Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2
Abstract
1. Introduction
2. Results
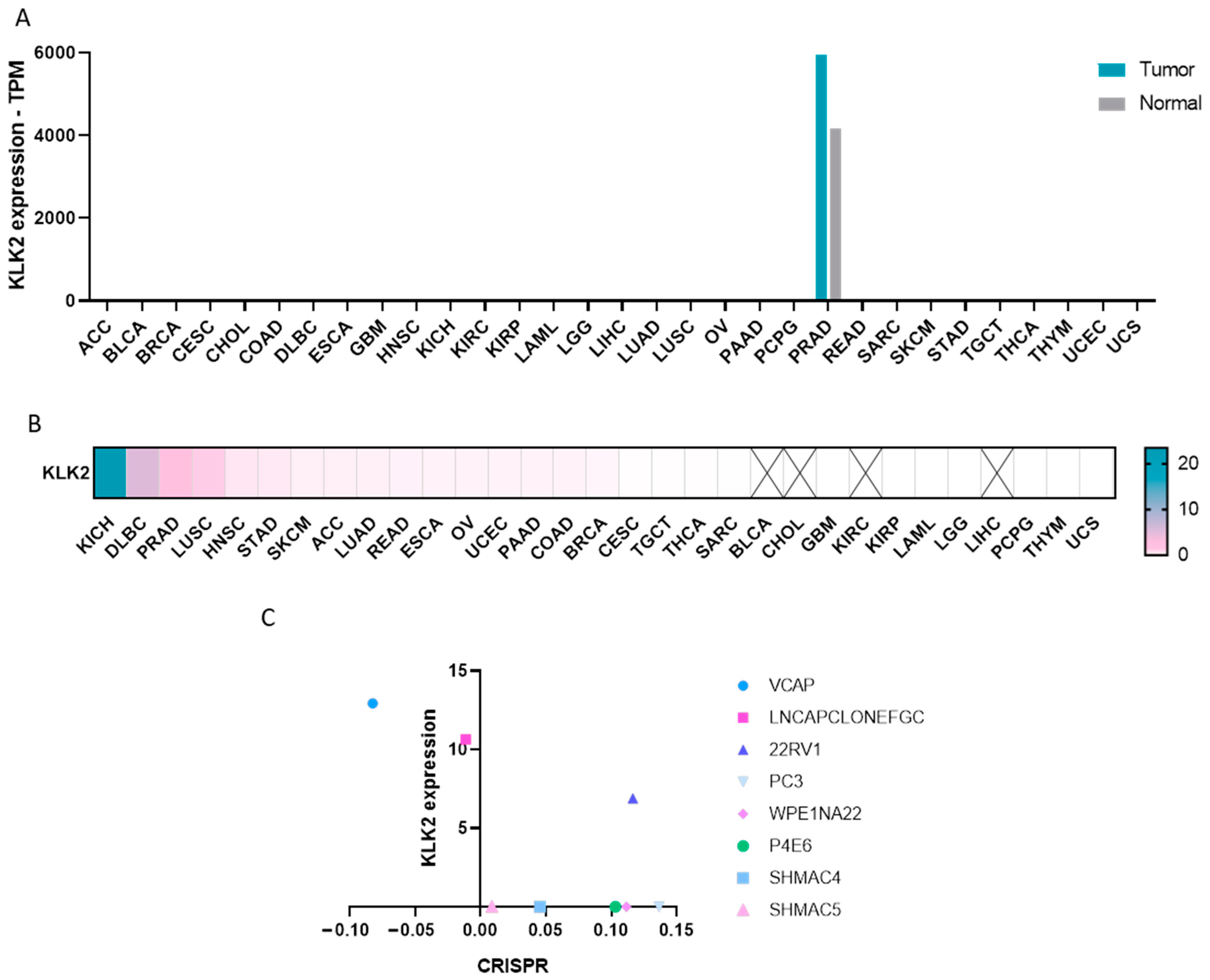
2.1. Expression of KLK2 in Cancer
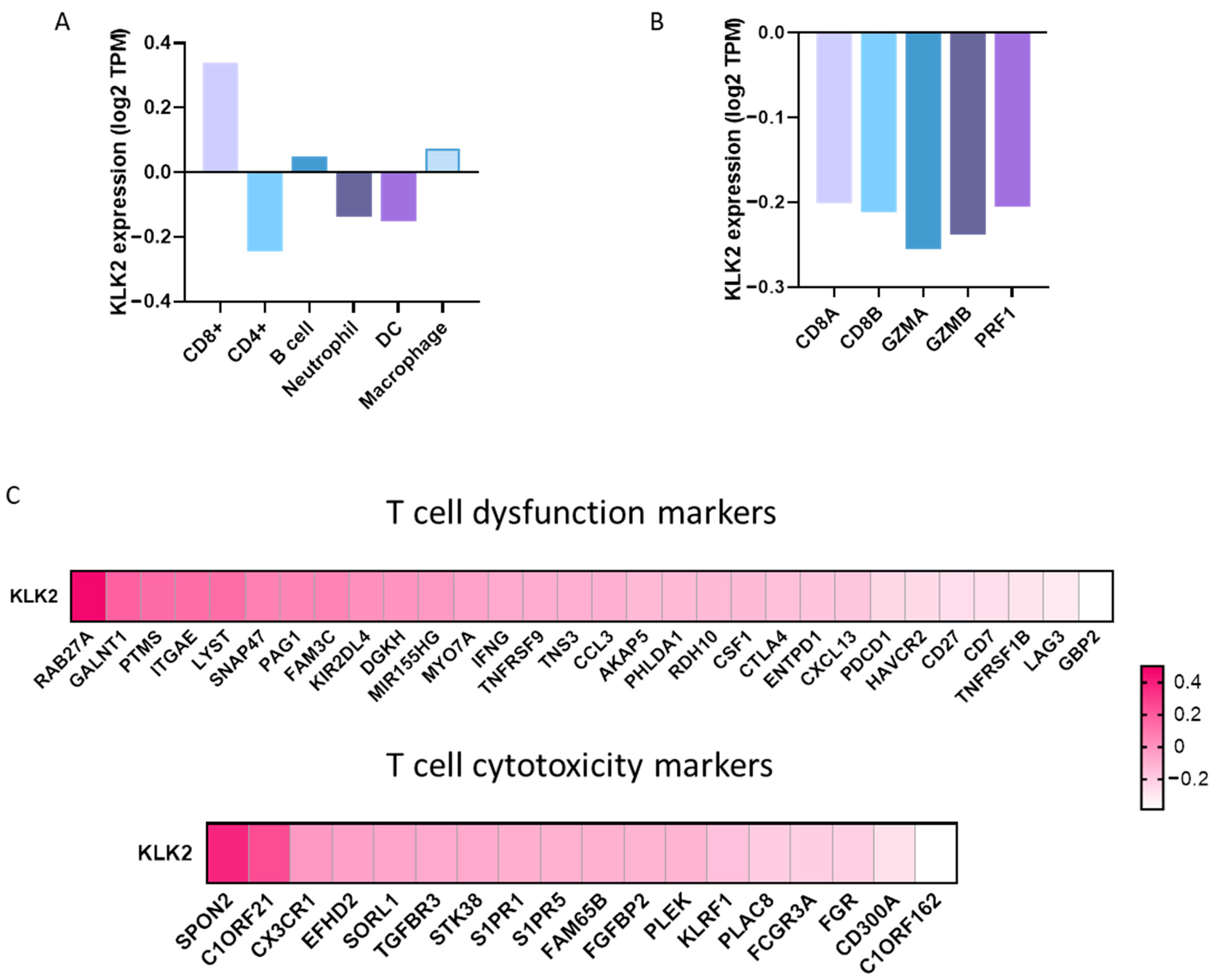
2.2. Expression of Immune Populations in PRAD with a High Expression of KLK2
2.3. Genomic Alterations in PRAD and Expression of KLK2
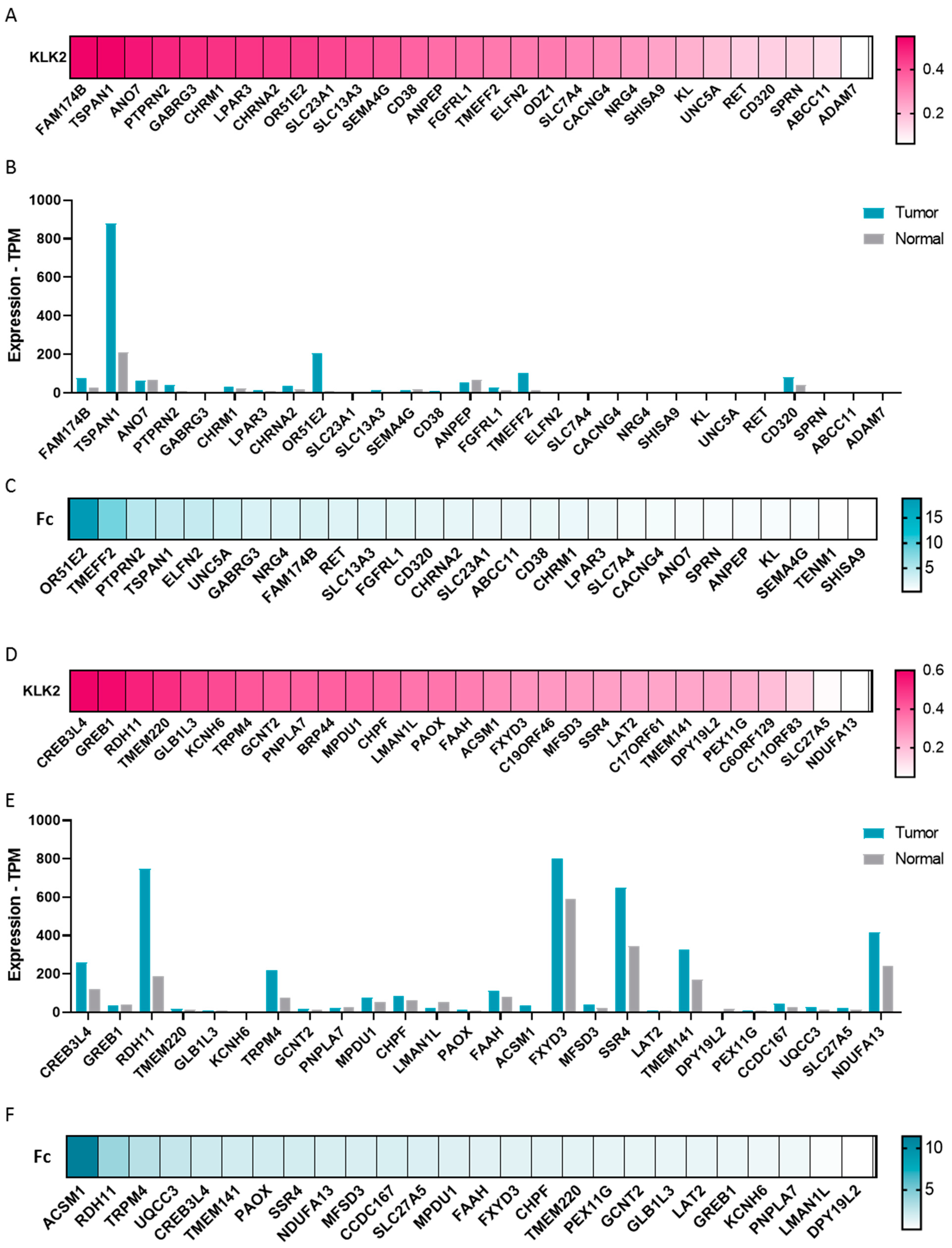
2.4. Transcriptomic Profile in KLK2 PRAD
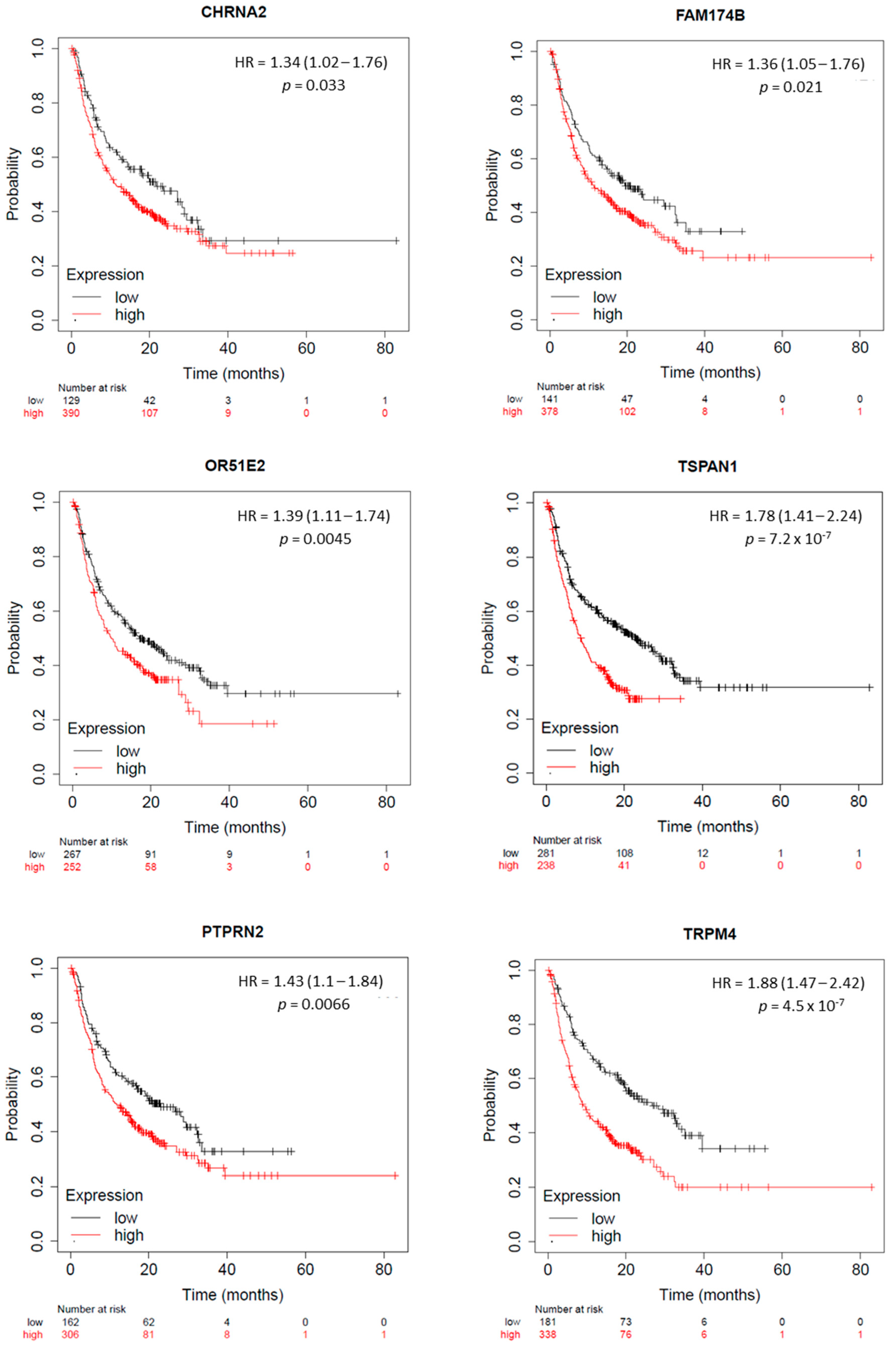
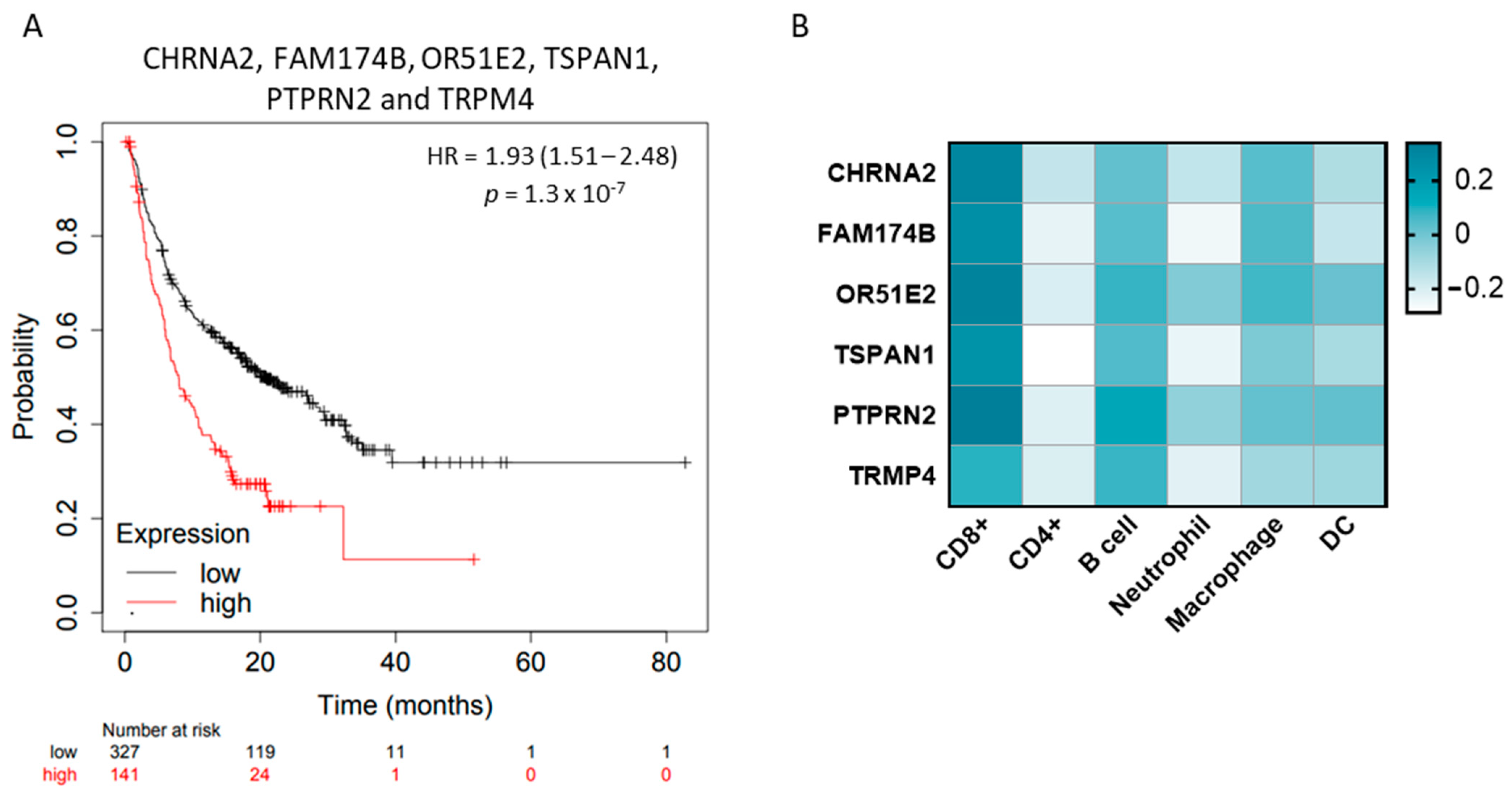
2.5. KLK2 Co-Upregulated Genes and the Clinical Outcome and Response to ICIs
3. Discussion
4. Materials and Methods
4.1. Gene Expression and Genomic Alteration Analysis
4.2. CRISPR Dependency Score
4.3. Association between Tumor Immune Infiltrates and Gene Expression
4.4. Genes Expression in PRAD with Overexpression of KLK2
4.5. Surfaceome
4.6. Outcome, Prognosis Analysis, and Biochemical Recurrence
4.7. Protein–Protein Interaction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Ocana, A.; Garcia-Alonso, S.; Amir, E.; Pandiella, A. Refining Early Antitumoral Drug Development. Trends Pharmacol. Sci. 2018, 39, 922–925. [Google Scholar] [CrossRef] [PubMed]
- Rudolph, J.; Settleman, J.; Malek, S. Emerging Trends in Cancer Drug Discovery-From Drugging the “Undruggable” to Overcoming Resistance. Cancer Discov. 2021, 11, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Jimenez, C.; Sanvicente, A.; Diaz-Tejeiro, C.; Moreno, V.; Lopez de Sa, A.; Calvo, E.; Martinez-Lopez, J.; Perez-Segura, P.; Ocana, A. Uncovering therapeutic opportunities in the clinical development of antibody-drug conjugates. Clin. Transl. Med. 2023, 13, e1329. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.T.W.; Harris, P.W.R.; Brimble, M.A.; Kavianinia, I. An Insight into FDA Approved Antibody-Drug Conjugates for Cancer Therapy. Molecules 2021, 26, 5847. [Google Scholar] [CrossRef] [PubMed]
- Fuca, G.; Spagnoletti, A.; Ambrosini, M.; de Braud, F.; Di Nicola, M. Immune cell engagers in solid tumors: Promises and challenges of the next generation immunotherapy. ESMO Open 2021, 6, 100046. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.C.; Calvo-Jimenez, E.; Del Carmen, S.; Abad, M.; Ocana, A.; Pandiella, A. Surfaceome analyses uncover CD98hc as an antibody drug-conjugate target in triple negative breast cancer. J. Exp. Clin. Cancer Res. 2022, 41, 106. [Google Scholar] [CrossRef]
- Alcaraz-Sanabria, A.; Cabanas Morafraile, E.; Fernandez-Hinojal, G.; Velasco, G.; Perez-Segura, P.; Pandiella, A.; Gyorffy, B.; Ocana, A. Transcriptomic Mapping of Non-Small Cell Lung Cancer K-RAS p.G12C Mutated Tumors: Identification of Surfaceome Targets and Immunologic Correlates. Front. Immunol. 2021, 12, 786069. [Google Scholar] [CrossRef] [PubMed]
- Avgeris, M.; Mavridis, K.; Scorilas, A. Kallikrein-related peptidase genes as promising biomarkers for prognosis and monitoring of human malignancies. Biol. Chem. 2010, 391, 505–511. [Google Scholar] [CrossRef]
- A Study of JNJ-78278343, a T-Cell-Redirecting Agent Targeting Human Kallikrein 2 (KLK2), for Advanced Prostate Cancer. Available online: https://www.clinicaltrials.gov/study/NCT04898634?term=KLK2&rank=1 (accessed on 5 February 2024).
- Kimura, T. East meets West: Ethnic differences in prostate cancer epidemiology between East Asians and Caucasians. Chin J Cancer 2012, 31, 421–429. [Google Scholar] [CrossRef]
- McHugh, J.; Saunders, E.J.; Dadaev, T.; McGrowder, E.; Bancroft, E.; Kote-Jarai, Z.; Eeles, R. Prostate cancer risk in men of differing genetic ancestry and approaches to disease screening and management in these groups. Br. J. Cancer 2022, 126, 1366–1373. [Google Scholar] [CrossRef]
- Maher, J.; Davies, D.M. CAR-Based Immunotherapy of Solid Tumours-A Survey of the Emerging Targets. Cancers 2023, 15, 1171. [Google Scholar] [CrossRef]
- A Study of JNJ-78278343 in Combination with JNJ-63723283 (Cetrelimab) for Metastatic Castration-Resistant Prostate Cancer. Available online: https://www.clinicaltrials.gov/study/NCT05818683?term=KLK2%20&rank=4 (accessed on 5 February 2024).
- A Study of JNJ-69086420, an Actinium-225-Labeled Antibody Targeting Human Kallikrein-2 (hK2) for Advanced Prostate Cancer. Available online: https://www.clinicaltrials.gov/study/NCT04644770?term=KLK2%20&rank=6 (accessed on 5 February 2024).
- Perner, S.; Demichelis, F.; Beroukhim, R.; Schmidt, F.H.; Mosquera, J.M.; Setlur, S.; Tchinda, J.; Tomlins, S.A.; Hofer, M.D.; Pienta, K.G.; et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006, 66, 8337–8341. [Google Scholar] [CrossRef]
- Paoloni-Giacobino, A.; Chen, H.; Peitsch, M.C.; Rossier, C.; Antonarakis, S.E. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics 1997, 44, 309–320. [Google Scholar] [CrossRef]
- Lin, B.; Ferguson, C.; White, J.T.; Wang, S.; Vessella, R.; True, L.D.; Hood, L.; Nelson, P.S. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999, 59, 4180–4184. [Google Scholar]
- Yoshimoto, M.; Joshua, A.M.; Chilton-Macneill, S.; Bayani, J.; Selvarajah, S.; Evans, A.J.; Zielenska, M.; Squire, J.A. Three-color FISH analysis of TMPRSS2/ERG fusions in prostate cancer indicates that genomic microdeletion of chromosome 21 is associated with rearrangement. Neoplasia 2006, 8, 465–469. [Google Scholar] [CrossRef]
- Vancolen, S.; Sebire, G.; Robaire, B. Influence of androgens on the innate immune system. Andrology 2023, 11, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- GeneCards: The Human Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KLK2 (accessed on 12 July 2023).
- Samarkina, A.; Youssef, M.K.; Ostano, P.; Ghosh, S.; Ma, M.; Tassone, B.; Proust, T.; Chiorino, G.; Levesque, M.P.; Goruppi, S.; et al. Androgen receptor is a determinant of melanoma targeted drug resistance. Nat. Commun. 2023, 14, 6498. [Google Scholar] [CrossRef] [PubMed]
- Khosh Kish, E.; Choudhry, M.; Gamallat, Y.; Buharideen, S.M.; Dhananjaya, D.; Bismar, T.A. The Expression of Proto-Oncogene ETS-Related Gene (ERG) Plays a Central Role in the Oncogenic Mechanism Involved in the Development and Progression of Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 4772. [Google Scholar] [CrossRef] [PubMed]
- Simao, D.C.; Zarrabi, K.K.; Mendes, J.L.; Luz, R.; Garcia, J.A.; Kelly, W.K.; Barata, P.C. Bispecific T-Cell Engagers Therapies in Solid Tumors: Focusing on Prostate Cancer. Cancers 2023, 15, 1412. [Google Scholar] [CrossRef]
- Fucikova, J.; Kepp, O.; Kasikova, L.; Petroni, G.; Yamazaki, T.; Liu, P.; Zhao, L.; Spisek, R.; Kroemer, G.; Galluzzi, L. Detection of immunogenic cell death and its relevance for cancer therapy. Cell Death Dis. 2020, 11, 1013. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. Available online: http://gepia2.cancer-pku.cn/#index (accessed on 5 February 2024).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Gao, C. cBioportal. Available online: https://www.cbioportal.org (accessed on 12 July 2023).
- UALCAN: The University of Alabama at Birmingham Cancer Data Analysis Portal. Available online: https://ualcan.path.uab.edu/ (accessed on 21 November 2023).
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Dependency Map (DepMap). Available online: https://depmap.org/portal/ (accessed on 20 September 2023).
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef]
- Shi, B.; Ding, J.; Qi, J.; Gu, Z. Characteristics and prognostic value of potential dependency genes in clear cell renal cell carcinoma based on a large-scale CRISPR-Cas9 and RNAi screening database DepMap. Int. J. Med. Sci. 2021, 18, 2063–2075. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Taiwen, L. Tumor Immune Estimation Resource (TIMER 2.0). Available online: http://timer.cistrome.org/ (accessed on 15 July 2023).
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Bausch-Fluck, D.; Goldmann, U.; Muller, S.; van Oostrum, M.; Muller, M.; Schubert, O.T.; Wollscheid, B. The in silico human surfaceome. Proc. Natl. Acad. Sci. USA 2018, 115, E10988–E10997. [Google Scholar] [CrossRef]
- Damaris Bausch-Fluck, U.G.; Müller, S.; van Oostrum, M.; Müller, M.; Olga, T. Schubert and Bernd Wollscheid. The In Silico Human Surfaceome. Available online: https://wlab.ethz.ch/surfaceome/ (accessed on 10 September 2023).
- TCGA-PRAD. Available online: https://portal.gdc.cancer.gov/projects/TCGA-PRAD (accessed on 5 September 2023).
- Kovacs, S.A.; Gyorffy, B. Transcriptomic datasets of cancer patients treated with immune-checkpoint inhibitors: A systematic review. J. Transl. Med. 2022, 20, 249. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B. The Kaplan–Meier Plotter Online Tool. Available online: https://kmplot.com/analysis/ (accessed on 10 October 2023).
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- STRING. Available online: https://string-db.org/ (accessed on 11 October 2023).

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paniagua-Herranz, L.; Moreno, I.; Nieto-Jiménez, C.; Garcia-Lorenzo, E.; Díaz-Tejeiro, C.; Sanvicente, A.; Doger, B.; Pedregal, M.; Ramón, J.; Bartolomé, J.; et al. Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2. Int. J. Mol. Sci. 2024, 25, 2222. https://doi.org/10.3390/ijms25042222
Paniagua-Herranz L, Moreno I, Nieto-Jiménez C, Garcia-Lorenzo E, Díaz-Tejeiro C, Sanvicente A, Doger B, Pedregal M, Ramón J, Bartolomé J, et al. Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2. International Journal of Molecular Sciences. 2024; 25(4):2222. https://doi.org/10.3390/ijms25042222
Chicago/Turabian StylePaniagua-Herranz, Lucía, Irene Moreno, Cristina Nieto-Jiménez, Esther Garcia-Lorenzo, Cristina Díaz-Tejeiro, Adrián Sanvicente, Bernard Doger, Manuel Pedregal, Jorge Ramón, Jorge Bartolomé, and et al. 2024. "Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2" International Journal of Molecular Sciences 25, no. 4: 2222. https://doi.org/10.3390/ijms25042222
APA StylePaniagua-Herranz, L., Moreno, I., Nieto-Jiménez, C., Garcia-Lorenzo, E., Díaz-Tejeiro, C., Sanvicente, A., Doger, B., Pedregal, M., Ramón, J., Bartolomé, J., Manzano, A., Gyorffy, B., Gutierrez-Uzquiza, Á., Pérez Segura, P., Calvo, E., Moreno, V., & Ocana, A. (2024). Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2. International Journal of Molecular Sciences, 25(4), 2222. https://doi.org/10.3390/ijms25042222






