The Mechanism of Tigecycline Resistance in Acinetobacter baumannii under Sub-Minimal Inhibitory Concentrations of Tigecycline
Abstract
1. Introduction
2. Results
2.1. Sub-MIC Selection of Tigecycline Resistant Strains
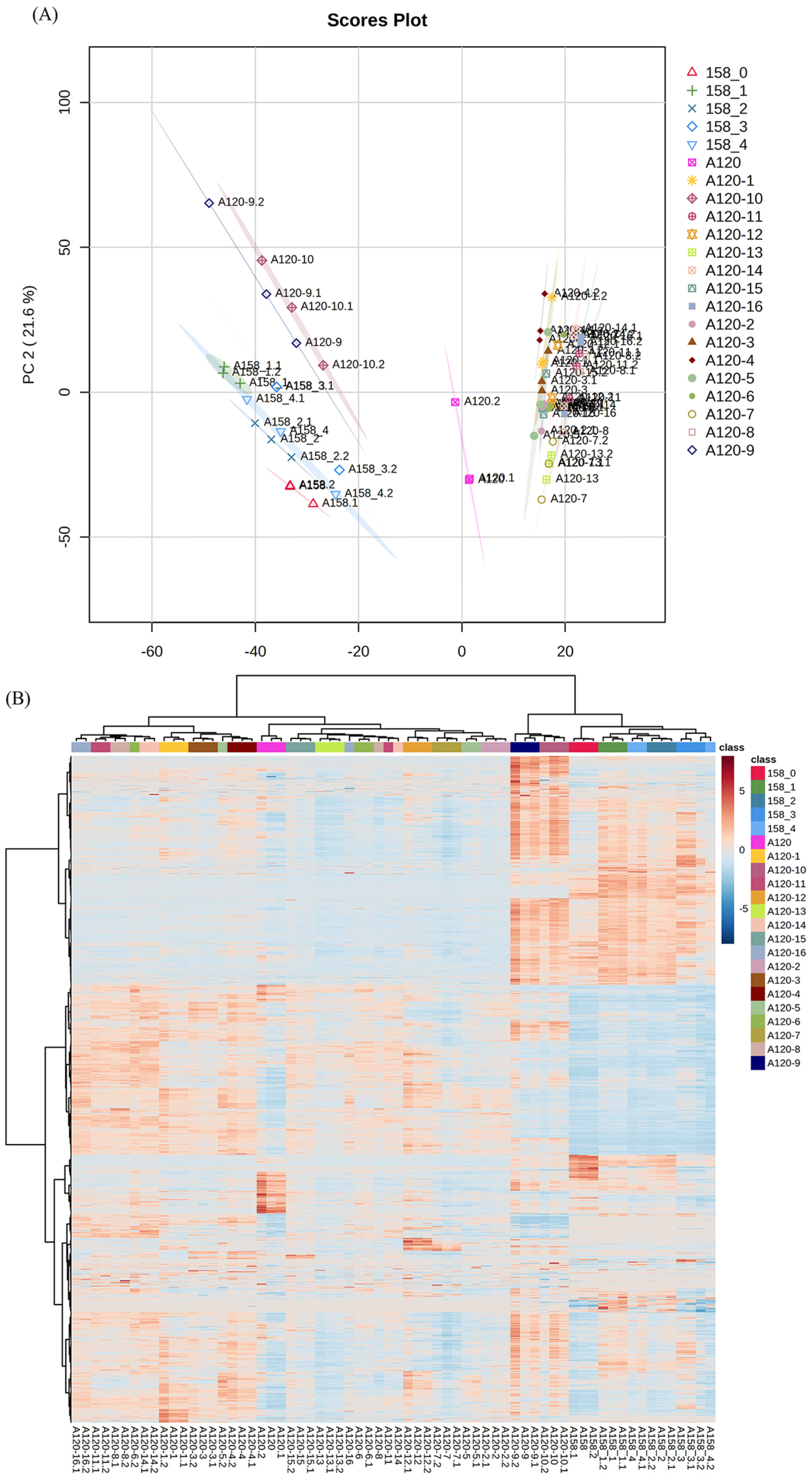
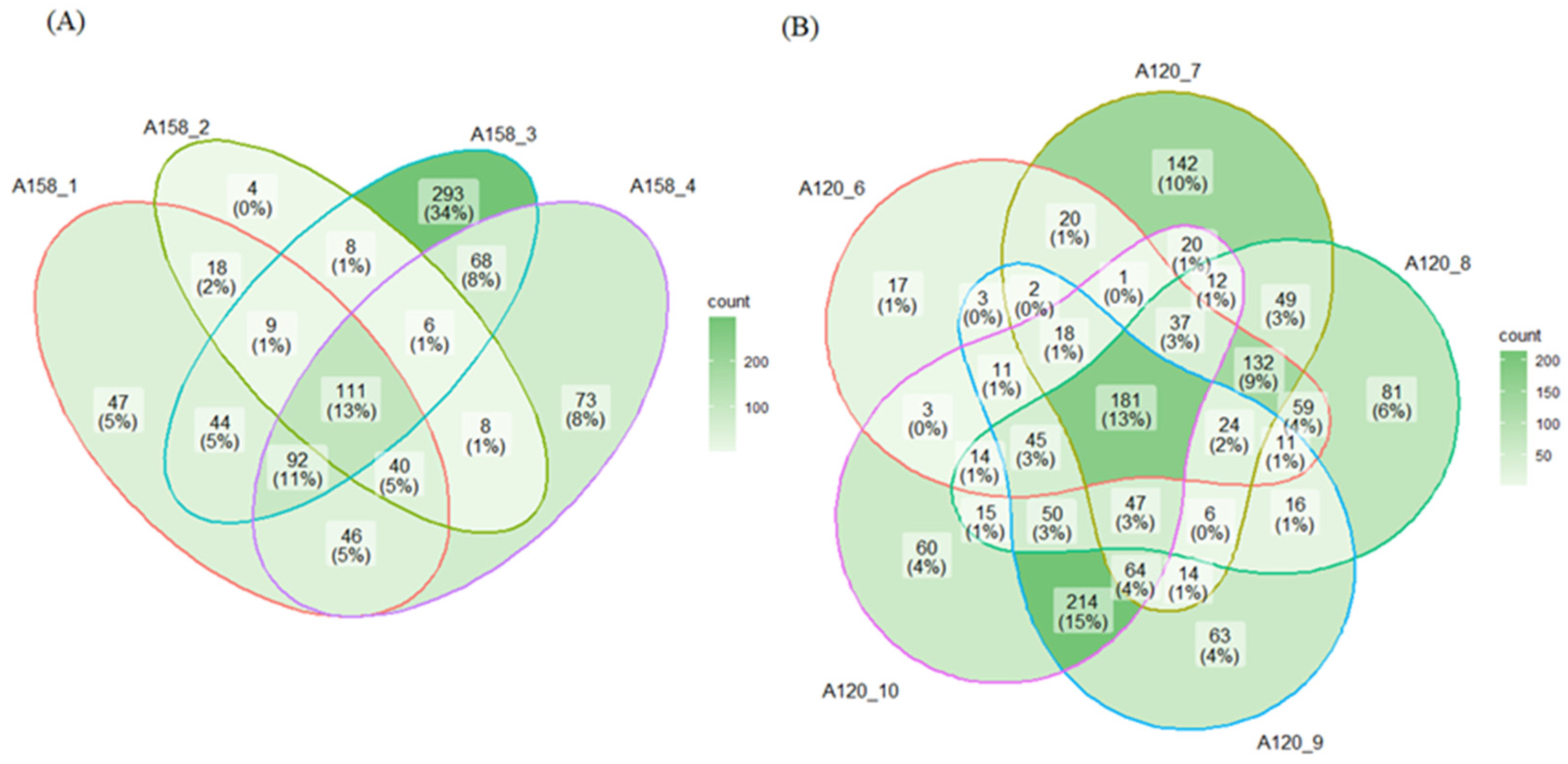
2.2. Proteomic Analysis
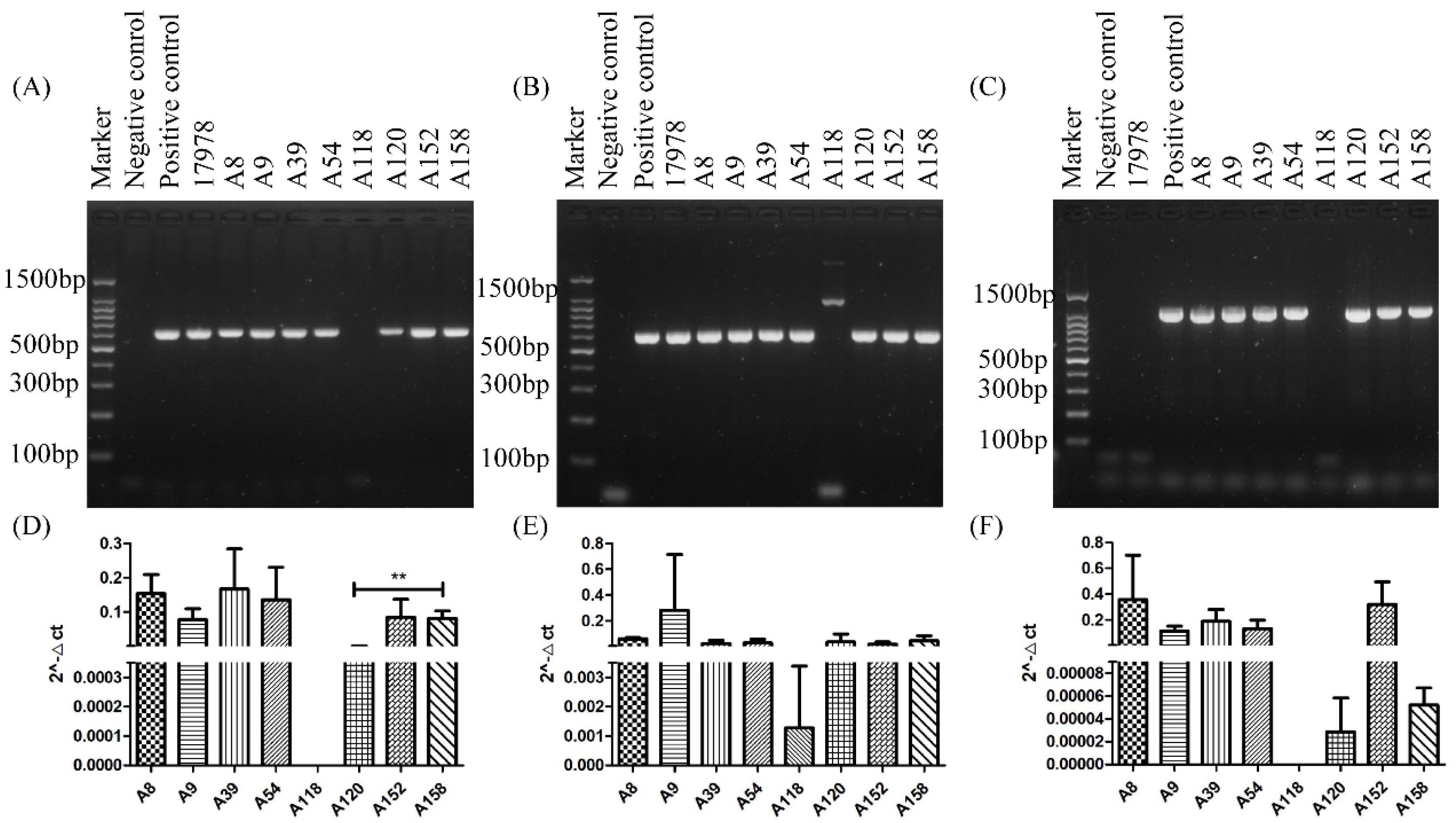
2.3. PCR and qRT-PCR of adeABC in Different Strains
2.4. Inhibition of Efflux Pump
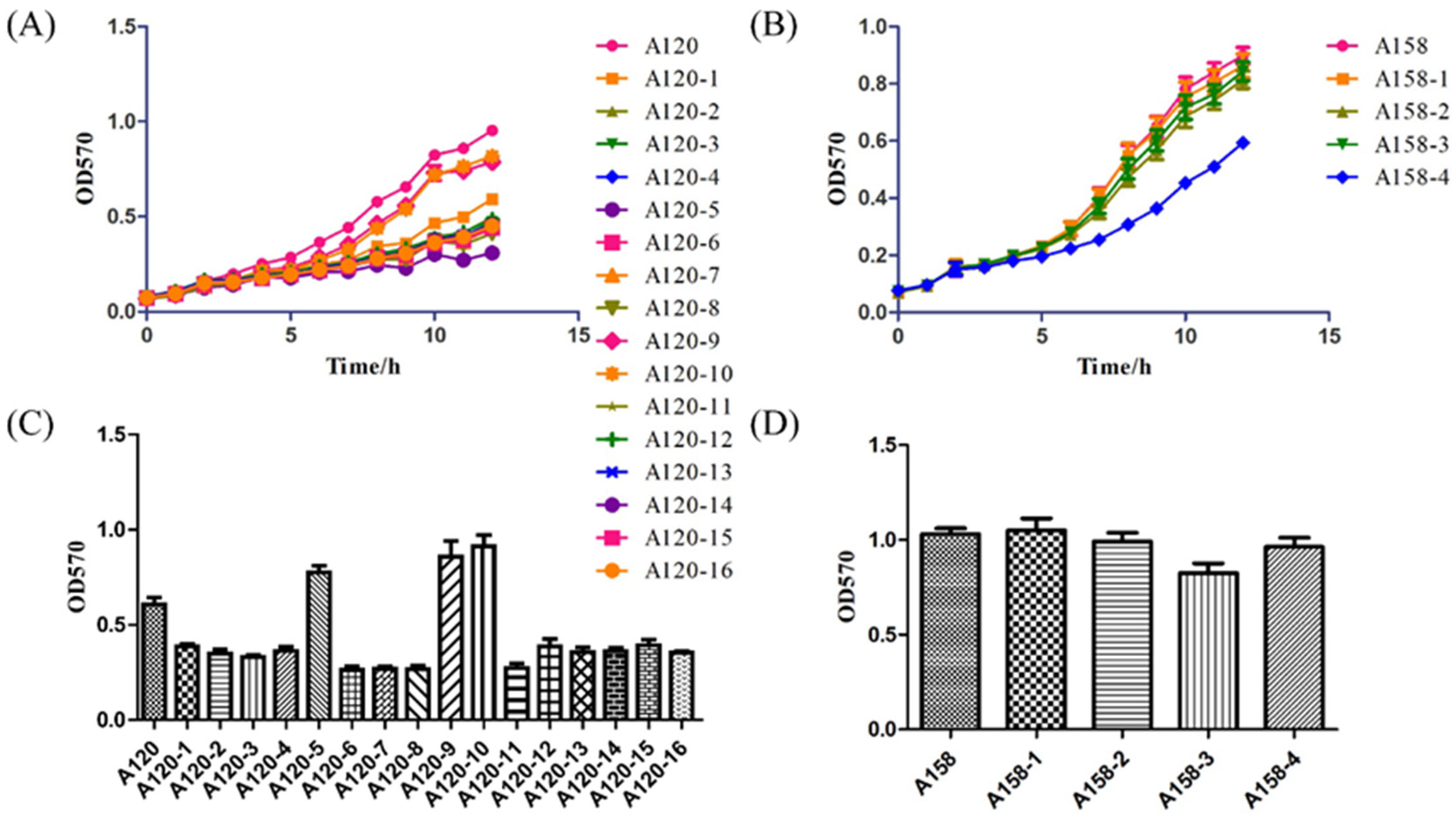
2.5. Growth Curve and Biofilm
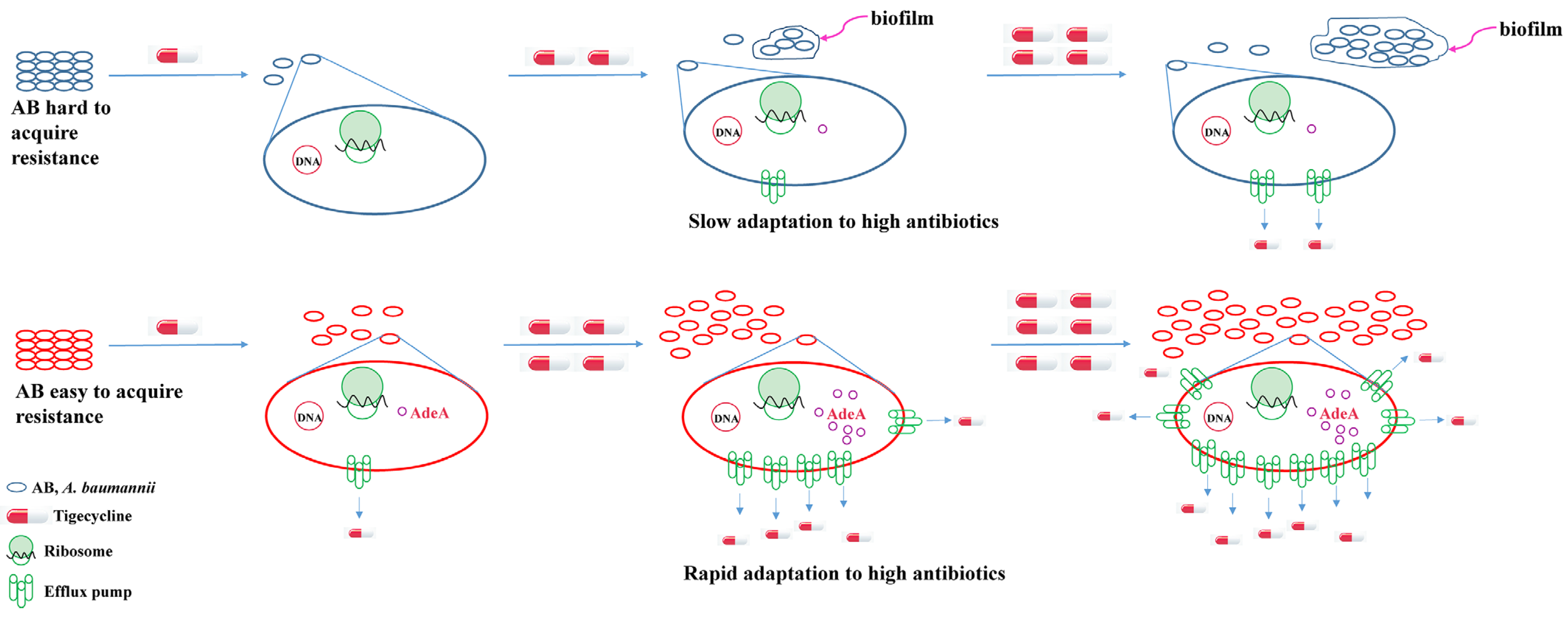
3. Discussion
4. Materials and Methods
4.1. Bacteria Strains and Antibiotic Susceptibility Test
4.2. Selection of Tigecycline-Resista17978nt Phenotype with Sub-MIC Tigecycline
4.3. Experimental Design and Statistical Rationale
4.4. Proteomic Analysis
4.5. qRT-PCR
4.6. Efflux Pump Inhibition Experiment
4.7. Growth Curve Measurement Experiment
4.8. Biofilm Formation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tu, Q.; Pu, M.; Li, Y.; Wang, Y.; Li, M.; Song, L.; Li, M.; An, X.; Fan, H.; Tong, Y. Acinetobacter Baumannii Phages: Past, Present and Future. Viruses 2023, 15, 673. [Google Scholar] [CrossRef] [PubMed]
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.M.; Hennon, S.W.; Feldman, M.F. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 2018, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, X.; Chen, L.; Duan, X.; Jiang, Z. In Vitro Activity of Various Antibiotics in Combination with Tigecycline Against Acinetobacter baumannii: A Systematic Review and Meta-Analysis. Microb. Drug Resist. 2017, 23, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cai, Y.; Liu, X.; Bai, N.; Liang, B.; Wang, R. The emergence of clinical resistance to tigecycline. Int. J. Antimicrob. Agents 2013, 41, 110–116. [Google Scholar] [CrossRef]
- Jamal, S.; Al Atrouni, A.; Rafei, R.; Dabboussi, F.; Hamze, M.; Osman, M. Molecular mechanisms of antimicrobial resistance in Acinetobacter baumannii, with a special focus on its epidemiology in Lebanon. J. Glob. Antimicrob. Resist. 2018, 15, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Yu, Y.; Hua, X. Resistance mechanisms of tigecycline in Acinetobacter baumannii. Front. Cell Infect. Microbiol. 2023, 13, 1141490. [Google Scholar] [CrossRef]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug. Resist. Updat. 2012, 15, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Orlen, H.; Hughes, D. Weak mutators can drive the evolution of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 2006, 50, 3454–3456. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Huang, Y.; Wu, W.; Zhu, J.; Zhang, T. Analysis of Efflux Pump System and Other Drug Resistance Related Gene Mutations in Tigecycline-Resistant Acinetobacter baumannii. Comput. Math. Methods Med. 2023, 2023, 8611542. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.R.; Pagal, S.; Amar, A.K.; Panda, L.; Dezvi, C.S.; Shashikala, P.; Kanungo, R.; Prashanth, K. Coexistence of blaNDM-1, blaOXA-51, blaOXA-23, and armA in conjunction with novel mutations detected in RND efflux pump regulators in tigecycline resistant clinical isolates of Acinetobacter baumannii. Pathog. Dis. 2022, 80, ftac020. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Liu, X.; Luo, Q.; Liu, B.F. Mass spectrometry in systems biology: An overview. Mass Spectrom. Rev. 2008, 27, 635–660. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Sun, R.; Xue, Z.; Guo, T. Mass Spectrometry-based Proteomics of Epithelial Ovarian Cancers: A Clinical Perspective. Mol. Cell Proteom. 2023, 22, 100578. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.J.; Shamsuzzaman, S.M. Distribution of adeB and NDM-1 genes in multidrug resistant Acinetobacter baumannii isolated from infected wound of patients admitted in a tertiary care hospital in Bangladesh. Malays. J. Pathol. 2017, 39, 277–283. [Google Scholar]
- Wieczorek, P.; Sacha, P.; Hauschild, T.; Zorawski, M.; Krawczyk, M.; Tryniszewska, E. Multidrug resistant Acinetobacter baumannii--the role of AdeABC (RND family) efflux pump in resistance to antibiotics. Folia Histochem. Cytobiol. 2008, 46, 257–267. [Google Scholar] [CrossRef]
- Lucaßen, K.; Müller, C.; Wille, J.; Xanthopoulou, K.; Hackel, M.; Seifert, H.; Higgins, P.G. Prevalence of RND efflux pump regulator variants associated with tigecycline resistance in carbapenem-resistant Acinetobacter baumannii from a worldwide survey. J. Antimicrob. Chemother. 2021, 76, 1724–1730. [Google Scholar] [CrossRef] [PubMed]
- Ardebili, A.; Lari, A.R.; Talebi, M. Correlation of ciprofloxacin resistance with the AdeABC efflux system in Acinetobacter baumannii clinical isolates. Ann. Lab. Med. 2014, 34, 433–438. [Google Scholar] [CrossRef] [PubMed]


| Strain | AdeA (Abundance Ratio) | AdeB (Abundance Ratio) | Strain | AdeA (Abundance Ratio) | AdeB (Abundance Ratio) |
|---|---|---|---|---|---|
| A158-1 | 48.209 | - | A120-7 | - | - |
| A158-2 | 24.205 | - | A120-8 | - | - |
| A158-3 | 99.772 | 18.84 | A120-9 | 100 | 14.195 |
| A158-4 | 27.582 | - | A120-10 | 100 | - |
| A120-1 | - | - | A120-11 | - | 15.276 |
| A120-2 | - | - | A120-12 | - | 26.619 |
| A120-3 | - | - | A120-13 | - | 0.107 |
| A120-4 | - | 21.67 | A120-14 | - | 20.785 |
| A120-5 | - | 27.919 | A120-15 | - | 0.113 |
| A120-6 | - | - | A120-16 | - | 41.994 |
| Strain | Tigecycline MIC (ng/μL) | Tigecycline + CCCP MIC (ng/μL) | Strain | Tigecycline MIC (ng/μL) | Tigecycline + CCCP MIC (ng/μL) |
|---|---|---|---|---|---|
| ATCC 25922 | 0.25 | - | A120-11 | 8 | 8 |
| A120 | 0.5 | 0.5 | A120-12 | 8 | 8 |
| A120-1 | 2 | 2 | A120-13 | 8 | 8 |
| A120-2 | 4 | 2 | A120-14 | 16 | 8 |
| A120-3 | 8 | 8 | A120-15 | 8 | 8 |
| A120-4 | 16 | 16 | A120-16 | 16 | 8 |
| A120-5 | 16 | 16 | A158 | 0.5 | 0.25 |
| A120-6 | 1 | 1 | A158-1 | 16 | 8 |
| A120-7 | 4 | 4 | A158-2 | 16 | 8 |
| A120-8 | 4 | 4 | A158-3 | 32 | 16 |
| A120-9 | 64 | 32 | A158-4 | 128 | 64 |
| A120-10 | 256 | 256 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Liu, J.; Lu, Q.; Wang, P.; Zou, Q. The Mechanism of Tigecycline Resistance in Acinetobacter baumannii under Sub-Minimal Inhibitory Concentrations of Tigecycline. Int. J. Mol. Sci. 2024, 25, 1819. https://doi.org/10.3390/ijms25031819
Liu C, Liu J, Lu Q, Wang P, Zou Q. The Mechanism of Tigecycline Resistance in Acinetobacter baumannii under Sub-Minimal Inhibitory Concentrations of Tigecycline. International Journal of Molecular Sciences. 2024; 25(3):1819. https://doi.org/10.3390/ijms25031819
Chicago/Turabian StyleLiu, Cunwei, Jia Liu, Qinghui Lu, Ping Wang, and Qinghua Zou. 2024. "The Mechanism of Tigecycline Resistance in Acinetobacter baumannii under Sub-Minimal Inhibitory Concentrations of Tigecycline" International Journal of Molecular Sciences 25, no. 3: 1819. https://doi.org/10.3390/ijms25031819
APA StyleLiu, C., Liu, J., Lu, Q., Wang, P., & Zou, Q. (2024). The Mechanism of Tigecycline Resistance in Acinetobacter baumannii under Sub-Minimal Inhibitory Concentrations of Tigecycline. International Journal of Molecular Sciences, 25(3), 1819. https://doi.org/10.3390/ijms25031819






