The Inhibition Effect of Epigallocatechin-3-Gallate on the Co-Aggregation of Amyloid-β and Human Islet Amyloid Polypeptide Revealed by Replica Exchange Molecular Dynamics Simulations
Abstract
1. Introduction
2. Results and Discussion
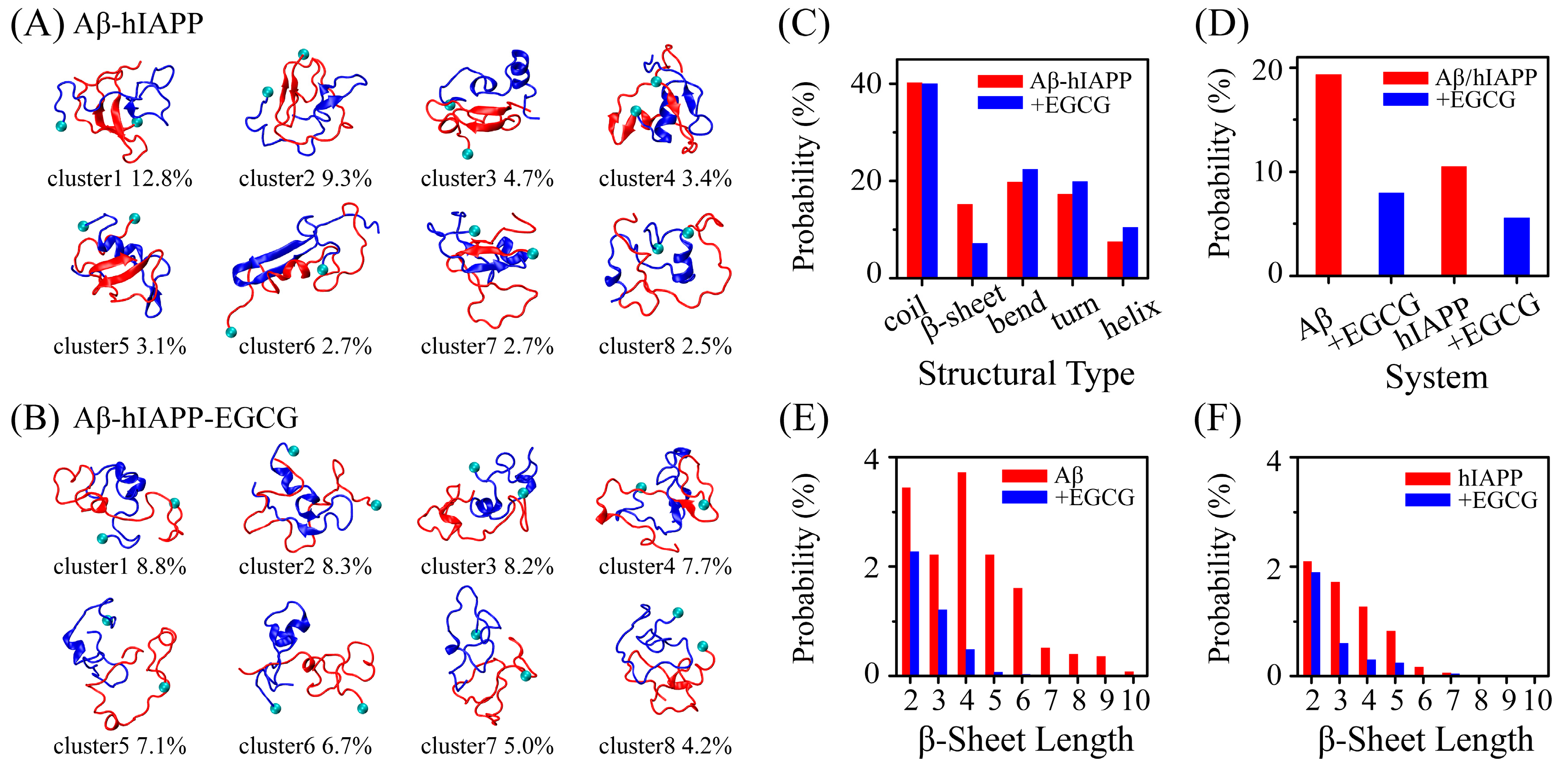
2.1. EGCG Significantly Diminishes the β-Sheet Propensity of Aβ-hIAPP Heterodimers
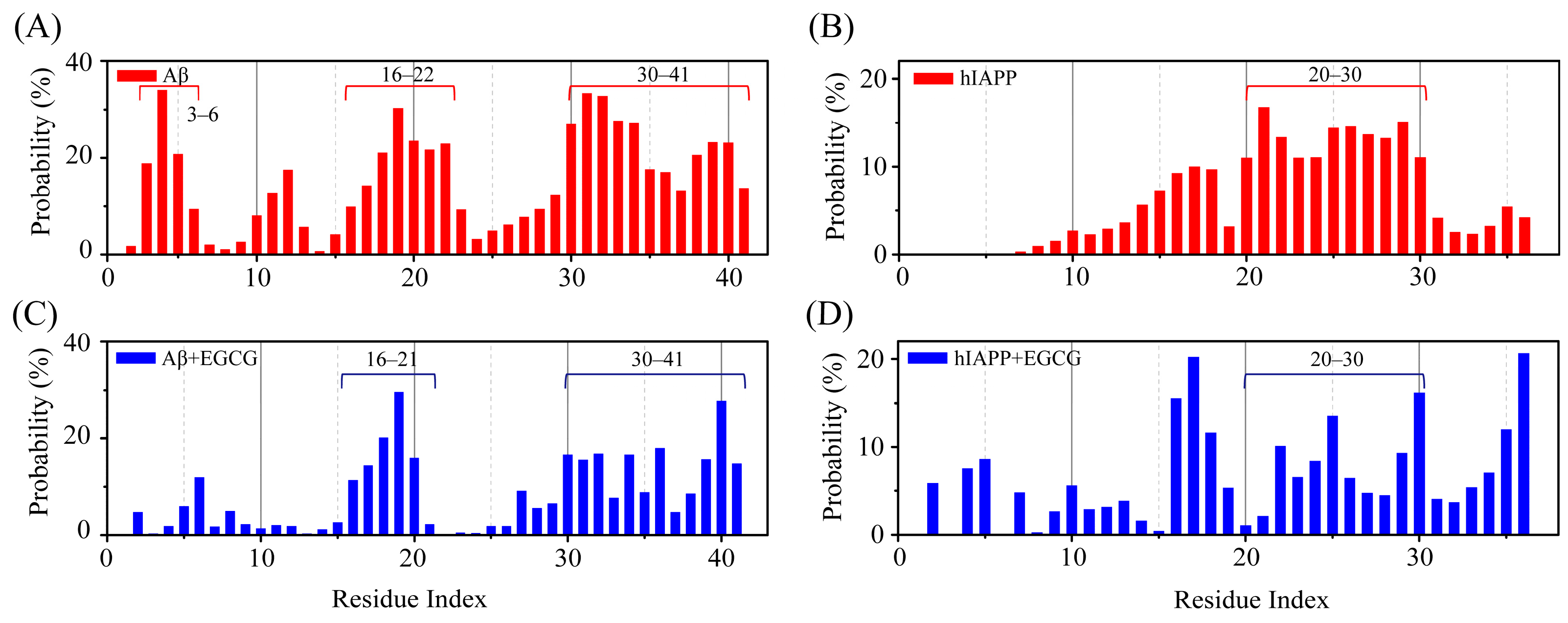
2.2. EGCG Attenuates the Aggregation Propensity of Both Aβ and hIAPP in Their Amyloid Core Regions
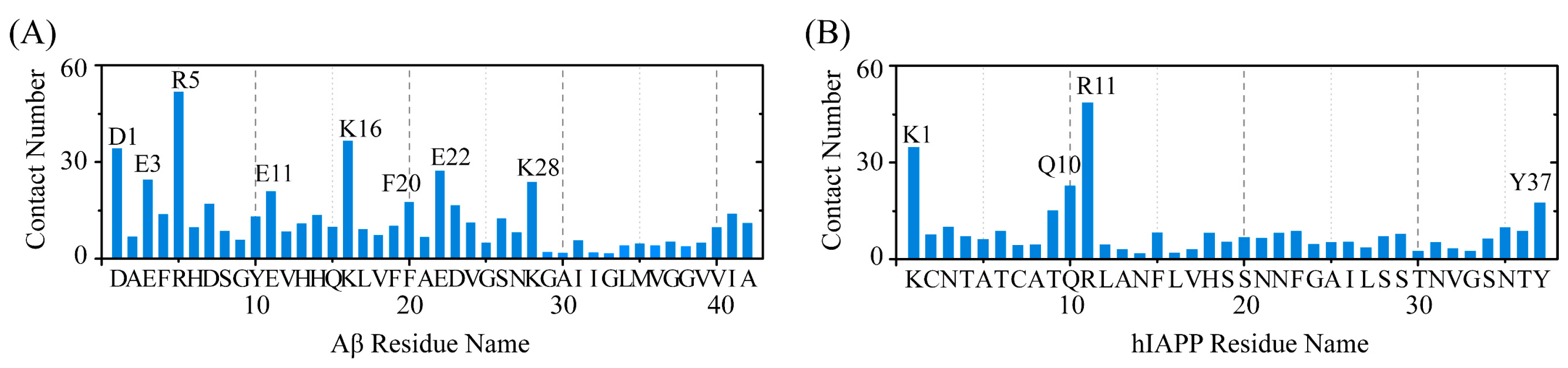
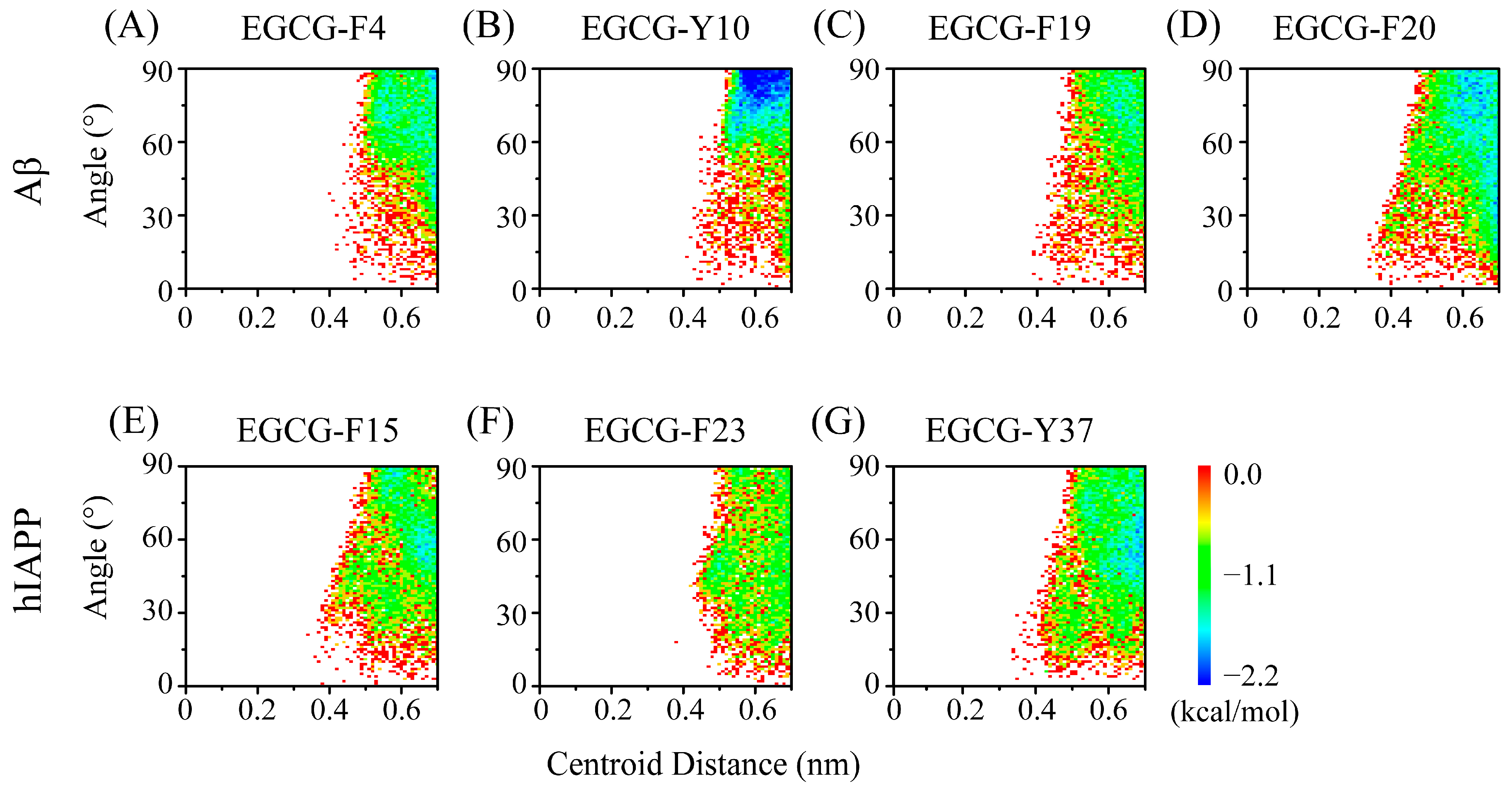
2.3. Polar and Aromatic Residues Serve as the Primary Binding Sites for EGCG within Both Aβ and hIAPP
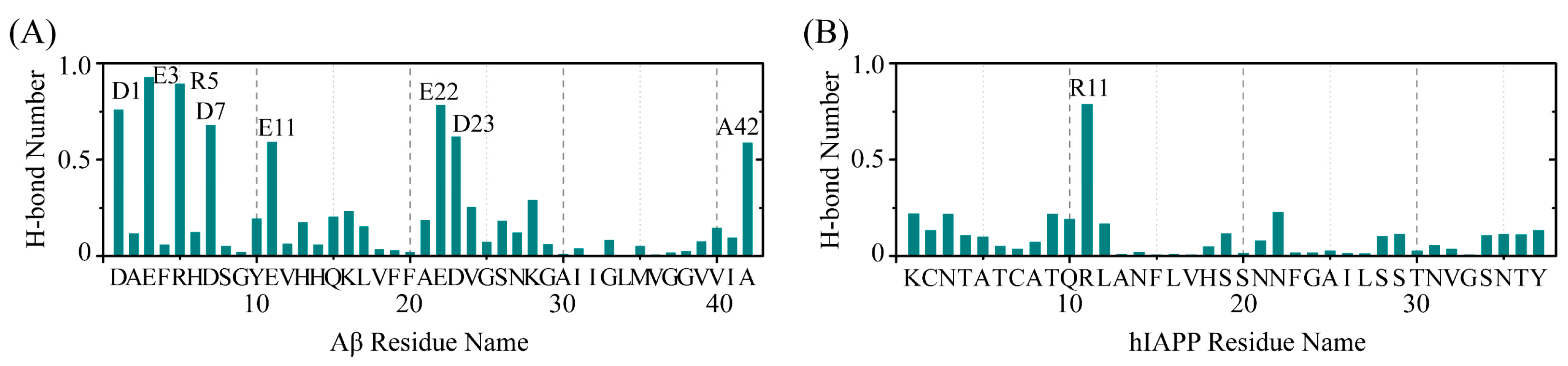
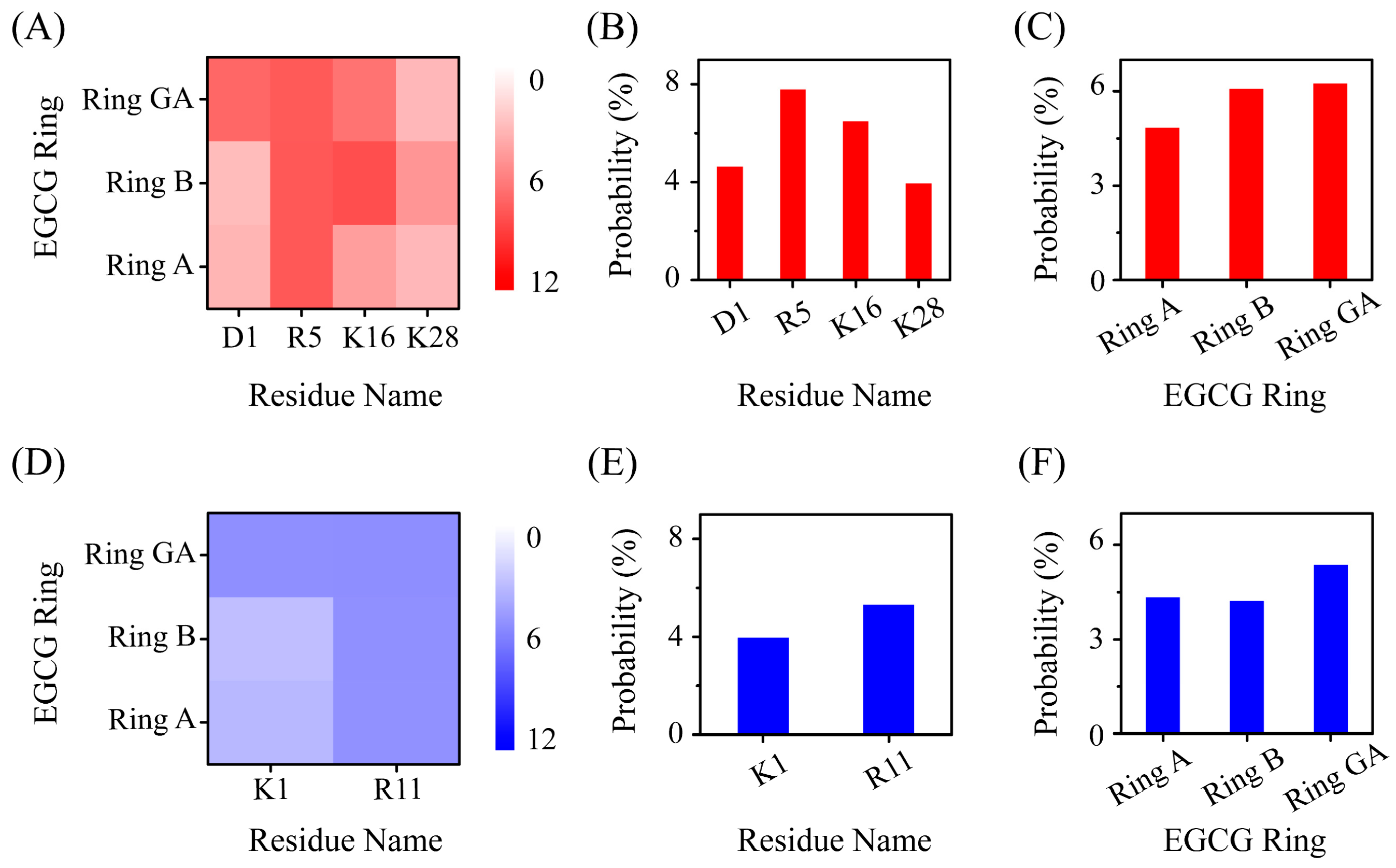
2.4. The Interactions between EGCG and Aβ-hIAPP Heterodimers Primarily Involve H-Bond Interaction, π–π Stacking and Cation–π Interaction
2.5. EGCG Attenuates Both Inter-Chain Interaction between Aβ and hIAPP, as Well as the Intra-Chain Long-Range Interaction of Their Respective Peptides within Aβ-hIAPP Heterodimers
3. Materials and Methods
3.1. System Modeling
3.2. Simulation Details
3.3. Analysis Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hartl, F.U. Protein misfolding diseases. Annu. Rev. Biochem. 2017, 86, 21–26. [Google Scholar] [CrossRef]
- Dawkins, E.; Derks, R.J.E.; Schifferer, M.; Trambauer, J.; Winkler, E.; Simons, M.; Paquet, D.; Giera, M.; Kamp, F.; Steiner, H. Membrane lipid remodeling modulates gamma-secretase processivity. J. Biol. Chem. 2023, 299, 103027. [Google Scholar] [CrossRef]
- Colvin, M.T.; Silvers, R.; Ni, Q.Z.; Can, T.V.; Sergeyev, I.; Rosay, M.; Donovan, K.J.; Michael, B.; Wall, J.; Linse, S.; et al. Atomic Resolution Structure of Monomorphic Abeta42 Amyloid Fibrils. J. Am. Chem. Soc. 2016, 138, 9663–9674. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Sun, Y.; Guo, Y.; Chen, Y.; Yu, B.; Zhang, H.; Wu, J.; Yu, X.; Kong, W.; Wu, H. Comparison of neurotoxicity of different aggregated forms of Aβ40, Aβ42 and Aβ43 in cell cultures. J. Pept. Sci. 2017, 23, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Bishoyi, A.K.; Roham, P.H.; Rachineni, K.; Save, S.; Hazari, M.A.; Sharma, S.; Kumar, A. Human islet amyloid polypeptide (hIAPP)-a curse in type II diabetes mellitus: Insights from structure and toxicity studies. Biol. Chem. 2021, 402, 133–153. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.L.; Hubble, J.P.; Lyons, K.; Paolo, A.; Troster, A.I.; Hassanein, R.E.; Koller, W.C. Risk factors for dementia in Parkinson’s disease: Effect of education. Neuroepidemiology 1996, 15, 20–25. [Google Scholar] [CrossRef]
- Ott, A.; Stolk, R.P.; van Harskamp, F.; Pols, H.A.; Hofman, A.; Breteler, M.M. Diabetes mellitus and the risk of dementia: The Rotterdam Study. Neurology 1999, 53, 1937–1942. [Google Scholar] [CrossRef] [PubMed]
- Peila, R.; Rodriguez, B.L.; Launer, L.J.; Honolulu-Asia Aging, S. Type 2 diabetes, APOE gene, and the risk for dementia and related pathologies: The Honolulu-Asia Aging Study. Diabetes 2002, 51, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Janson, J.; Laedtke, T.; Parisi, J.E.; O’Brien, P.; Petersen, R.C.; Butler, P.C. Increased Risk of Type 2 Diabetes in Alzheimer Disease. Diabetes 2004, 53, 474. [Google Scholar] [CrossRef]
- Ristow, M. Neurodegenerative disorders associated with diabetes mellitus. Int. J. Mol. Med. 2004, 82, 510–529. [Google Scholar] [CrossRef]
- Hu, G.; Jousilahti, P.; Bidel, S.; Antikainen, R.; Tuomilehto, J. Type 2 diabetes and the risk of Parkinson’s disease. Diabetes Care 2007, 30, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Tsigelny, I.F.; Crews, L.; Desplats, P.; Shaked, G.M.; Sharikov, Y.; Mizuno, H.; Spencer, B.; Rockenstein, E.; Trejo, M.; Platoshyn, O.; et al. Mechanisms of hybrid oligomer formation in the pathogenesis of combined Alzheimer’s and Parkinson’s diseases. PLoS ONE 2008, 3, e3135. [Google Scholar] [CrossRef] [PubMed]
- Miklossy, J.; Qing, H.; Radenovic, A.; Kis, A.; Vileno, B.; Làszló, F.; Miller, L.; Martins, R.N.; Waeber, G.; Mooser, V. Beta amyloid and hyperphosphorylated tau deposits in the pancreas in type 2 diabetes. Neurobiol. Aging 2010, 31, 1503–1515. [Google Scholar] [CrossRef] [PubMed]
- Santiago, J.A.; Potashkin, J.A. System-based approaches to decode the molecular links in Parkinson’s disease and diabetes. Neurobiol. Dis. 2014, 72, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Horvath, I.; Wittung-Stafshede, P. Cross-talk between amyloidogenic proteins in type-2 diabetes and Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2016, 113, 12473–12477. [Google Scholar] [CrossRef]
- Bharadwaj, P.; Wijesekara, N.; Liyanapathirana, M.; Newsholme, P.; Ittner, L.; Fraser, P.; Verdile, G. The Link between Type 2 Diabetes and Neurodegeneration: Roles for Amyloid-β, Amylin, and Tau Proteins. J. Alzheimers Dis. 2017, 59, 421–432. [Google Scholar] [CrossRef]
- Athanasaki, A.; Melanis, K.; Tsantzali, I.; Stefanou, M.I.; Ntymenou, S.; Paraskevas, S.G.; Kalamatianos, T.; Boutati, E.; Lambadiari, V.; Voumvourakis, K.I. Type 2 diabetes mellitus as a risk factor for Alzheimer’s disease: Review and meta-analysis. Biomedicines 2022, 10, 778. [Google Scholar] [CrossRef]
- Moreno-Gonzalez, I.; Edwards Iii, G.; Salvadores, N.; Shahnawaz, M.; Diaz-Espinoza, R.; Soto, C. Molecular interaction between type 2 diabetes and Alzheimer’s disease through cross-seeding of protein misfolding. Mol. Psychiatry 2017, 22, 1327–1334. [Google Scholar] [CrossRef]
- Raimundo, A.F.; Ferreira, S.; Martins, I.C.; Menezes, R. Islet Amyloid Polypeptide: A Partner in Crime With Abeta in the Pathology of Alzheimer’s Disease. Front. Mol. Neurosci. 2020, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, G.D.; Bild, V.; Ababei, D.C.; Rusu, R.N.; Cobzaru, A.; Paduraru, L.; Bulea, D. Link Between Diabetes and Alzheimer’s Disease due to the Shared Amyloid Aggregation and Deposition Involving both Neurodegenerative Changes and Neurovascular Damages. J. Clin. Med. 2020, 9, 1713. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Tang, Y.; Zhan, C.; Wei, G. Green tea extract EGCG plays a dual role in Abeta(42) protofibril disruption and membrane protection: A molecular dynamic study. Chem. Phys. Lipids 2021, 234, 105024. [Google Scholar] [CrossRef]
- Fernandes, L.; Cardim-Pires, T.R.; Foguel, D.; Palhano, F.L. Green Tea Polyphenol Epigallocatechin-Gallate in Amyloid Aggregation and Neurodegenerative Diseases. Front. Neurosci. 2021, 15, 718188. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Najafi, M.; Samarghandian, S.; Mohammadinejad, R.; Ahn, K.S. Resveratrol targeting tau proteins, amyloid-beta aggregations, and their adverse effects: An updated review. Phytother. Res. 2020, 34, 2867–2888. [Google Scholar] [CrossRef]
- Yokoyama, T.; Kusaka, K.; Mizuguchi, M.; Nabeshima, Y.; Fujiwara, S. Resveratrol Derivatives Inhibit Transthyretin Fibrillization: Structural Insights into the Interactions between Resveratrol Derivatives and Transthyretin. J. Med. Chem. 2023, 66, 15511–15523. [Google Scholar] [CrossRef]
- Jakubowski, J.M.; Orr, A.A.; Le, D.A.; Tamamis, P. Interactions between Curcumin Derivatives and Amyloid-beta Fibrils: Insights from Molecular Dynamics Simulations. J. Chem. Inf. Model. 2020, 60, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Chainoglou, E.; Hadjipavlou-Litina, D. Curcumin in Health and Diseases: Alzheimer’s Disease and Curcumin Analogues, Derivatives, and Hybrids. Int. J. Mol. Sci. 2020, 21, 1975. [Google Scholar] [CrossRef] [PubMed]
- Al Adem, K.; Shanti, A.; Srivastava, A.; Homouz, D.; Thomas, S.A.; Khair, M.; Stefanini, C.; Chan, V.; Kim, T.Y.; Lee, S. Linking Alzheimer’s Disease and Type 2 Diabetes: Characterization and Inhibition of Cytotoxic Abeta and IAPP Hetero-Aggregates. Front. Mol. Biosci. 2022, 9, 842582. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qu, L.; Cui, Z.; Lu, F.; Li, L.; Liu, F. Citrus Flavonoid Hesperetin Inhibits alpha-Synuclein Fibrillogenesis, Disrupts Mature Fibrils, and Reduces Their Cytotoxicity: In Vitro and In Vivo Studies. J. Agric. Food Chem. 2023, 71, 16174–16183. [Google Scholar] [CrossRef] [PubMed]
- Tavanti, F.; Pedone, A.; Menziani, M.C. Insights into the Effect of Curcumin and (-)-Epigallocatechin-3-Gallate on the Aggregation of Abeta(1–40) Monomers by Means of Molecular Dynamics. Int. J. Mol. Sci. 2020, 21, 5462. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, W.; Dong, X.; Sun, Y. Epigallocatechin gallate-derived carbonized polymer dots: A multifunctional scavenger targeting Alzheimer’s beta-amyloid plaques. Acta Biomater. 2023, 157, 524–537. [Google Scholar] [CrossRef]
- Franko, A.; Rodriguez Camargo, D.C.; Boddrich, A.; Garg, D.; Rodriguez Camargo, A.; Rathkolb, B.; Janik, D.; Aichler, M.; Feuchtinger, A.; Neff, F.; et al. Epigallocatechin gallate (EGCG) reduces the intensity of pancreatic amyloid fibrils in human islet amyloid polypeptide (hIAPP) transgenic mice. Sci. Rep. 2018, 8, 1116. [Google Scholar] [CrossRef]
- Kakinen, A.; Adamcik, J.; Wang, B.; Ge, X.; Mezzenga, R.; Davis, T.P.; Ding, F.; Ke, P.C. Nanoscale inhibition of polymorphic and ambidextrous IAPP amyloid aggregation with small molecules. Nano Res. 2018, 11, 3636–3647. [Google Scholar] [CrossRef]
- Bieschke, J.; Russ, J.; Friedrich, R.P.; Ehrnhoefer, D.E.; Wobst, H.; Neugebauer, K.; Wanker, E.E. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. USA 2010, 107, 7710–7715. [Google Scholar] [CrossRef]
- Kaku, T.; Tsukakoshi, K.; Ikebukuro, K. Cytotoxic Abeta Protofilaments Are Generated in the Process of Abeta Fibril Disaggregation. Int. J. Mol. Sci. 2021, 22, 12780. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.X.; Ma, G.L.; Zhang, Q.; Chen, C.H.; He, Y.M.; Xu, L.H.; Zhou, G.R.; Li, Z.H.; Yang, H.J.; Zhou, P. Inhibitory Mechanism of Epigallocatechin Gallate on Fibrillation and Aggregation of Amidated Human Islet Amyloid Polypeptide. Chemphyschem 2017, 18, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Lei, J.; Sun, Y.; Zhang, Q.; Wei, G. Conformational Ensemble of hIAPP Dimer: Insight into the Molecular Mechanism by which a Green Tea Extract inhibits hIAPP Aggregation. Sci. Rep. 2016, 6, 33076. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, J.; Derreumaux, P.; Mu, Y. Molecular mechanism of the inhibition of EGCG on the Alzheimer Abeta(1–42) dimer. J. Phys. Chem. B 2013, 117, 3993–4002. [Google Scholar] [CrossRef]
- Sakono, M.; Zako, T. Amyloid oligomers: Formation and toxicity of Aβ oligomers. FEBS J. 2010, 277, 1348–1358. [Google Scholar] [CrossRef]
- Meier, J.J.; Kayed, R.; Lin, C.-Y.; Gurlo, T.; Haataja, L.; Jayasinghe, S.; Langen, R.; Glabe, C.G.; Butler, P.C. Inhibition of human IAPP fibril formation does not prevent β-cell death: Evidence for distinct actions of oligomers and fibrils of human IAPP. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E1317–E1324. [Google Scholar] [CrossRef]
- Benilova, I.; Karran, E.; De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Wang, L.; Singh, S.; Ling, Y.L.; Buchanan, L.; Zanni, M.T.; Skinner, J.L.; de Pablo, J.J. Stable and metastable states of human amylin in solution. Biophys. J. 2010, 99, 2208–2216. [Google Scholar] [CrossRef]
- Burley, S.K.; Petsko, G.A. Aromatic-aromatic interaction: A mechanism of protein structure stabilization. Science 1985, 229, 23–28. [Google Scholar] [CrossRef]
- Burley, S.K.; Petsko, G.A. Weakly polar interactions in proteins. Adv. Protein Chem. 1988, 39, 125–189. [Google Scholar]
- Zhan, C.; Chen, Y.; Tang, Y.; Wei, G. Green tea extracts EGCG and EGC display distinct mechanisms in disrupting Aβ42 protofibril. ACS Chem. Neurosci. 2020, 11, 1841–1851. [Google Scholar] [CrossRef]
- Porat, Y.; Abramowitz, A.; Gazit, E. Inhibition of amyloid fibril formation by polyphenols: Structural similarity and aromatic interactions as a common inhibition mechanism. Chem. Biol. Drug Des. 2006, 67, 27–37. [Google Scholar] [CrossRef]
- Sinha, S.; Du, Z.; Maiti, P.; Klärner, F.-G.; Schrader, T.; Wang, C.; Bitan, G. Comparison of three amyloid assembly inhibitors: The sugar scyllo-inositol, the polyphenol epigallocatechin gallate, and the molecular tweezer CLR01. ACS Chem. Neurosci. 2012, 3, 451–458. [Google Scholar] [CrossRef]
- Li, X.; Lao, Z.; Zou, Y.; Dong, X.; Li, L.; Wei, G. Mechanistic Insights into the Co-Aggregation of Abeta and hIAPP: An All-Atom Molecular Dynamic Study. J. Phys. Chem. B 2021, 125, 2050–2060. [Google Scholar] [CrossRef] [PubMed]
- Ehrnhoefer, D.E.; Bieschke, J.; Boeddrich, A.; Herbst, M.; Masino, L.; Lurz, R.; Engemann, S.; Pastore, A.; Wanker, E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008, 15, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Pithadia, A.; Brender, J.R.; Fierke, C.A.; Ramamoorthy, A. Inhibition of IAPP Aggregation and Toxicity by Natural Products and Derivatives. J. Diabetes Res. 2016, 2016, 2046327. [Google Scholar] [CrossRef] [PubMed]
- Marquetti, I.; Desai, S. Orientation effects on the nanoscale adsorption behavior of bone morphogenetic protein-2 on hydrophilic silicon dioxide. RSC Adv. 2019, 9, 906–916. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhan, C.; Li, X.; Pan, T.; Yao, Y.; Tan, Y.; Wei, G. Five similar anthocyanidin molecules display distinct disruptive effects and mechanisms of action on Abeta(1–42) protofibril: A molecular dynamic simulation study. Int. J. Biol. Macromol. 2024, 256 Pt 2, 128467. [Google Scholar] [CrossRef]
- Yang, Z.; Yao, Y.; Zhou, Y.; Li, X.; Tang, Y.; Wei, G. EGCG attenuates alpha-synuclein protofibril-membrane interactions and disrupts the protofibril. Int. J. Biol. Macromol. 2023, 230, 123194. [Google Scholar] [CrossRef] [PubMed]
- Dupradeau, F.Y.; Pigache, A.; Zaffran, T.; Savineau, C.; Lelong, R.; Grivel, N.; Lelong, D.; Rosanski, W.; Cieplak, P. The R.E.D. tools: Advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 2010, 12, 7821–7839. [Google Scholar] [CrossRef] [PubMed]
- Mo, Y.; Brahmachari, S.; Lei, J.; Gilead, S.; Tang, Y.; Gazit, E.; Wei, G. The inhibitory effect of hydroxylated carbon nanotubes on the aggregation of human islet amyloid polypeptide revealed by a combined computational and experimental study. ACS Chem. Neurosci. 2018, 9, 2741–2752. [Google Scholar] [CrossRef]
- Li, X.; Lei, J.; Qi, R.; Xie, L.; Wei, G. Mechanistic insight into E22Q-mutation-induced antiparallel-to-parallel β-sheet transition of Aβ16−22 fibrils: An all-atom simulation study. Phys. Chem. Chem. Phys. 2019, 21, 15686–15694. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Sun, Y.; Chen, Y.; Lei, J.; Wei, G. Molecular dynamics simulations reveal the mechanism of graphene oxide nanosheet inhibition of Aβ1–42 peptide aggregation. Phys. Chem. Chem. Phys. 2019, 21, 10981–10991. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. Softwarex 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Van Der Spoel, D. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef]
- Lindorfflarsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct. Funct. Bioinf. 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Postma, J.P.; van Gunsteren, W.F.; Hermans, J. Interaction models for water in relation to protein hydration. In Intermolecular Forces; Springer: Berlin/Heidelberg, Germany, 1981; pp. 331–342. [Google Scholar]
- Chen, Y.; Li, X.; Zhan, C.; Lao, Z.; Li, F.; Dong, X.; Wei, G. A Comprehensive Insight into the Mechanisms of Dopamine in Disrupting Aβ Protofibrils and Inhibiting Aβ Aggregation. ACS Chem. Neurosci. 2021, 12, 4007–4019. [Google Scholar] [CrossRef]
- Dong, X.; Qi, R.; Qiao, Q.; Li, X.; Li, F.; Wan, J.; Zhang, Q.; Wei, G. Heparin remodels the microtubule-binding repeat R3 of Tau protein towards fibril-prone conformations. Phys. Chem. Chem. Phys. 2021, 23, 20406–20418. [Google Scholar] [CrossRef]
- Piana, S.; Lindorff-Larsen, K.; Shaw, D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011, 100, L47–L49. [Google Scholar] [CrossRef]
- Hoffmann, K.Q.; McGovern, M.; Chiu, C.C.; de Pablo, J.J. Secondary Structure of Rat and Human Amylin across Force Fields. PLoS ONE 2015, 10, e0134091. [Google Scholar] [CrossRef]
- Somavarapu, A.K.; Kepp, K.P. The Dependence of Amyloid-beta Dynamics on Protein Force Fields and Water Models. Chemphyschem 2015, 16, 3278–3289. [Google Scholar] [CrossRef] [PubMed]
- Carballo-Pacheco, M.; Strodel, B. Comparison of force fields for Alzheimer’s A beta42: A case study for intrinsically disordered proteins. Protein Sci. 2017, 26, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Johnson, W.L.; Goddard, W.A. Ergodicity and Convergence of Fluctuations in Parrinello-Rahman Molecular Dynamics. MRS Online Proc. Libr. 1992, 291, 285–290. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle—An Analytical Version of the Shake and Rattle Algorithm for Rigid Water Models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- Hess, B. P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 2008, 4, 116–122. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Li, X.; Dong, X.; Wei, G.; Margittai, M.; Nussinov, R.; Ma, B. The distinct structural preferences of tau protein repeat domains. Chem. Commun. 2018, 54, 5700–5703. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Tang, Y.; Lao, Z.; Lei, J.; Wei, G. Common cancer mutations R175H and R273H drive the p53 DNA-binding domain towards aggregation-prone conformations. Phys. Chem. Chem. Phys. 2020, 22, 9225–9232. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, Z.; Chen, Y.; Zhang, S.; Wei, G.; Zhang, L. Dissecting the Molecular Mechanisms of the Co-Aggregation of Abeta40 and Abeta42 Peptides: A REMD Simulation Study. J. Phys. Chem. B 2023, 127, 4050–4060. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure-Pattern-Recognition of Hydrogen-Bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; Van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]

| System | Aβ-hIAPP | Aβ-hIAPP-EGCG |
|---|---|---|
| Number of replicas | 48 | |
| Temperature range | 308.20–400.00 K | |
| Simulation time | 500 ns | 600 ns |
| Number of atoms | 40,522 | 40,447 |
| Number of water molecules | 13,094 | 12,899 |
| Box size | 7.5 × 7.5 × 7.5 nm3 | |
| Number of EGCG | --- | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Zhang, Y.; Yang, Z.; Zhang, S.; Zhang, L. The Inhibition Effect of Epigallocatechin-3-Gallate on the Co-Aggregation of Amyloid-β and Human Islet Amyloid Polypeptide Revealed by Replica Exchange Molecular Dynamics Simulations. Int. J. Mol. Sci. 2024, 25, 1636. https://doi.org/10.3390/ijms25031636
Li X, Zhang Y, Yang Z, Zhang S, Zhang L. The Inhibition Effect of Epigallocatechin-3-Gallate on the Co-Aggregation of Amyloid-β and Human Islet Amyloid Polypeptide Revealed by Replica Exchange Molecular Dynamics Simulations. International Journal of Molecular Sciences. 2024; 25(3):1636. https://doi.org/10.3390/ijms25031636
Chicago/Turabian StyleLi, Xuhua, Yu Zhang, Zhiwei Yang, Shengli Zhang, and Lei Zhang. 2024. "The Inhibition Effect of Epigallocatechin-3-Gallate on the Co-Aggregation of Amyloid-β and Human Islet Amyloid Polypeptide Revealed by Replica Exchange Molecular Dynamics Simulations" International Journal of Molecular Sciences 25, no. 3: 1636. https://doi.org/10.3390/ijms25031636
APA StyleLi, X., Zhang, Y., Yang, Z., Zhang, S., & Zhang, L. (2024). The Inhibition Effect of Epigallocatechin-3-Gallate on the Co-Aggregation of Amyloid-β and Human Islet Amyloid Polypeptide Revealed by Replica Exchange Molecular Dynamics Simulations. International Journal of Molecular Sciences, 25(3), 1636. https://doi.org/10.3390/ijms25031636







