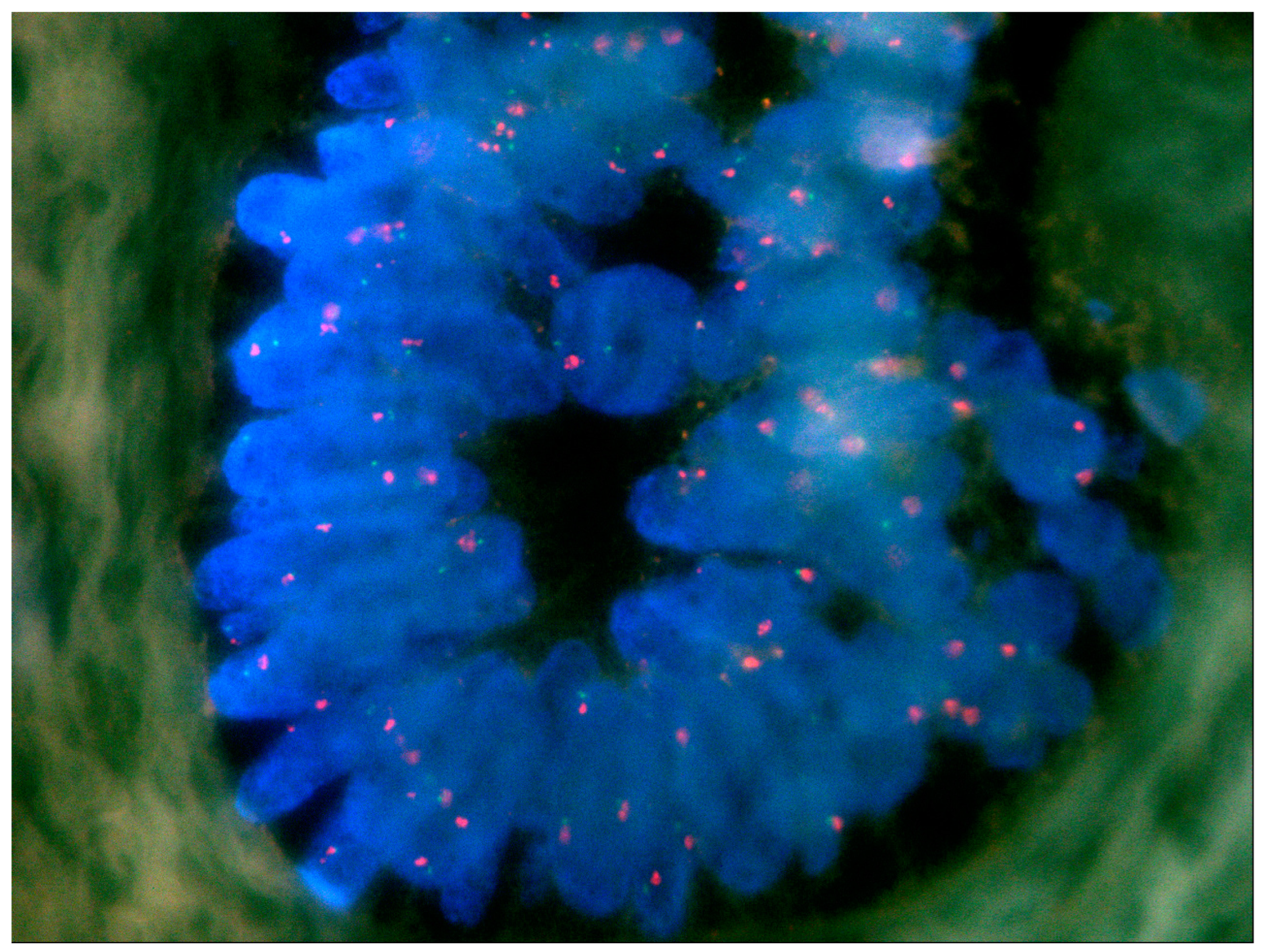
2.2. FISH
3. Discussion
4. Materials and Methods
4.1. Immunohistochemistry
4.2. FISH
4.3. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Avelar, R.L.; Primo, B.T.; Pinheiro-Nogueira, C.B.; Studart-Soares, E.C.; de Oliveira, R.B.; Romulo de Medeiros, J.; Hernandez, P.A. Worldwide incidence of odontogenic tumors. J. Craniofac. Surg. 2011, 22, 2118–2123. [Google Scholar] [CrossRef]
- Siriwardena, B.; Crane, H.; O’Neill, N.; Abdelkarim, R.; Brierley, D.J.; Franklin, C.D.; Farthing, P.M.; Speight, P.M.; Hunter, K.D. Odontogenic tumors and lesions treated in a single specialist oral and maxillofacial pathology unit in the United Kingdom in 1992–2016. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 127, 151–166. [Google Scholar] [CrossRef]
- da Silva, L.A.M.; Filho, S.R.C.; Saraiva, M.J.D.; Maia, C.R.; Santos, C.; Santos, P.P.A. Clinical, Radiographic and Histopathological Analysis of Craniopharyngiomas and Ameloblastomas: A Systematic Review. Head Neck Pathol. 2022, 16, 1195–1222. [Google Scholar] [CrossRef]
- Boffano, P.; Cavarra, F.; Tricarico, G.; Masu, L.; Brucoli, M.; Ruslin, M.; Forouzanfar, T.; Ridwan-Pramana, A.; Rodriguez-Santamarta, T.; Rui Ranz, M.; et al. The epidemiology and management of ameloblastomas: A European multicenter study. J. Craniomaxillofac. Surg. 2021, 49, 1107–1112. [Google Scholar] [CrossRef]
- El-Naggar, A.K.; Chan, J.K.C.; Grandis, J.R.; Takata, T.; Slootweg, P.J. Odontogenic and maxillofacial bone tumors. In WHO Classification of Head and Neck Tumours, 4th ed.; IARC: Lyon, France, 2017; pp. 215–219. [Google Scholar]
- McClary, A.C.; West, R.B.; Pollack, J.R.; Fischbein, N.J.; Holsinger, C.F.; Sunwoo, J.; Colevas, A.D.; Sirjani, D. Ameloblastoma: A clinical review and trends in management. Eur. Arch. Otorhinolaryngol. 2015, 273, 1649–1661. [Google Scholar] [CrossRef] [PubMed]
- Hendra, F.N.; Helder, M.N.; Ruslin, M.; Van Cann, E.M.; Forouzanfar, T. A network meta-analysis assessing the effectiveness of various radical and conservative surgical approaches regarding recurrence in treating solid/multicystic ameloblastomas. Sci. Rep. 2023, 13, 8445. [Google Scholar] [CrossRef]
- Laborde, A.; Nicot, R.; Wojcik, T.; Ferri, J.; Raoul, G. Ameloblastoma of the jaws: Management and recurrence rate. Eur. Ann. Otorhinolaryngol. Head Neck Dis. 2017, 134, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Armocida, D.; Berra, L.V.; Pucci, R.; Battisti, A.; Della Monaca, M.; Valentini, V.; Santoro, A. Ameloblastoma and Intracranial Involvement: The Current Challenge of the Radical Surgical Treatment. Comprehensive Review of the Literature and Institution experience. J. Maxillofac. Oral. Surg. 2022, 21, 34–43. [Google Scholar] [CrossRef]
- Sweeney, R.T.; McClary, A.C.; Myers, B.R.; Biscocho, J.; Neahring, L.; Kwei, K.A.; Qu, K.; Gong, X.; Ng, T.; Jones, C.D.; et al. Identification of recurrent SMO and BRAF mutations in ameloblastomas. Nat. Genet. 2014, 46, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Gultekin, S.E.; Aziz, R.; Heydt, C.; Senguven, B.; Zoller, J.; Safi, A.F.; Kreppel, M.; Buettner, R. The landscape of genetic alterations in ameloblastomas relates to clinical features. Virchows Arch. 2018, 472, 807–814. [Google Scholar] [CrossRef]
- Brown, N.A.; Rolland, D.; McHugh, J.B.; Weigelin, H.C.; Zhao, L.; Lim, M.S.; Elenitoba-Johnson, K.S.; Betz, B.L. Activating FGFR2-RAS-BRAF mutations in ameloblastoma. Clin. Cancer Res. 2014, 20, 5517–5526. [Google Scholar] [CrossRef]
- Kurppa, K.J.; Caton, J.; Morgan, P.R.; Ristimaki, A.; Ruhin, B.; Kellokoski, J.; Elenius, K.; Heikinheimo, K. High frequency of BRAF V600E mutations in ameloblastoma. J. Pathol. 2014, 232, 492–498. [Google Scholar] [CrossRef]
- Soltani, M.; Tabatabaiefar, M.A.; Mohsenifar, Z.; Pourreza, M.R.; Moridnia, A.; Shariati, L.; Razavi, S.M. Genetic study of the BRAF gene reveals new variants and high frequency of the V600E mutation among Iranian ameloblastoma patients. J. Oral. Pathol. Med. 2018, 47, 86–90. [Google Scholar] [CrossRef]
- Diniz, M.G.; Gomes, C.C.; Guimaraes, B.V.; Castro, W.H.; Lacerda, J.C.; Cardoso, S.V.; de Faria, P.R.; Dias, F.L.; Eisenberg, A.L.; Loyola, A.M.; et al. Assessment of BRAFV600E and SMOF412E mutations in epithelial odontogenic tumours. Tumour Biol. 2015, 36, 5649–5653. [Google Scholar] [CrossRef]
- Oh, K.Y.; Kim, J.H.; Cho, S.D.; Yoon, H.J.; Lee, J.I.; Hong, S.D. BRAF V600E and previously unidentified KRAS G12C mutations in odontogenic tumors may affect MAPK activation differently depending on tumor type. Genes Chromosomes Cancer 2022, 61, 481–490. [Google Scholar] [CrossRef] [PubMed]
- do Canto, A.M.; da Silva Marcelino, B.M.R.; Schussel, J.L.; Wastner, B.F.; Sassi, L.M.; Correa, L.; de Freitas, R.R.; Hasseus, B.; Kjeller, G.; Junior, C.A.L.; et al. Immunohistochemical analysis of BRAF V600E mutation in ameloblastomas. Clin. Oral. Investig. 2019, 23, 779–784. [Google Scholar] [CrossRef]
- Kaye, F.J.; Ivey, A.M.; Drane, W.E.; Mendenhall, W.M.; Allan, R.W. Clinical and radiographic response with combined BRAF-targeted therapy in stage 4 ameloblastoma. J. Natl. Cancer Inst. 2015, 107, 378. [Google Scholar] [CrossRef] [PubMed]
- Faden, D.L.; Algazi, A. Durable treatment of ameloblastoma with single agent BRAFi Re: Clinical and radiographic response with combined BRAF-targeted therapy in stage 4 ameloblastoma. J. Natl. Cancer Inst. 2017, 109, djw190. [Google Scholar] [CrossRef] [PubMed]
- Broudic-Guibert, M.; Blay, J.Y.; Vazquez, L.; Evrard, A.; Karanian, M.; Taieb, S.; Hoog-Labouret, N.; Oukhatar, C.M.A.; Boustany-Grenier, R.; Arnaud, A. Persistent response to vemurafenib in metastatic ameloblastoma with BRAF mutation: A case report. J. Med. Case Rep. 2019, 13, 245. [Google Scholar] [CrossRef]
- Fernandes, G.S.; Girardi, D.M.; Bernardes, J.P.G.; Fonseca, F.P.; Fregnani, E.R. Clinical benefit and radiological response with BRAF inhibitor in a patient with recurrent ameloblastoma harboring V600E mutation. BMC Cancer 2018, 18, 887. [Google Scholar] [CrossRef]
- Brunet, M.; Khalifa, E.; Italiano, A. Enabling Precision Medicine for Rare Head and Neck Tumors: The Example of BRAF/MEK Targeting in Patients with Metastatic Ameloblastoma. Front. Oncol. 2019, 9, 1204. [Google Scholar] [CrossRef] [PubMed]
- Abramson, Z.; Dayton, O.L.; Drane, W.E.; Mendenhall, W.M.; Kaye, F.J. Managing stage 4 ameloblastoma with dual BRAF/MEK inhibition: A case report with 8-year clinical follow-up. Oral Oncol. 2022, 128, 105854. [Google Scholar] [CrossRef] [PubMed]
- Hirschhorn, A.; Campino, G.A.; Vered, M.; Greenberg, G.; Yacobi, R.; Yahalom, R.; Barshack, I.; Toren, A.; Amariglio, N.; Rechavi, G. Upfront rational therapy in BRAF V600E mutated pediatric ameloblastoma promotes ad integrum mandibular regeneration. J. Tissue Eng. Regen. Med. 2021, 15, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Pollack, J.R.; Kaplan, M.J.; Colevas, A.D.; West, R.B. BRAF inhibitor treatment of primary BRAF-mutant ameloblastoma with pathologic assessment of response. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2016, 122, e5–e7. [Google Scholar] [CrossRef] [PubMed]
- Grynberg, S.; Vered, M.; Shapira-Frommer, R.; Asher, N.; Ben-Betzalel, G.; Stoff, R.; Steinberg, Y.; Amariglio, N.; Greenberg, G.; Barshack, I.; et al. Neoadjuvant BRAF Targeted Therapy for Ameloblastoma of the Mandible: An Organ Preservation Approach. J. Natl. Cancer Inst. 2023, djad232. [Google Scholar] [CrossRef]
- Carvalhais, J.; Aguiar, M.; Araujo, V.; Araujo, N.; Gomez, R. p53 and MDM2 expression in odontogenic cysts and tumours. Oral Dis. 1999, 5, 218–222. [Google Scholar] [CrossRef]
- Sandra, F.; Nakamura, N.; Kanematsu, T.; Hirata, M.; Ohishi, M. The role of MDM2 in the proliferative activity of ameloblastoma. Oral Oncol. 2002, 38, 153–157. [Google Scholar] [CrossRef]
- Kumamoto, H.; Izutsu, T.; Ohki, K.; Takahashi, N.; Ooya, K. p53 gene status and expression of p53, MDM2, and p14 proteins in ameloblastomas. J. Oral Pathol. Med. 2004, 33, 292–299. [Google Scholar] [CrossRef]
- Sharifi-Sistani, N.; Zartab, H.; Babakoohi, S.; Saghravanian, N.; Jamshidi, S.; Esmaili, H.; Mohtasham, N.; Zamanzadeh, M.; Abbaszadeh-Bidokhty, H. Immunohistochemical comparison of the expression of p53 and MDM2 proteins in ameloblastomas and keratocystic odontogenic tumors. J. Craniofac. Surg. 2011, 22, 1652–1656. [Google Scholar] [CrossRef]
- Krishna, A.; Kaveri, H.; Naveen Kumar, R.K.; Kumaraswamy, K.L.; Shylaja, S.; Murthy, S. Overexpression of MDM2 protein in ameloblastomas as compared to adenomatoid odontogenic tumor. J. Cancer Res. Ther. 2012, 8, 232–237. [Google Scholar] [CrossRef]
- Singh, A.; Jain, A.; Shetty, D.C.; Rathore, A.S.; Juneja, S. Immunohistochemical expression of p53 and murine double minute 2 protein in odontogenic keratocyst versus variants of ameloblastoma. J. Cancer Res. Ther. 2020, 16, 521–529. [Google Scholar] [CrossRef]
- Udeabor, S.E.; Adisa, A.O.; Lawal, A.O.; Barbeck, M.; Booms, P.; Sader, R.A.; Ghanaati, S. PTCH-1 and MDM2 expression in ameloblastoma from a West African sub-population: Implication for chemotherapeutics. Pan Afr. Med. J. 2015, 20, 140. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, J.J. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010, 24, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Fakharzadeh, S.S.; Trusko, S.P.; George, D.L. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991, 10, 1565–1569. [Google Scholar] [CrossRef] [PubMed]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Honda, R.; Tanaka, H.; Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997, 420, 25–27. [Google Scholar] [CrossRef] [PubMed]
- Kubbutat, M.H.; Jones, S.N.; Vousden, K.H. Regulation of p53 stability by Mdm2. Nature 1997, 387, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Lehman, J.A.; Mayo, L.D. Integration of DNA damage and repair with murine double-minute 2 (Mdm2) in tumorigenesis. Int. J. Mol. Sci. 2012, 13, 16373–16386. [Google Scholar] [CrossRef] [PubMed]
- Juven-Gershon, T.; Oren, M. Mdm2: The ups and downs. Mol. Med. 1999, 5, 71–83. [Google Scholar] [CrossRef]
- Fu, W.; Ma, Q.; Chen, L.; Li, P.; Zhang, M.; Ramamoorthy, S.; Nawaz, Z.; Shimojima, T.; Wang, H.; Yang, Y.; et al. MDM2 acts downstream of p53 as an E3 ligase to promote FOXO ubiquitination and degradation. J. Biol. Chem. 2009, 284, 13987–14000. [Google Scholar] [CrossRef]
- Yang, J.Y.; Zong, C.S.; Xia, W.; Yamaguchi, H.; Ding, Q.; Xie, X.; Lang, J.Y.; Lai, C.C.; Chang, C.J.; Huang, W.C.; et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 2008, 10, 138–148. [Google Scholar] [CrossRef]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef]
- Levav-Cohen, Y.; Goldberg, Z.; Tan, K.H.; Alsheich-Bartok, O.; Zuckerman, V.; Haupt, S.; Haupt, Y. The p53-Mdm2 Loop: A Critical Juncture of Stress Response. In Mutant p53 and MDM2 in Cancer; Deb, S.P., Deb, S., Eds.; Springer: Dordrecht, The Netherland, 2014; Volume 85, pp. 161–186. [Google Scholar]
- Vogelstein, B.; Lane, D.; Levine, A.J. Surfing the p53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef]
- Pilotti, S.; Della Torre, G.; Lavarino, C.; Di Palma, S.; Sozzi, G.; Minoletti, F.; Rao, S.; Pasquini, G.; Azzarelli, A.; Rilke, F.; et al. Distinct mdm2/p53 expression patterns in liposarcoma subgroups: Implications for different pathogenetic mechanisms. J. Pathol. 1997, 181, 14–24. [Google Scholar] [CrossRef]
- Eischen, C.M.; Lozano, G. The Mdm network and its regulation of p53 activities: A rheostat of cancer risk. Hum. Mutat. 2014, 35, 728–737. [Google Scholar] [CrossRef] [PubMed]
- Ware, P.L.; Snow, A.N.; Gvalani, M.; Pettenati, M.J.; Qasem, S.A. MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: Characterizing progression to high-grade tumors. Am. J. Clin. Pathol. 2014, 141, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Tisato, V.; Voltan, R.; Gonelli, A.; Secchiero, P.; Zauli, G. MDM2/X inhibitors under clinical evaluation: Perspectives for the management of hematological malignancies and pediatric cancer. J. Hematol. Oncol. 2017, 10, 133. [Google Scholar] [CrossRef] [PubMed]
- Gembarska, A.; Luciani, F.; Fedele, C.; Russell, E.A.; Dewaele, M.; Villar, S.; Zwolinska, A.; Haupt, S.; de Lange, J.; Yip, D.; et al. MDM4 is a key therapeutic target in cutaneous melanoma. Nat. Med. 2012, 18, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Polsky, D.; Bastian, B.C.; Hazan, C.; Melzer, K.; Pack, J.; Houghton, A.; Busam, K.; Cordon-Cardo, C.; Osman, I. HDM2 protein overexpression, but not gene amplification, is related to tumorigenesis of cutaneous melanoma. Cancer Res. 2001, 61, 7642–7646. [Google Scholar] [PubMed]
- Lu, M.; Breyssens, H.; Salter, V.; Zhong, S.; Hu, Y.; Baer, C.; Ratnayaka, I.; Sullivan, A.; Brown, N.R.; Endicott, J.; et al. Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer Cell 2013, 23, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Thiel, A.; Ristimaki, A. Toward a Molecular Classification of Colorectal Cancer: The Role of BRAF. Front. Oncol. 2013, 3, 281. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Njauw, C.N.; Taylor, M.; Neel, V.; Flaherty, K.T.; Tsao, H. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J. Investig. Dermatol. 2012, 132, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.; Garcia, L.F.; Mannella, V.; Gammon, L.; Borg, T.M.; Maffucci, T.; Scatolini, M.; Chiorino, G.; Vergani, E.; Rodolfo, M.; et al. Targeting p63 Upregulation Abrogates Resistance to MAPK Inhibitors in Melanoma. Cancer Res. 2020, 80, 2676–2688. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.; Goldblum, J.R.; Turner, S.; Tubbs, R.R.; Wang, W.L.; Lazar, A.J.; Rubin, B.P. Detection of MDM2 gene amplification or protein expression distinguishes sclerosing mesenteritis and retroperitoneal fibrosis from inflammatory well-differentiated liposarcoma. Mod. Pathol. 2009, 22, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Pfister, C.; Larue, H.; Moore, L.; Lacombe, L.; Veilleux, C.; Tetu, B.; Meyer, F.; Fradet, Y. Tumorigenic pathways in low-stage bladder cancer based on p53, MDM2 and p21 phenotypes. Int. J. Cancer 2000, 89, 100–104. [Google Scholar] [CrossRef]
- Lianes, P.; Orlow, I.; Zhang, Z.F.; Oliva, M.R.; Sarkis, A.S.; Reuter, V.E.; Cordon-Cardo, C. Altered patterns of MDM2 and TP53 expression in human bladder cancer. J. Natl. Cancer Inst. 1994, 86, 1325–1330. [Google Scholar] [CrossRef] [PubMed]
- Schmitz-Drager, B.J.; Kushima, M.; Goebell, P.; Jax, T.W.; Gerharz, C.D.; Bultel, H.; Schulz, W.A.; Ebert, T.; Ackermann, R. p53 and MDM2 in the development and progression of bladder cancer. Eur. Urol. 1997, 32, 487–493. [Google Scholar]
- Hu, S. Unraveling the Trasncriptome of Odontogenic Tumors. PhD Thesis, The University of North Carolina at Chapel Hill, Chapel Hill, NC, USA, 2016. [Google Scholar]
- Diniz, M.G.; Duarte, A.P.; Villacis, R.A.; Guimaraes, B.V.A.; Duarte, L.C.P.; Rogatto, S.R.; Gomez, R.S.; Gomes, C.C. Rare copy number alterations and copy-neutral loss of heterozygosity revealed in ameloblastomas by high-density whole-genome microarray analysis. J. Oral. Pathol. Med. 2017, 46, 371–376. [Google Scholar] [CrossRef]
- Capoulade, C.; Bressac-de Paillerets, B.; Lefrere, I.; Ronsin, M.; Feunteun, J.; Tursz, T.; Wiels, J. Overexpression of MDM2, due to enhanced translation, results in inactivation of wild-type p53 in Burkitt’s lymphoma cells. Oncogene 1998, 16, 1603–1610. [Google Scholar] [CrossRef]
- Bueso-Ramos, C.E.; Manshouri, T.; Haidar, M.A.; Yang, Y.; McCown, P.; Ordonez, N.; Glassman, A.; Sneige, N.; Albitar, M. Abnormal expression of MDM-2 in breast carcinomas. Breast Cancer Res. Treat. 1996, 37, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Rathinavelu, P.; Malave, A.; Raney, S.R.; Hurst, J.; Roberson, C.T.; Rathinavelu, A. Expression of mdm-2 oncoprotein in the primary and metastatic sites of mammary tumor (GI-101) implanted athymic nude mice. Cancer Biochem. Biophys. 1999, 17, 133–146. [Google Scholar]
- Shiina, H.; Igawa, M.; Shigeno, K.; Yamasaki, Y.; Urakami, S.; Yoneda, T.; Wada, Y.; Honda, S.; Nagasaki, M. Clinical significance of mdm2 and p53 expression in bladder cancer. A comparison with cell proliferation and apoptosis. Oncology 1999, 56, 239–247. [Google Scholar] [CrossRef]
- Cordon-Cardo, C.; Latres, E.; Drobnjak, M.; Oliva, M.R.; Pollack, D.; Woodruff, J.M.; Marechal, V.; Chen, J.; Brennan, M.F.; Levine, A.J. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994, 54, 794–799. [Google Scholar]
- Landers, J.E.; Cassel, S.L.; George, D.L. Translational enhancement of mdm2 oncogene expression in human tumor cells containing a stabilized wild-type p53 protein. Cancer Res. 1997, 57, 3562–3568. [Google Scholar]
- Landers, J.E.; Haines, D.S.; Strauss, J.F., 3rd; George, D.L. Enhanced translation: A novel mechanism of mdm2 oncogene overexpression identified in human tumor cells. Oncogene 1994, 9, 2745–2750. [Google Scholar] [PubMed]
- Yam, C.H.; Siu, W.Y.; Arooz, T.; Chiu, C.H.; Lau, A.; Wang, X.Q.; Poon, R.Y. MDM2 and MDMX inhibit the transcriptional activity of ectopically expressed SMAD proteins. Cancer Res. 1999, 59, 5075–5078. [Google Scholar] [PubMed]
- Martins-de-Barros, A.V.; Anjos, R.S.D.; Silva, C.C.G.; Silva, E.; Araujo, F.; Carvalho, M.V. Diagnostic accuracy of immunohistochemistry compared with molecular tests for detection of BRAF V600E mutation in ameloblastomas: Systematic review and meta-analysis. J. Oral. Pathol. Med. 2022, 51, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Karathanasi, V.; Tosios, K.I.; Nikitakis, N.G.; Piperi, E.; Koutlas, I.; Trimis, G.; Sklavounou, A. TGF-beta1, Smad-2/-3, Smad-1/-5/-8, and Smad-4 signaling factors are expressed in ameloblastomas, adenomatoid odontogenic tumors, and calcifying cystic odontogenic tumors: An immunohistochemical study. J. Oral Pathol. Med. 2013, 42, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Shibata, T.; Nakata, D.; Chiba, I.; Yamashita, T.; Abiko, Y.; Tada, M.; Moriuchi, T. Detection of TP53 mutation in ameloblastoma by the use of a yeast functional assay. J. Oral Pathol. Med. 2002, 31, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Bartels, S.; Adisa, A.; Aladelusi, T.; Lemound, J.; Stucki-Koch, A.; Hussein, S.; Kreipe, H.; Hartmann, C.; Lehmann, U.; Hussein, K. Molecular defects in BRAF wild-type ameloblastomas and craniopharyngiomas-differences in mutation profiles in epithelial-derived oropharyngeal neoplasms. Virchows Arch. 2018, 472, 1055–1059. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T. Small-molecule antagonists of p53-MDM2 binding: Research tools and potential therapeutics. Cell Cycle 2004, 3, 419–421. [Google Scholar] [CrossRef]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Ji, Z.; Kumar, R.; Taylor, M.; Rajadurai, A.; Marzuka-Alcala, A.; Chen, Y.E.; Njauw, C.N.; Flaherty, K.; Jonsson, G.; Tsao, H. Vemurafenib synergizes with nutlin-3 to deplete survivin and suppresses melanoma viability and tumor growth. Clin. Cancer Res. 2013, 19, 4383–4391. [Google Scholar] [CrossRef]
- Saiki, A.Y.; Caenepeel, S.; Yu, D.; Lofgren, J.A.; Osgood, T.; Robertson, R.; Canon, J.; Su, C.; Jones, A.; Zhao, X.; et al. MDM2 antagonists synergize broadly and robustly with compounds targeting fundamental oncogenic signaling pathways. Oncotarget 2014, 5, 2030–2043. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.F.; Lu, P.H.; Tseng, C.H.; Wang, Y.P.; Lee, J.J.; Chiang, C.P. Factors affecting the accuracy of anti-BRAF V600E immunohistochemistry results in ameloblastomas. J. Oral. Pathol. Med. 2023, 52, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Cappuzzo, F.; Hirsch, F.R.; Rossi, E.; Bartolini, S.; Ceresoli, G.L.; Bemis, L.; Haney, J.; Witta, S.; Danenberg, K.; Domenichini, I.; et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J. Natl. Cancer Inst. 2005, 97, 643–655. [Google Scholar] [CrossRef]
- Weaver, J.; Downs-Kelly, E.; Goldblum, J.R.; Turner, S.; Kulkarni, S.; Tubbs, R.R.; Rubin, B.P.; Skacel, M. Fluorescence in situ hybridization for MDM2 gene amplification as a diagnostic tool in lipomatous neoplasms. Mod. Pathol. 2008, 21, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Barril, N.; Oliveira, P.R.; Tajara, E.H. Monosomy 22 and del(10)(p12) in an ameloblastoma previously diagnosed as an adenoid cystic carcinoma of the salivary gland. Cancer Genet. Cytogenet. 1996, 91, 74–76. [Google Scholar] [CrossRef]
- Jaaskelainen, K.; Jee, K.J.; Leivo, I.; Saloniemi, I.; Knuutila, S.; Heikinheimo, K. Cell proliferation and chromosomal changes in human ameloblastoma. Cancer Genet. Cytogenet. 2002, 136, 31–37. [Google Scholar] [CrossRef]
- Stenman, G.; Sandros, J.; Mark, J.; Happonen, R.P. Observations by G-banding in benign odontogenic tumors. Cancer Genet. Cytogenet. 1986, 19, 253–259. [Google Scholar] [CrossRef]
- Toida, M.; Balazs, M.; Treszl, A.; Rakosy, Z.; Kato, K.; Yamazaki, Y.; Matsui, T.; Suwa, T.; Hatakeyama, D.; Makita, H.; et al. Analysis of ameloblastomas by comparative genomic hybridization and fluorescence in situ hybridization. Cancer Genet. Cytogenet. 2005, 159, 99–104. [Google Scholar] [CrossRef] [PubMed]

| Gender | Age | Jaw | Histological Subtype | MDM2 | BRAFV600E | p53 |
|---|---|---|---|---|---|---|
| M | 52 | maxilla | reticular | 3 | 1 | - |
| M | 33 | mandible | follicular | 3 | 1 | - |
| F | 37 | mandible | follicular | 3 | 1 | + |
| M | 45 | maxilla | reticular | 3 | 1 | - |
| F | 55 | maxilla | follicular | 3 | 0 | - |
| M | 61 | maxilla | follicular | 3 | 1 | - |
| M | 46 | mandible | follicular | 2 | 0 | - |
| M | 49 | mandible | follicular | 2 | 0 | - |
| M | 24 | mandible | follicular | 3 | 0 | - |
| F | 77 | mandible | follicular | 3 | 1 | - |
| M | 67 | mandible | follicular | 3 | 1 | - |
| F | 31 | mandible | follicular | 3 | 0 | - |
| F | 55 | mandible | follicular | 3 | 0 | - |
| M | 43 | maxilla | follicular | 3 | 0 | - |
| F | 41 | maxilla | follicular | 2 | 0 | - |
| M | 38 | mandible | follicular | 3 | 0 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tosios, K.I.; Kalogirou, E.-M.; Koutlas, I.G. Association of MDM2 Overexpression in Ameloblastomas with MDM2 Amplification and BRAFV600E Expression. Int. J. Mol. Sci. 2024, 25, 2238. https://doi.org/10.3390/ijms25042238
Tosios KI, Kalogirou E-M, Koutlas IG. Association of MDM2 Overexpression in Ameloblastomas with MDM2 Amplification and BRAFV600E Expression. International Journal of Molecular Sciences. 2024; 25(4):2238. https://doi.org/10.3390/ijms25042238
Chicago/Turabian StyleTosios, Konstantinos I., Eleni-Marina Kalogirou, and Ioannis G. Koutlas. 2024. "Association of MDM2 Overexpression in Ameloblastomas with MDM2 Amplification and BRAFV600E Expression" International Journal of Molecular Sciences 25, no. 4: 2238. https://doi.org/10.3390/ijms25042238
APA StyleTosios, K. I., Kalogirou, E.-M., & Koutlas, I. G. (2024). Association of MDM2 Overexpression in Ameloblastomas with MDM2 Amplification and BRAFV600E Expression. International Journal of Molecular Sciences, 25(4), 2238. https://doi.org/10.3390/ijms25042238






