OsLRR-RLP2 Gene Regulates Immunity to Magnaporthe oryzae in Japonica Rice
Abstract
1. Introduction
2. Results
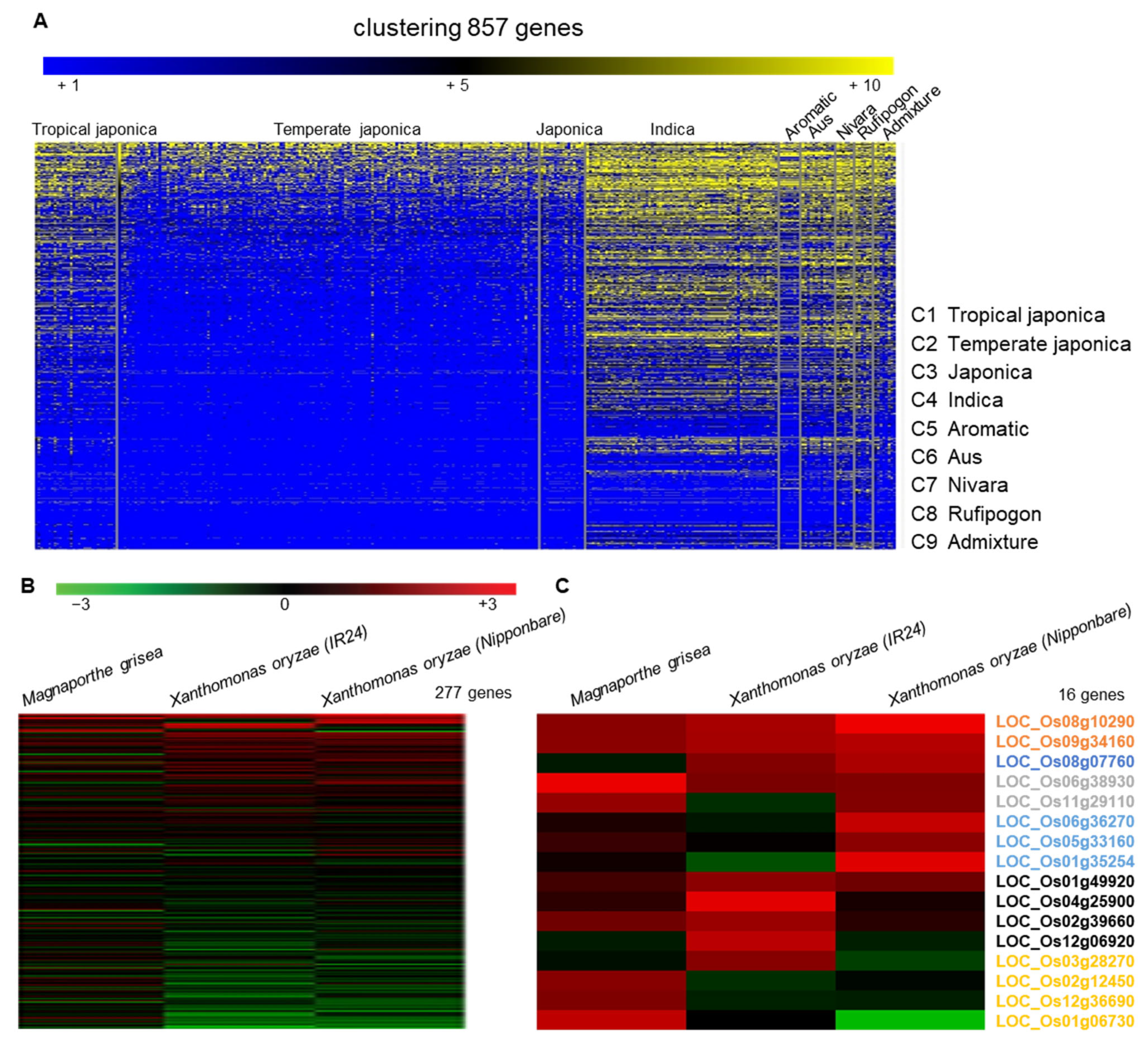
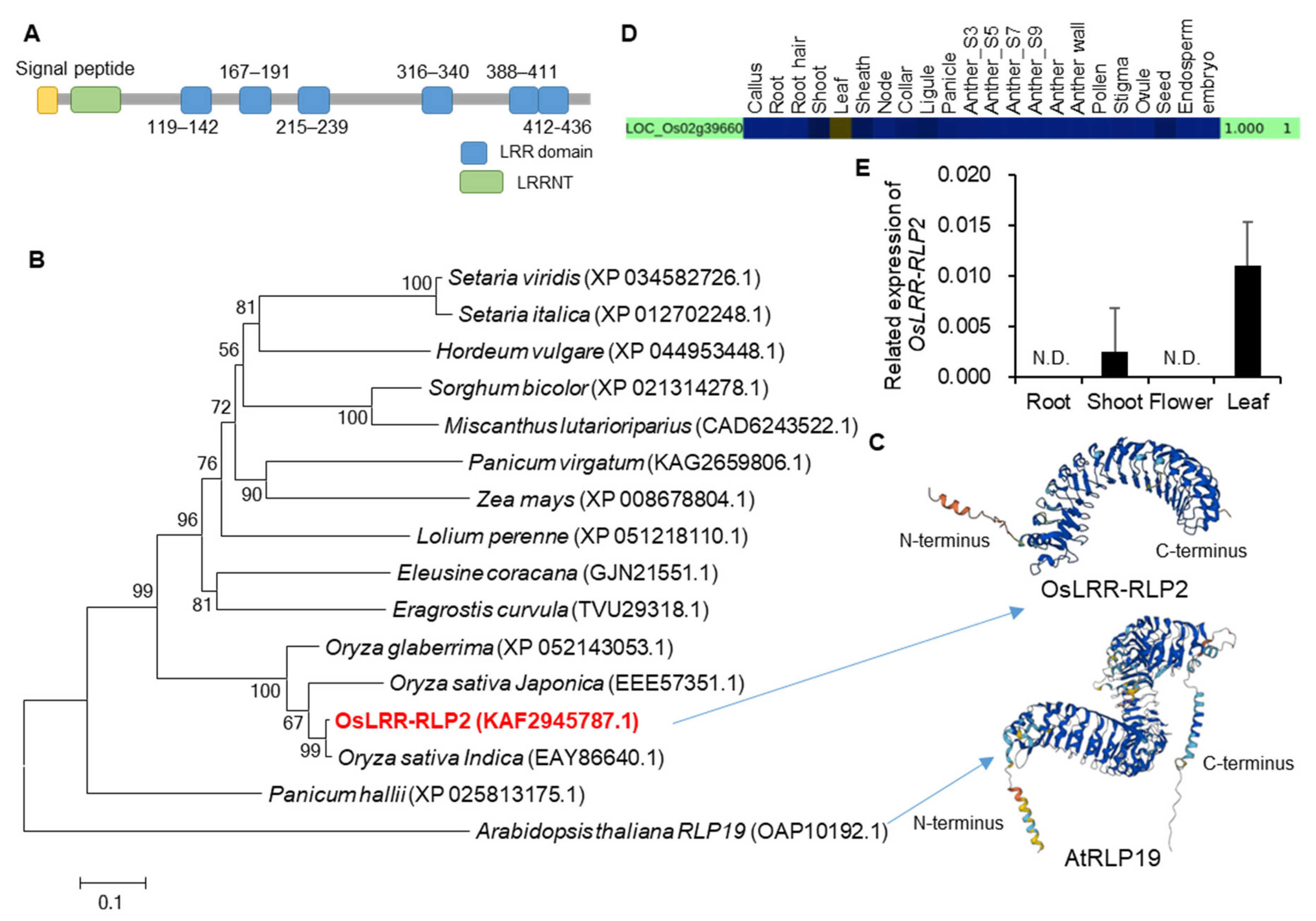
2.1. Selection of LRR Candidates and Phylogenetic Analysis
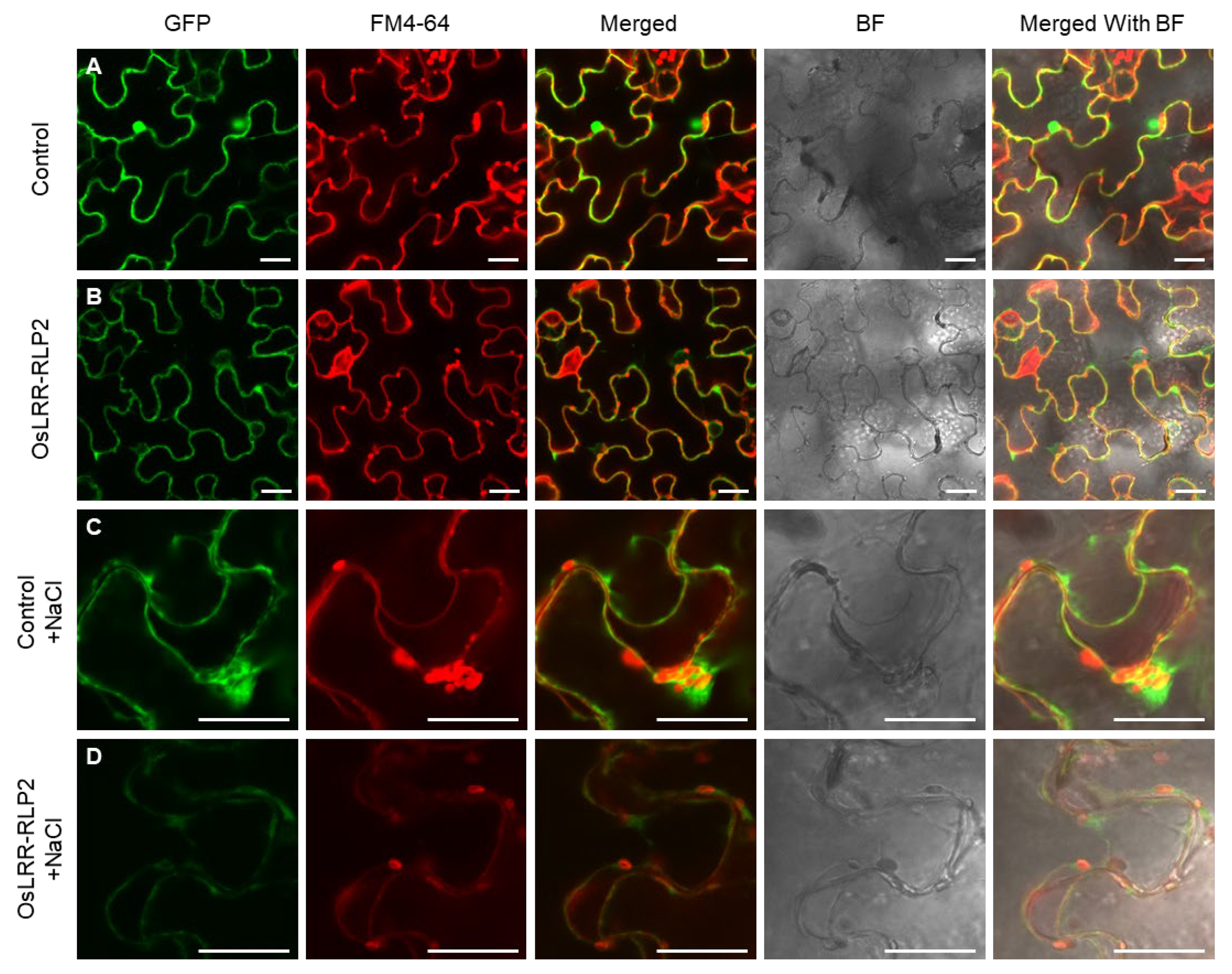
2.2. Expression and Localization Analysis of OsLRR-RLP
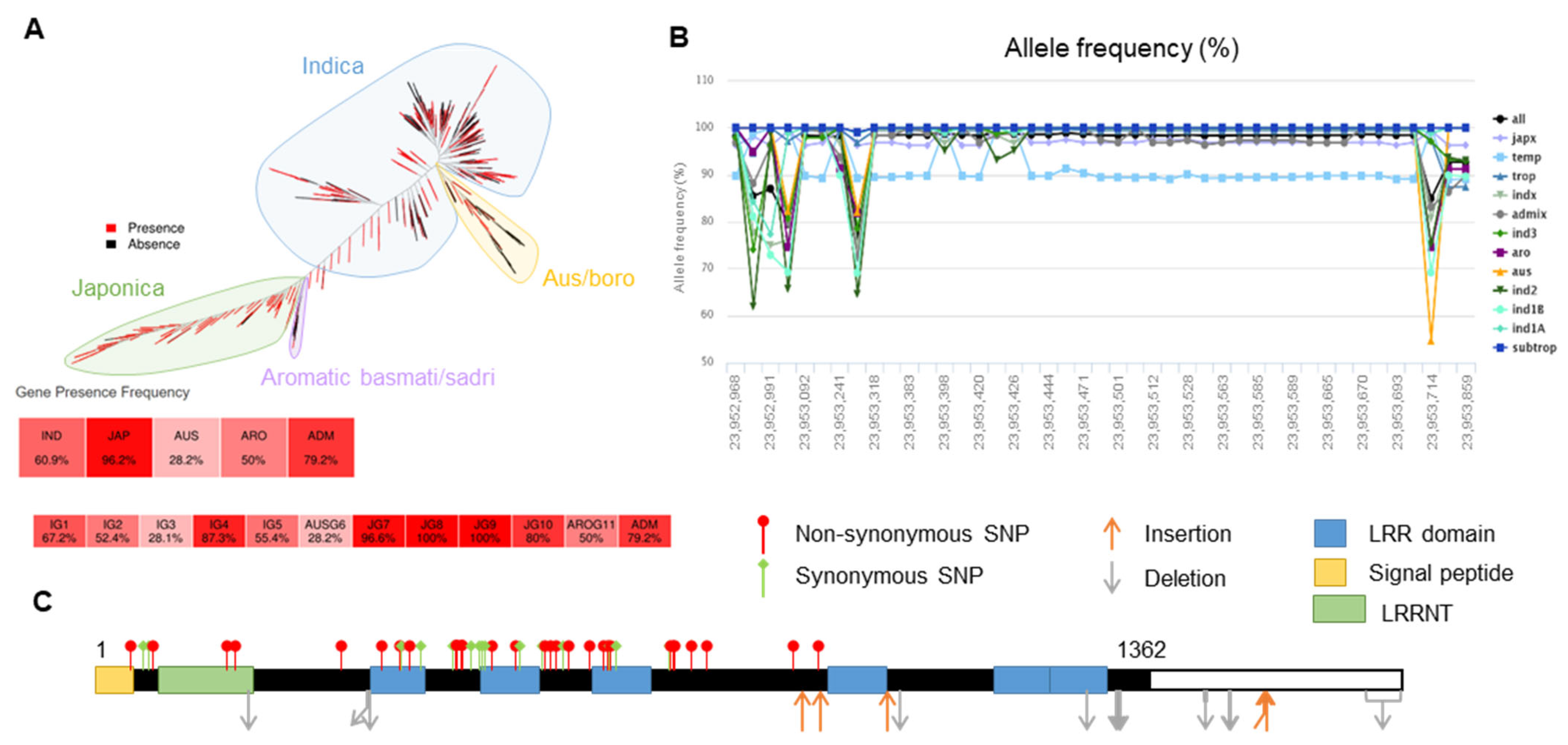
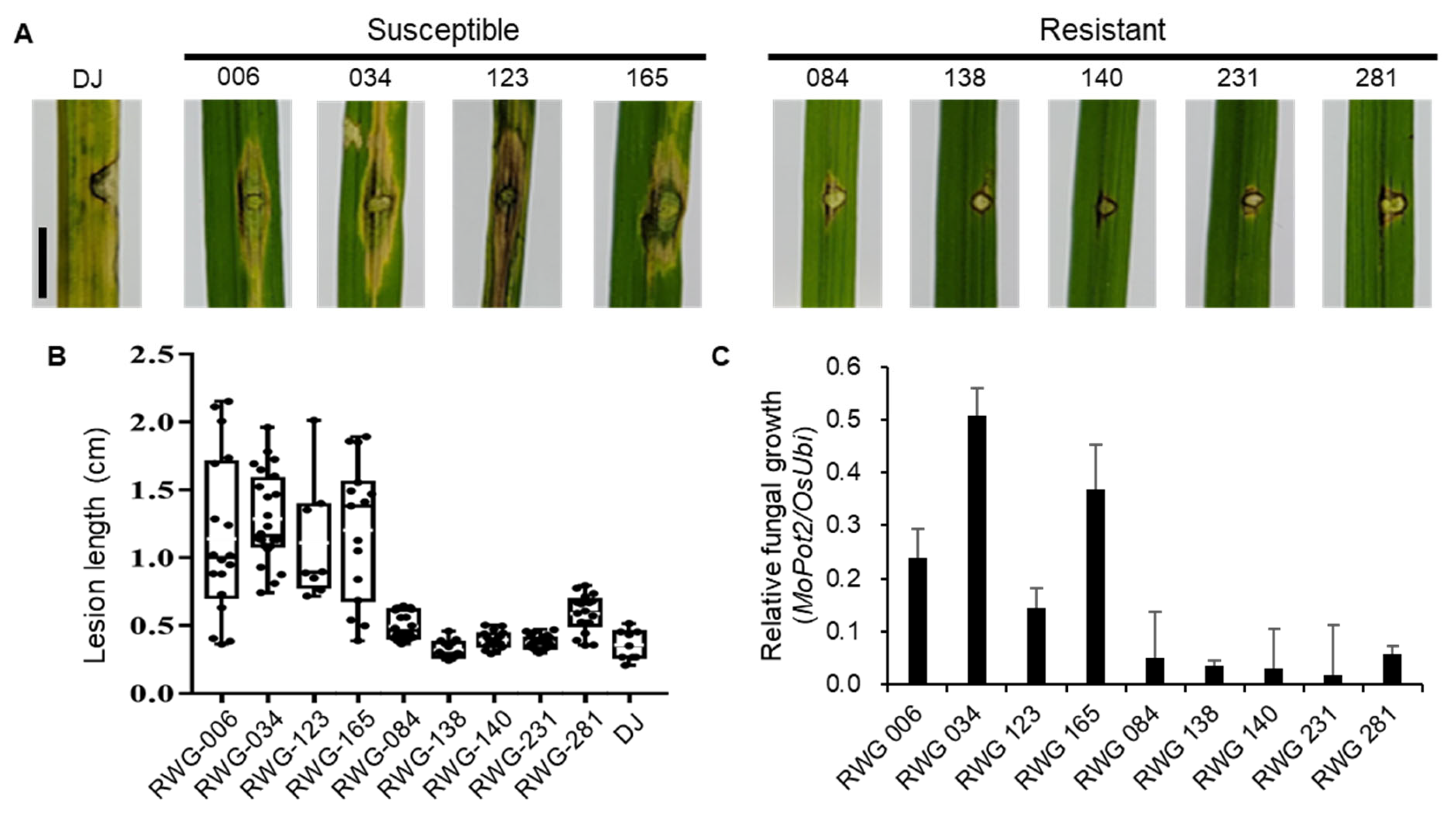
2.3. Haplotype Analysis of OsLRR-RLP2 Sequence and Validation of M. oryzae Response
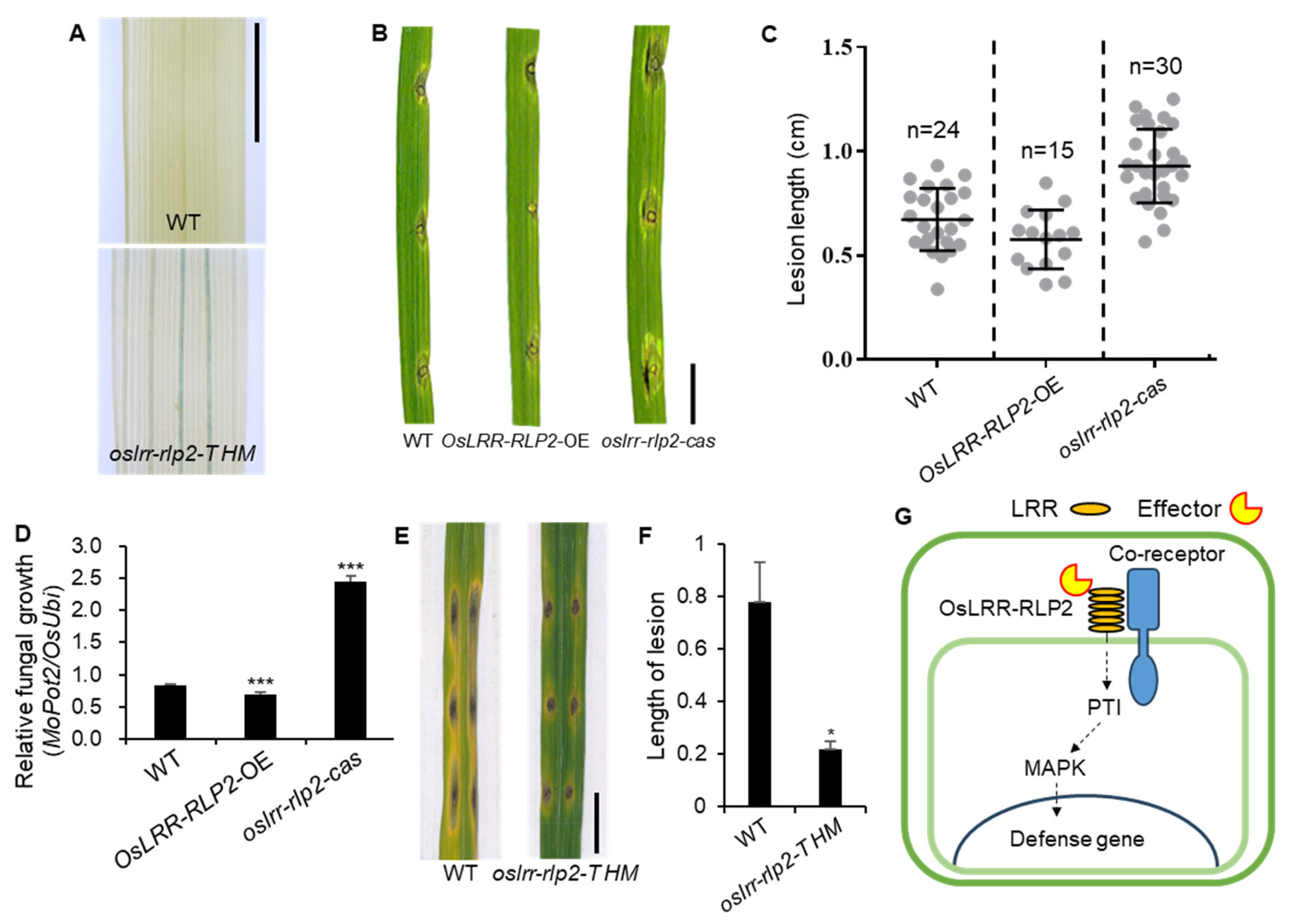
2.4. Functional Analysis of OsLRR-RLP2 Involving Rice Blast Disease
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Blast Inoculation
4.2. Gene Identification and Sequence Analysis
4.3. Construction of Vectors and Plant Transformation
4.4. DNA Isolation and Genotypic Analysis
4.5. RNA Isolation, cDNA Synthesis, and qRT-PCR
4.6. Generation of Fusion Proteins and Subcellular Localization Analysis
4.7. Histochemical GUS Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CRISPR/Cas9 | Clustered regularly interspaced palindromic repeats/CRISPR-associated protein 9 |
| CT | Cycle value |
| CTAB | Classical cetyltrimethylammonium bromide |
| DJ | Dongjin |
| DPI | Days post inoculation |
| ETI | Effector-triggered immunity |
| GFP | Green fluorescent protein |
| GUS | β-glucuronidase |
| HM | Homozygous |
| HR | Hypersensitive response |
| Indels | Insertions/deletions |
| LMO | Living modified organism |
| LRR | Leucine-rich repeat |
| LRRNT | LRR N-terminus domain |
| MAPK | Mitogen-activated protein kinases |
| MoPot2 | Magnaporthe oryzae Pot2 |
| NLP | Necrosis- and Ethylene-inducing peptide 1-like protein |
| NLR | Nucleotide-binding and leucine-rich repeat receptor |
| OE | Overexpression |
| PAMP | Pathogen-associated molecular pattern |
| PRR | Pattern recognition receptor |
| PTI | Pattern-triggered immunity |
| qRT-PCR | Quantitative reverse transcription PCR |
| R gene | Resistance gene |
| RFP | Red fluorescent protein |
| RLK | Receptor-like kinase |
| RLP | Receptor-like protein |
| SNP | Single-nucleotide polymorphisms |
| TM | Transmembrane |
| Ubi | Ubiquitin |
| WT | Wild-type |
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology; Elsevier: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Chen, W.-L.; Lin, Y.-B.; Ng, F.-L.; Liu, C.-Y.; Lin, Y.-W. RiceTalk: Rice blast detection using Internet of Things and artificial intelligence technologies. IEEE Internet Things J. 2019, 7, 1001–1010. [Google Scholar] [CrossRef]
- Wang, X.; Lee, S.; Wang, J.; Ma, J.; Bianco, T.; Jia, Y. Current advances on genetic resistance to rice blast disease. Rice-Germplasm Genet. Improv. 2014, 23, 195–217. [Google Scholar]
- Skamnioti, P.; Gurr, S.J. Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Couto, D.; Zipfel, C. Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 2016, 16, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Tsuda, K.; Parker, J.E. Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 2015, 66, 487–511. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Boutrot, F.; Zipfel, C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef]
- Wang, G.; Ellendorff, U.; Kemp, B.; Mansfield, J.W.; Forsyth, A.; Mitchell, K.; Bastas, K.; Liu, C.-M.; Woods-Tör, A.; Zipfel, C. A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 2008, 147, 503–517. [Google Scholar] [CrossRef]
- Jones, D.A.; Thomas, C.M.; Hammond-Kosack, K.E.; Balint-Kurti, P.J.; Jones, J.D. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 1994, 266, 789–793. [Google Scholar] [CrossRef]
- Nadeau, J.A.; Sack, F.D. Control of stomatal distribution on the Arabidopsis leaf surface. Science 2002, 296, 1697–1700. [Google Scholar] [CrossRef]
- Macho, A.P.; Zipfel, C. Plant PRRs and the activation of innate immune signaling. Mol. Cell 2014, 54, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, U.; Lau, K.; Hothorn, M. The structural basis of ligand perception and signal activation by receptor kinases. Annu. Rev. Plant Biol. 2017, 68, 109–137. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Fritz-Laylin, L.K.; Krishnamurthy, N.; Tör, M.; Sjölander, K.V.; Jones, J.D. Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol. 2005, 138, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Jehle, A.K.; Lipschis, M.; Albert, M.; Fallahzadeh-Mamaghani, V.; Fürst, U.; Mueller, K.; Felix, G. The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 2013, 25, 2330–2340. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.M.; Jones, D.A.; Parniske, M.; Harrison, K.; Balint-Kurti, P.J.; Hatzixanthis, K.; Jones, J.D. Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 1997, 9, 2209–2224. [Google Scholar] [PubMed]
- Zhang, H.; Chen, C.; Li, L.; Tan, X.; Wei, Z.; Li, Y.; Li, J.; Yan, F.; Chen, J.; Sun, Z. A rice LRR receptor-like protein associates with its adaptor kinase OsSOBIR1 to mediate plant immunity against viral infection. Plant Biotechnol. J. 2021, 19, 2319–2332. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Huang, X.; Tian, Z.; Wing, R.A.; Han, B. The genomics of Oryza species provides insights into rice domestication and heterosis. Annu. Rev. Plant Biol. 2019, 70, 639–665. [Google Scholar] [CrossRef]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic structure and diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef]
- Escolà, G.; González-Miguel, V.M.; Campo, S.; Catala-Forner, M.; Domingo, C.; Marqués, L.; San Segundo, B. Development and Genome-Wide Analysis of a Blast-Resistant japonica Rice Variety. Plants 2023, 12, 3536. [Google Scholar] [CrossRef] [PubMed]
- Enkhbayar, P.; Kamiya, M.; Osaki, M.; Matsumoto, T.; Matsushima, N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins Struct. Funct. Bioinform. 2004, 54, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Zhang, H.; Zeng, Y.; Kim, B.; Kwon, S.-W. Discovery of Genomic Regions and Candidate Genes for Awn Length Using QTL-seq in Rice (Oryza sativa L.). Plant Breed. Biotech. 2023, 11, 271–277. [Google Scholar] [CrossRef]
- Boller, T.; Felix, G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009, 60, 379–406. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed]
- van Ooijen, G.; van den Burg, H.A.; Cornelissen, B.J.; Takken, F.L. Structure and function of resistance proteins in solanaceous plants. Annu. Rev. Phytopathol. 2007, 45, 43–72. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, Y.; Zhang, X.; Chen, Z.; Xia, Y.; Wang, L.; Sun, Y.; Zhang, M.; Xiao, Y.; Han, Z. Plant receptor-like protein activation by a microbial glycoside hydrolase. Nature 2022, 610, 335–342. [Google Scholar] [CrossRef]
- Albert, I.; Zhang, L.; Bemm, H.; Nürnberger, T. Structure-function analysis of immune receptor at RLP23 with its ligand nlp20 and coreceptors at SOBIR1 and At BAK1. Mol. Plant-Microbe Interact. 2019, 32, 1038–1046. [Google Scholar] [CrossRef]
- Zhang, W.; Fraiture, M.; Kolb, D.; Löffelhardt, B.; Desaki, Y.; Boutrot, F.F.; Tör, M.; Zipfel, C.; Gust, A.A.; Brunner, F. Arabidopsis receptor-like protein30 and receptor-like kinase suppressor of BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 2013, 25, 4227–4241. [Google Scholar] [CrossRef]
- Yuan, M.; Ngou, B.P.M.; Ding, P.; Xin, X.-F. PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 2021, 62, 102030. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Sharma, P.; de Silva, F.G.; Ronald, P. The Xanthomonas oryzae pv.◊ oryzae raxP and raxQ genes encode an ATP sulphurylase and adenosine-5′-phosphosulphate kinase that are required for AvrXa21 avirulence activity. Mol. Microbiol. 2002, 44, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Zhang, A.; Ren, Y.; Wu, F.; Wang, G.; Xu, Y.; Lei, C.; Zhu, S.; Pan, T. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 2019, 29, 820–831. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Yamada, K.; Ishikawa, K.; Yoshimura, S.; Hayashi, N.; Uchihashi, K.; Ishihama, N.; Kishi-Kaboshi, M.; Takahashi, A.; Tsuge, S. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 2013, 13, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Liebrand, T.W.; van den Burg, H.A.; Joosten, M.H. Two for all: Receptor-associated kinases SOBIR1 and BAK1. Trends Plant Sci. 2014, 19, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Li, Z.; Feng, D.; Xiong, F.; Liu, J.; Li, J.F.; Wang, M.; Wang, J.; Liu, B.; Wang, H.B. Os CERK 1 and Os RLCK 176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014, 80, 1072–1084. [Google Scholar] [CrossRef] [PubMed]
- Jehle, A.K.; Fürst, U.; Lipschis, M.; Albert, M.; Felix, G. Perception of the novel MAMP eMax from different Xanthomonas species requires the Arabidopsis receptor-like protein ReMAX and the receptor kinase SOBIR. Plant Signal. Behav. 2013, 8, e27408. [Google Scholar] [CrossRef] [PubMed]
- Liebrand, T.W.; van den Berg, G.C.; Zhang, Z.; Smit, P.; Cordewener, J.H.; America, A.H.; Sklenar, J.; Jones, A.M.; Tameling, W.I.; Robatzek, S. Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef]
- Min, C.W.; Jang, J.W.; Lee, G.H.; Gupta, R.; Yoon, J.; Park, H.J.; Cho, H.S.; Park, S.R.; Kwon, S.-W.; Cho, L.-H. TMT-based quantitative membrane proteomics identified PRRs potentially involved in the perception of MSP1 in rice leaves. J. Proteom. 2022, 267, 104687. [Google Scholar] [CrossRef]
- Singh, J.; Gupta, S.K.; Devanna, B.; Singh, S.; Upadhyay, A.; Sharma, T.R. Blast resistance gene Pi54 over-expressed in rice to understand its cellular and sub-cellular localization and response to different pathogens. Sci. Rep. 2020, 10, 5243. [Google Scholar] [CrossRef]
- Wang, G.-F.; Balint-Kurti, P.J. Cytoplasmic and nuclear localizations are important for the hypersensitive response conferred by maize autoactive Rp1-D21 protein. Mol. Plant-Microbe Interact. 2015, 28, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Liu, J.; Chang, C.; Zhang, L.; Maekawa, T.; Wang, Q.; Xiao, W.; Liu, Y.; Chai, J.; Takken, F.L. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012, 8, e1002752. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xun, Q.; Guo, Y.; Zhang, J.; Cheng, K.; Shi, T.; He, K.; Hou, S.; Gou, X.; Li, J. Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant 2016, 9, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.Á.; Benhamou, N.; Datnoff, L.E.; Jones, J.B.; Bélanger, R.R. Ultrastructural and cytochemical aspects of silicon-mediated rice blast resistance. Phytopathology 2003, 93, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Dolatabadian, A.; Bayer, P.E.; Tirnaz, S.; Hurgobin, B.; Edwards, D.; Batley, J. Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 2020, 18, 969–982. [Google Scholar] [CrossRef] [PubMed]
- Petre, B.; Hacquard, S.; Duplessis, S.; Rouhier, N. Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front. Plant Sci. 2014, 5, 111. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xiao, N.; Yu, L.; Pan, C.; Li, Y.; Zhang, X.; Liu, G.; Dai, Z.; Pan, X.; Li, A. Combination patterns of major R genes determine the level of resistance to the M. oryzae in rice (Oryza sativa L.). PLoS ONE 2015, 10, e0126130. [Google Scholar] [CrossRef]
- An, S.; Park, S.; Jeong, D.-H.; Lee, D.-Y.; Kang, H.-G.; Yu, J.-H.; Hur, J.; Kim, S.-R.; Kim, Y.-H.; Lee, M. Generation and analysis of end sequence database for T-DNA tagging lines in rice. Plant Physiol. 2003, 133, 2040–2047. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef]
- Lee, S.; Jeon, J.-S.; Jung, K.-H.; An, G. Binary vectors for efficient transformation of rice. J. Plant Biol. 1999, 42, 310–316. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sparkes, I.A.; Runions, J.; Kearns, A.; Hawes, C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006, 1, 2019–2025. [Google Scholar] [CrossRef]
- Moon, S.; Chandran, A.K.N.; Kim, Y.-J.; Gho, Y.; Hong, W.-J.; An, G.; Lee, C.; Jung, K.-H. Rice RHC encoding a putative cellulase is essential for normal root hair elongation. J. Plant Biol. 2019, 62, 82–91. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-J.; Jang, J.W.; Pham, T.; Tuyet, V.; Kim, J.-H.; Park, C.W.; Gho, Y.-S.; Kim, E.-J.; Kwon, S.-W.; Jeon, J.-S.; et al. OsLRR-RLP2 Gene Regulates Immunity to Magnaporthe oryzae in Japonica Rice. Int. J. Mol. Sci. 2024, 25, 2216. https://doi.org/10.3390/ijms25042216
Kim H-J, Jang JW, Pham T, Tuyet V, Kim J-H, Park CW, Gho Y-S, Kim E-J, Kwon S-W, Jeon J-S, et al. OsLRR-RLP2 Gene Regulates Immunity to Magnaporthe oryzae in Japonica Rice. International Journal of Molecular Sciences. 2024; 25(4):2216. https://doi.org/10.3390/ijms25042216
Chicago/Turabian StyleKim, Hyo-Jeong, Jeong Woo Jang, Thuy Pham, Van Tuyet, Ji-Hyun Kim, Chan Woo Park, Yun-Shil Gho, Eui-Jung Kim, Soon-Wook Kwon, Jong-Seong Jeon, and et al. 2024. "OsLRR-RLP2 Gene Regulates Immunity to Magnaporthe oryzae in Japonica Rice" International Journal of Molecular Sciences 25, no. 4: 2216. https://doi.org/10.3390/ijms25042216
APA StyleKim, H.-J., Jang, J. W., Pham, T., Tuyet, V., Kim, J.-H., Park, C. W., Gho, Y.-S., Kim, E.-J., Kwon, S.-W., Jeon, J.-S., Kim, S. T., Jung, K.-H., & Kim, Y.-J. (2024). OsLRR-RLP2 Gene Regulates Immunity to Magnaporthe oryzae in Japonica Rice. International Journal of Molecular Sciences, 25(4), 2216. https://doi.org/10.3390/ijms25042216






