Abstract
In the artificial breeding of Pampus argenteus (Euphrasen, 1788), female fish spawn before male release sperm, which indicates rapid ovarian development. In fish, aromatase is responsible for converting androgens into estrogens and estrogen plays a crucial role in ovarian development. In this study, we aimed to investigate the potential role of brain-type and ovarian-type aromatase to study the rapid ovarian development mechanism. The results showed that cyp19a1a was mainly expressed in the ovary and could be classified as the ovarian type, whereas cyp19a1b could be considered as the brain type for its expression was mainly in the brain. During ovarian development, the expression of cyp19a1a in the ovary significantly increased from stage IV to stage V and Cyp19a1a signals were present in the follicle cells, while cyp19a1b expression in the pituitary gland decreased from stage IV to stage V. To further investigate the function of Cyp19a1a, recombinant Cyp19a1a (rCyp19a1a) was produced and specific anti-Cyp19a1a antiserum was obtained. The expressions of cyp19a1a, estrogen receptors 2 alpha (esr2a), and androgen receptor alpha (arα) were significantly upregulated in the presence of rCyp19a1a. Meanwhile, cyp19a1a was expressed significantly after E2 treatment in both ovarian and testicular tissue culture. Taken together, we found two forms of aromatase in silver pomfret. The ovarian-type aromatase might play an important role in ovarian differentiation and maturation, and participate in E2 synthesis through co-regulation with esr2a. The brain-type aromatase cyp19a1b might be involved in the regulation of both brain and gonadal development.
1. Introduction
Pampus argenteus is widely distributed in the Indian Ocean, North Sea, Oman Sea, and Persian Gulf, as well as from the Bohai Sea to the South China Sea in China [1,2,3]. P. argenteus is a commercially important fish in the aquaculture industry worldwide, and its artificial breeding technique has been studied in many countries [4,5]. With the breakthrough of P. argenteus aquaculture technology, artificial cultivation is gradually moving towards large-scale farming. However, in the farming production process, challenges such as high mortality rates, poor development, differences in meat quality, and disparities compared to wild P. argenteus still exist. The cultivation of P. argenteus also presents the phenomenon of females spawning before male fish release sperm, which increases the difficulty of fry rearing. In addition, the body size of female-cultured P. argenteus is larger than that of males at harvest, which reveals that it is important to study the gonad differentiation and development mechanism in artificial breeding of this species. The results will provide a theoretical basis for addressing related farming challenges of artificial cultivation and the establishment of mono-sex female production technology in P. argenteus.
Cytochrome P450 aromatase is a key steroidogenic enzyme that catalyzes the conversion of androgens to estrogens [6,7]. Two homologous genes encoding aromatase have been identified in fish, which are cyp19a1a and cyp19a1b. The cyp19a1a gene is primarily expressed in the gonads, whereas cyp19a1b is expressed in the brain [8]. Cyp19a1a, as a key enzyme for estradiol synthesis in teleosts, is crucial for gonadal differentiation, maintenance, and maturation [9]. Previous studies have shown that rapid downregulation of the cyp19a1a gene results in a significant decrease in the estradiol content in the serum of fish [10,11,12], and the gonads of protandrous hermaphroditic eels with a cyp19a1a knockout remain undifferentiated and do not develop into ovaries [13]. Meanwhile, Cyp19a1a is highly expressed in the yolk formation process [14,15,16], and has been proved to be involved in ovarian differentiation and vitellogenesis of oocytes in teleosts. In the pituitary gland of fish, cyp19a1b not only regulates the synthesis of estrogen through estrogen receptors but also controls the synthesis and secretion of follicle-stimulating hormone and luteinizing hormone by secretory cells, thereby regulating the differentiation, development, maturation, and spawning of gonads [17,18,19,20]. Both brain-type and ovarian-type aromatase play a regulatory role in the gonadal development of fish. However, there was no research on P. argenteus. Thus, this study aimed to study the ovarian development mechanism to explore the reason for rapid ovarian development in the artificial breeding of P. argenteus through investigating cyp19a1a and cyp19a1b gene expression patterns and potential functions during the reproductive cycle.
2. Results
2.1. Molecular Identification and Phylogenetics of the P. argenteus cyp19a1a and cyp19a1b Genes
The sequences of the cyp19a1a and cyp19a1b genes were determined via ovary and brain cDNA cloning, respectively. The sequence of cyp19a1a (1536 bp) contained an open reading frame encoding 521 amino acids, and the sequence of cyp19a1b (1139 bp) contained an open reading frame encoding 379 amino acids. The phylogenetic analysis showed that Cyp19a1a and Cyp19a1b in P. argenteus were clustered with their homologous genes in other fishes (Figure 1).
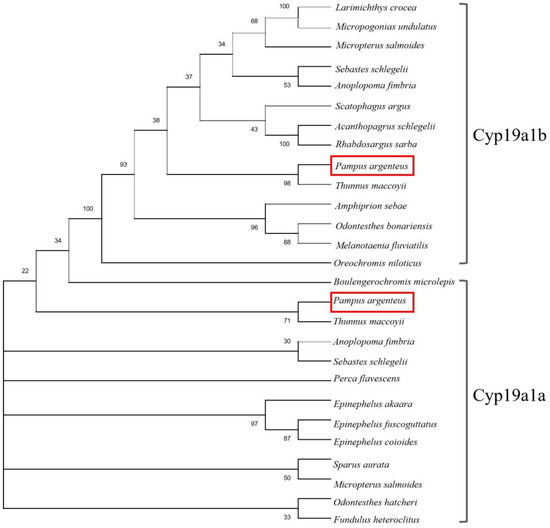
Figure 1.
Comparison of the P. argenteus Cyp19a1a and Cyp19a1b protein sequences in different fish. The evolutionary distances were computed using the Jones–Taylor–Thornton (JTT) matrix-based method and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.7). The confidence in the resulting tree was assessed through the bootstrap method with 1000 replications. The phylogenetic analysis of Cyp19a1a and Cyp19a1b was aligned using Clustal W. The GenBank accession numbers for sequence data analyzed with Cyp19a1a are as follows: Anoplopoma fimbria (AGH69789.1), Fundulus heteroclitus (AAR97268.1), Odontesthes hatcheri (ABK41198.1), Sebastes schlegelii (ACN39247.2), Epinephelus fuscoguttatus (AYQ58300.1), Epinephelus coioides (AAR97601.1), Epinephelus akaara (AY547354.1), Perca flavescens (ABL64075.1), Sparus aurata (AAL27699.1), Boulengerochromis microlepis (AGS14768.1), Micropterus salmoides (XP_038558015.1), Thunnus maccoyii (XP_042264966.1); and for Cyp19a1b: Micropogonias undulatus (AEL31294.1), Larimichthys crocea (ACO35042.1), Sebastes schlegelii (AGG53952.1), Rhabdosargus sarba (ABC70868.1), Acanthopagrus schlegelii (QEN95916.1), Scatophagus argus (AFV47578.2), Amphiprion sebae (AJP70568.1), Melanotaenia fluviatilis (AED99847.1), Odontesthes bonariensis (AAQ88434.2), Oreochromis niloticus (AAG18458.1), Micropterus salmoides (XP_038562227.1), Thunnus maccoyii (XP_042267530.1), Anoplopoma fimbria (XP_054476115.1). The two red frames indicated Cyp19a1b and Cyp19a1a in P. argenteus, respectively.
2.2. Expression Profiles of cyp19a1a and cyp19a1b in P. argenteus
Based on the histological characteristics, six different stages were classified in ovary and testis, respectively (Figure 2). The expression profiles of cyp19a1a and cyp19a1b were detected via qPCR. The qPCR results showed that cyp19a1a was significantly expressed in the ovary (Figure 3A), whereas cyp19a1b was significantly expressed in the pituitary, forebrain, and hypothalamus (Figure 3D). In the early development stage of P. argenteus, cyp19a1a was highly expressed from the yolk plug stage to the pre-hatching stage (Figure 3B). However, no significant change was found in cyp19a1b expression during the early development stage (Figure 3E). During gonadal development, cyp19a1a expression in ovaries was significantly higher than in testis (Figure 3C) with the highest expression at stage V in ovaries. The expression of cyp19a1b was significantly higher in males than in females at stage III (Figure 3F). In the pituitary gland, cyp19a1b expression decreased from stage III to stage V in males and stage IV to stage V in females.
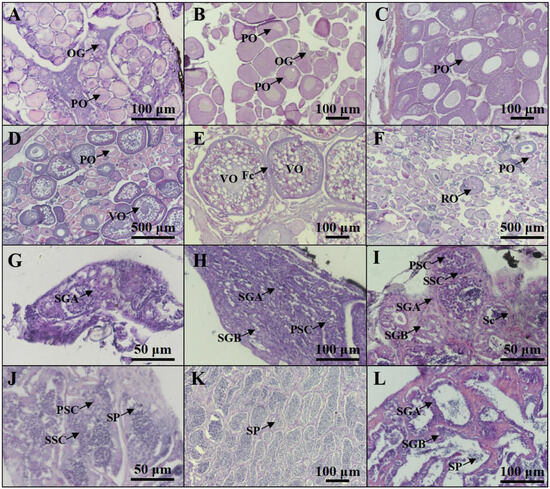
Figure 2.
Profiles of gonadal development in P. argenteus. The different stages of ovarian development include stage I (A), stage II (B), stage III (C), stage IV (D), stage V (E), and stage VI (F). The different stages of testicular development include stage I (G), stage II (H), stage III (I), stage IV (J), stage V (K), and stage VI (L). OG, oogonia; PO, primary oocyte; VO, vitellogenic oocyte; Fc, follicle cells; RO, regressed oocyte; SGA, spermatogonia type A; SGB, spermatogonia type B; PSC, primary spermatocyte; SSC, secondary spermatocyte; Sc, Sertoli cell; SP, sperm.
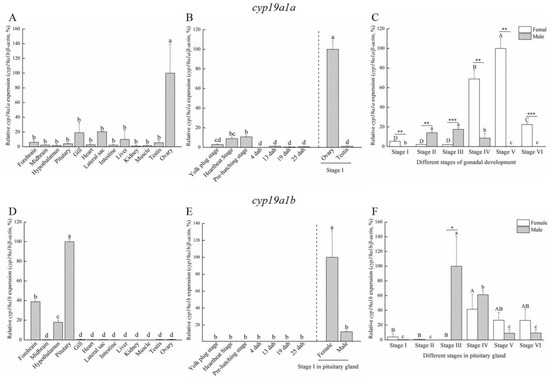
Figure 3.
The expression patterns of cyp19a1a and cyp19a1b in P. argenteus. (A) The tissue distribution of cyp19a1a expression. (B) cyp19a1a expression during the early development stage. (C) cyp19a1a expression during gonadal development. (D) The tissue distribution of cyp19a1b expression. (E) cyp19a1b expression during the early development stage. (F) cyp19a1b expression during gonadal development. Different uppercase letters indicate one-way ANOVA, followed by Duncan’s test (p < 0.05) in female group. Different lowercase letters indicate one-way ANOVA, followed by Duncan’s test (p < 0.05) in male group. Asterisk indicates Student’s t-test (p < 0.05). “*” indicates Student’s t-test (p < 0.05). “**” indicates Student’s t-test (p < 0.01). “***” indicates Student’s t-test (p < 0.01).
2.3. Western Blot Analysis and the Localization of Cyp19a1a
For the Western blot analysis, the signal was observed in ovarian and testicular tissues using the anti-β-Actin antibody. According to Figure 4, β-Actin in P. argenteus was about 42 kDa, and Cyp19a1a at 75 kDa was detected using the anti-Cyp19a1a antibody in ovarian tissue. IHC was then used to detect Cyp19a1a protein expression at different gonadal development stages (Figure 5). The results showed that the signal of Cyp19a1a was only present in the follicle cells at stage V in the ovary (Figure 5A–F). In the testis, the signals were detected in spermatogonia from stage II to stage IV, which was consistent with the qPCR results (Figure 5G–L).
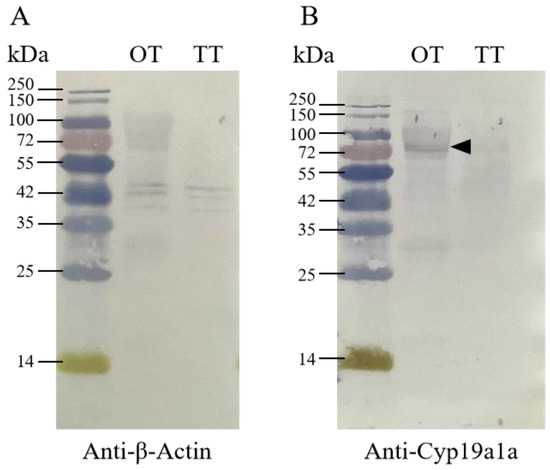
Figure 4.
The Cyp19a1a protein in the gonad of P. argenteus. (A) The β-Actin signals were detected via anti-β-Actin antibody in ovarian and testicular tissue. (B) Cyp19a1a signals were detected via anti-Cyp19a1a antibody in ovarian and testicular tissue. The black arrowhead indicates the signal of the anti-Cyp19a1a antibody. OT, ovarian tissue; TT, testicular tissue. The black arrowheads indicate the signals of the anti-Cyp19a1a antibody.
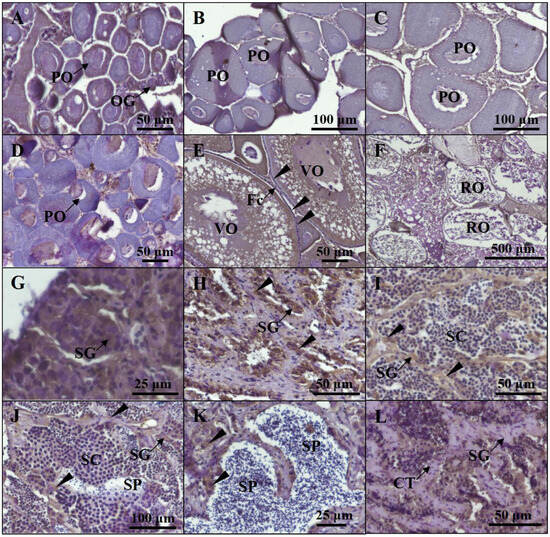
Figure 5.
The distribution of Cyp19a1a protein at different gonadal stages. The distribution of the Cyp19a1a protein was detected via IHC staining at different gonadal stages, including stage I (A), stage II (B), stage III (C), stage IV (D), stage V (E), and stage VI (F) in females and stage I (G), stage II (H), stage III (I), stage IV (J), stage V (K), and stage VI (L) in males. OG, oogonia; PO, primary oocyte; Fc, follicle cell; VO, vitellogenic oocyte; RO, regressed oocyte; SG, spermatogonia; SC, spermatocyte; SP, sperm; CT, connected tissue. The black arrowheads indicate the positive signals of the anti-Cyp19a1a antibody.
2.4. The Production of rCyp19a1a and the Role of rCyp19a1a in Gonadal Tissue Culture
rCyp19a1a was used to elucidate the function of Cyp19a1a through the in vitro ovarian and testicular tissue culture experiments. According to Western blotting, the Cyp19a1a signal was detected in rCyp19a1a cells, which was about 75 kDa (Figure 6). To further understand the role of Cyp19a1a in the gonads of P. argenteus, the in vitro system was used to study its effect on sex-related gene expression. The BrdU-incorporated cells were observed in the absence or presence of rCyp19a1a (Supplementary Figure S1). In cultured ovarian tissues, the qPCR results showed that the expression of cyp19a1a was significantly higher in both low and high doses of rCyp19a1a than in the absence of rCyp19a1a (Figure 7A). In addition, esr2a expression was higher in both low- and high-dose Cyp19a1a over-expressed groups (Figure 7C), while estrogen receptor 1 (esr1) was not significantly upregulated in these two doses in Cyp19a1a over-expressed groups (Figure 7B). However, rCyp19a1a treatment had no effect on the expression of arα, folliculogenesis specific bHLH transcription factor (figla), and hydroxysteroid 17-beta dehydrogenase 1 (hsd17β1) (Figure 7D–F). For cultured testicular tissues, rCyp19a1a treatment had no effect on the expression of cyp19a1a and esr1 (Figure 8A,B). However, esr2a and arα were significantly upregulated in the presence of the high-dose rCyp19a1a group (Figure 8C,D).
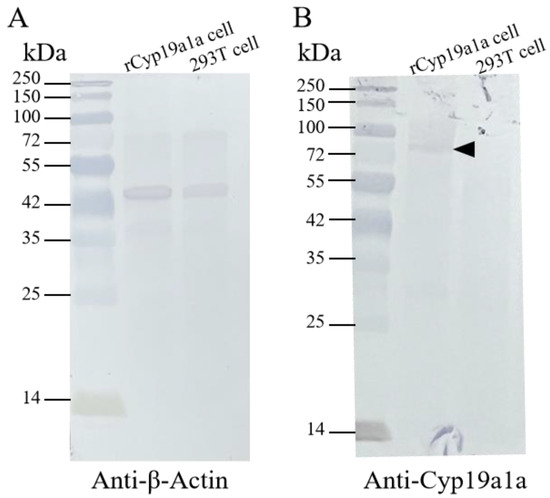
Figure 6.
Western blot of recombinant Cyp19a1a produced in HEK293 cells. (A) The β-Actin signals were detected via anti-β-Actin antibody in rCyp19a1a cells and 293T cells. (B) The rCyp19a1a signals were detected via anti-Cyp19a1a in the rCyp19a1a cells and 293T cells. The black arrowhead indicates the signals of the anti-Cyp19a1a antibody.
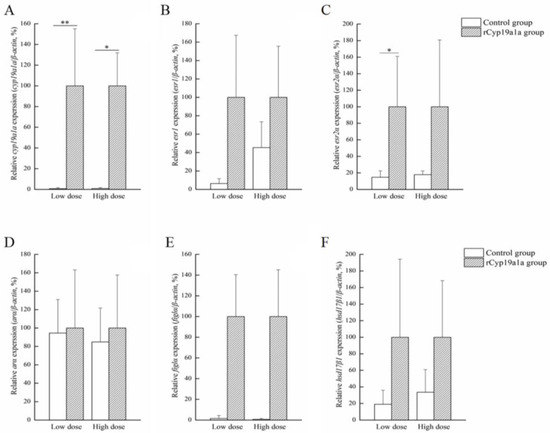
Figure 7.
Effects of rCyp19a1a on sex-related gene expression in ovarian tissue. The expression of cyp19a1a (A), esr1 (B), esr2a (C), arα (D), figla (E), and hsd17β1 (F) in cultured ovarian tissues. The expression of sex-related genes was analyzed using qPCR (n = 6 for each value). “*” indicates Student’s t-test (p < 0.05). “**” indicates Student’s t-test (p < 0.01).
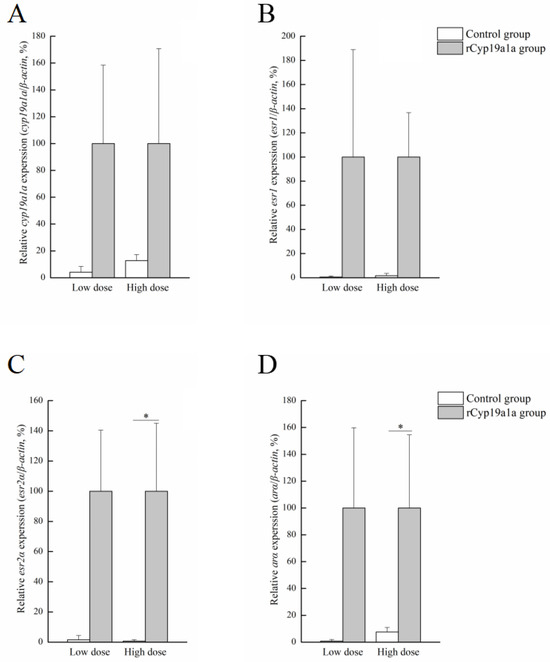
Figure 8.
Effects of rCyp19a1a on sex-related gene expression in testicular tissue. The expression of cyp19a1a (A), esr1 (B), esr2a (C), and arα (D) in cultured testicular tissues. The expression of sex-related genes was analyzed using qPCR (n = 6 for each value). “*” indicates Student’s t-test (p < 0.05).
2.5. The Influence of Different Hormones on Gonadal Tissue Culture
To analyze the proliferation activity of ovaries or testis in different hormones, IHC was used to detect the incorporation of BrdU. The BrdU-incorporated cells were observed in the different hormones treatment (Supplementary Figure S2). Furthermore, according to the results of BrdU IHC staining, the number of proliferated cells were counted (Supplementary Figure S3). According to the count, the different concentration groups with the highest cell proliferation count were selected as the optimal concentration. The 50 ng/mL E2 concentration was selected as the optimal concentration in the ovary (Supplementary Figure S2C). For the AI and MT concentrations, 1000 ng/mL and 120 ng/mL were selected, respectively (Supplementary Figure S2E,G). In testicular tissue culture, 100 ng/mL E2, 100 ng/mL AI, and 15 ng/mL MT concentrations had the highest numbers of proliferated cells (Supplementary Figure S2D,F,G). Therefore, the optimal concentrations of different hormones or medicine were selected to add to the culture medium. The results of different hormone treatments showed that cyp19a1a was expressed significantly in the E2 group than compared to the other groups in both ovarian tissue culture and testicular tissue culture (Figure 9). Also, in testicular tissue culture, excepted the E2 group, cyp19a1a expression in MT, E2+AI, and E2+MT groups was significantly higher than the control and AI groups (Figure 9B). Taken together, E2 may induce cyp19a1a expression in in vitro gonadal tissue culture.
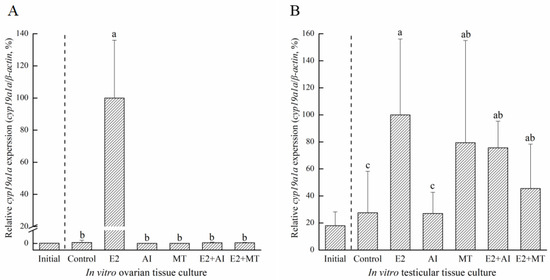
Figure 9.
Effects of different hormone treatments on cyp19a1a expression in ovarian (A) and testicular (B) tissues. Initial, cyp19a1a expression before the treatment experiment. E2, 50 ng/mL of β-estradiol; MT, 120 ng/mL of methyltestosterone solution; AI, 1000 ng/mL of anastrozole; E2+AI, 50 ng/mL of β-estradiol and 1000 ng/mL of anastrozole; E2+MT, 50 ng/mL of β-estradiol and 120 ng/mL of methyltestosterone solution. Superscript letters indicate one-way ANOVA, followed by Duncan’s test (p < 0.05). Superscript letters indicate one-way ANOVA, followed by Tukey’s test (p < 0.05).
3. Discussion
3.1. Duplicated Aromatase Genes in P. argenteus
The cyp19a1 genes in teleosts are different from most other vertebrates because they have two forms, namely cyp19a1a and cyp19a1b [21]. Up to now, these two forms of aromatase gene have been identified in many teleosts, such as rainbow trout (Oncorhynchus mykiss) [22,23], zebrafish (Danio rerio) [24], Nile tilapia (Oreochromis niloticus) [25], European sea bass (Dicentrarchus labrax) [26], red spotted pufferfish (Takifugu rubripes) [20], orange-spotted grouper (Epinephelus coioides) [27], slender wrasse (Halichoeres tenuispinis) [28], and Asian swamp eel (Monopterus albus) [29]. The duplicated cyp19a1 genes are assumed to be the result of the teleost-specific third round of whole-genome duplication (TWGD, 3R) [30]. Meanwhile, these two aromatase genes exhibit tissue-specific expression patterns, with cyp19a1a primarily expressed in the gonad and cyp19a1b predominantly expressed in the brain [31]. In this study, the cyp19a1a and cyp19a1b genes were identified in P. argenteus. According to the phylogenetic tree, Cyp19a1a and Cyp19a1b of P. argenteus clustered with their homologous genes in other fishes. However, although Cyp19a1a and Cyp19a1b were identified by the phylogenetic tree, the drawbacks of protein sequences, which were obtained from NCBI GenBank data, are that they may be influenced by errors in species identification. Also, GenBank records may lack detailed information regarding experimental methods, specimen sources, or other crucial experimental conditions. Protein sequences obtained from the NCBI database have certain limitations, such as some protein sequences were not identified as brain-type or ovarian-type aromatase. In this study, protein sequences annotated with ovarian aromatase and brain aromatase, or those with experimental results, were screened to minimize associated risks. Furthermore, cyp19a1a is mainly expressed in the gonad, whereas cyp19a1b is expressed in the brain, especially in the pituitary in this study. Therefore, the aromatase gene in P. argenteus was similar to that of most teleosts, exhibiting two forms which were brain type and ovarian type.
3.2. The Ovarian-Type Aromatase Might Play an Important Role in Ovarian Differentiation and Maturation
In teleosts, the gonadal aromatase gene cyp19a1a mainly expresses in the gonad and plays a role in early embryo development and gonadal sex differentiation, contributing to the initiation and maintenance of ovarian differentiation [9,32]. cyp19a1a also participates in gonadogenesis and gametogenesis in mature fish [33]. In P. argenteus, cyp19a1a was mainly expressed in the ovaries, which is similar to rainbow trout [23], Asian swamp eel [29], and Schizothorax prenanti [16]. In addition, a particularly high expression was detected at stage II and stage III during testicular development, which was consistent with the results in Scatophagus argus [34] and Heteropneustes fossilis [19]. In P. argenteus, high cyp19a1a expression in the ovary was observed in stage IV and stage V during gonadal development, which was in agreement with findings in rainbow trout [22], Pagrus major [35], Nile tilapia [36], Asian swamp eel [29], and S. prenanti [16], showing that cyp19a1a significantly increased at the stage of vitelline accumulation. Meanwhile, cyp19a1a barely expressed during the early developmental stages of P. argenteus. This result was different from Colossoma macropomum in which the expression of cyp19a1a increased when the body length of fish was 2 to 4 cm [37]. However, in P. argenteus, the expression of cyp19a1a was higher in early differentiated ovary than in undifferentiated gonad [38], which indicated that cyp19a1a may play a role in triggering the initiation of female sex differentiation. Also, cyp19a1a was closely associated with early gonadal differentiation in Nile tilapia [25,39], Oryzias luzonensis [40], and Cyprinus carpio [41]. Thus, the cyp19a1a of P. argenteus may play an important role in ovarian differentiation.
The regulation of ovarian maturation in fish by aromatase plays a crucial role in reproductive processes [42]. Aromatase is responsible for the conversion of androgens to estrogens, affecting various aspects of ovarian development, oocyte maturation, and the reproductive cycle. During the early stages of ovarian maturation, aromatase activity is relatively low, coinciding with elevated androgen levels [32]. As ovarian maturation progresses, there is a dynamic upregulation of aromatase expression, leading to an increased conversion of androgens to estrogens. This hormonal shift is essential for the development and maturation of oocytes within the ovaries [43]. In P. argenteus, cyp19a1a expression in ovaries was significantly higher than in testis, with the highest expression at stage V in ovaries during gonadal development, which indicated that the ovarian-type aromatase might play an important role in ovarian maturation in P. argenteus.
3.3. The Brain-Type Aromatase Might Be Involved in the Regulation of Both Brain and Gonadal Development
Aromatase activity in the brain is significantly more pronounced in teleosts compared to mammals. This difference has been proposed to contribute to the plasticity observed in the teleost brain [44,45]. The expression of cyp19a1b in teleost brain is related to brain cell proliferation and plays a crucial role in brain development, neurogenesis, and brain repair [46,47]. In the present study, cyp19a1b was expressed specifically in the brain, especially in the pituitary of P. argenteus. And during gonadal development, cyp19a1b was highly expressed at stage III to stage IV in the male pituitary, and stage IV to stage V in the female pituitary. This expression pattern in P. argenteus was similar to zebrafish [48], pufferfish [20], and the black porgy (Acanthopagrus schlegelii) [49]. In addition, it was found that the mutation of cyp19a1b increased the latency to the first oviposition in female zebrafish, indicating that cyp19a1b participates in the regulation of gonadal development. Taken together, cyp19a1b might be involved in brain development and also participate in the regulation of gonadal development in P. argenteus.
3.4. Cyp19a1a Participates in E2 Synthesis through Co-Regulation with esr2a
Aromatase is responsible for estrogen production, together with estrogens. It is generally considered to be essential for ovarian differentiation and plays a direct role in regulating ovary development and maturation [50,51]. In teleosts, such as Nile tilapia [52], zebrafish [53], and medaka (Oryzias luzonensis) [54], the knockout of cyp19a1a through genome editing methodologies leads to the development of testes. In numerous teleosts, genotypic males go into feminization after E2 treatment [55]. The methods of E2 or aromatase inhibitor treatment have been extensively employed for sex ratio control [56,57,58,59]. In this study, after the addition of different hormones, the expression of cyp19a1a was highest in the E2 addition group in the ovary and cyp19a1a expression was higher in E2 and MT groups in testes, which indicated that there existed a complex feedback regulation mechanism between aromatase and estrogen receptors. An increase in estrogen levels can stimulate the synthesis of aromatase. Also, in this study, cyp19a1a expression was significantly increased after rCyp19a1a treatment in both low and high doses in ovarian tissue, which indicated the effectiveness of Cyp19a1a in the ovary. esr2a was significantly increased after low-dose rCyp19a1a treatment. Although there were no significant differences in the femaleness-related genes, the expression of esr1, figla, and hsd17β1 genes increased after rCyp19a1a overexpression. In testicular culture, the expression of esr2a and ara was significantly increased. These data indicated that Cyp19a1a in P. argenteus was responsible for androgens converting into estrogens, which was similar to most teleosts. These data suggested that estrogen receptors regulated the expression of the aromatase gene directly or indirectly. The binding of estrogen could influence the transcription levels of the aromatase gene, thereby regulating the synthesis of estrogen. Studies indicated that estrogen receptors were widely expressed in ovarian tissue, divided into two main subtypes, esr1 and esr2 [60]. The expression patterns of these two subtypes undergo dynamic changes during different developmental stages and reproductive cycles [61]. Aromatase, through regulating the expression and activity of these estrogen receptors, directly influences the activation of the estrogen signaling pathway [62]. Thus, Cyp19a1a is involved in the synthesis of E2 through co-regulation with esr2a in P. argenteus.
4. Materials and Methods
4.1. Animal and Sample Collection
P. argenteus were reared at Xiangshan Bay, Zhejiang, China. All fish experiments were conducted in accordance with the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The Animal Care and Use Committee of Ningbo University approved the protocols (permit no. NBU20220079). We performed year-round sampling of cultured P. argenteus. The larvae of P. argenteus were collected from April 2022 to May 2022 and stored at −80 °C immediately. The gonads from stage I to stage VI of P. argenteus were sampled from July 2022 to August 2023. Monthly, random sampling was conducted with 20 fish. In total, 260 fish were collected. The body weight of sampled fish was from 4.1 g to 86.21 g, and the body length was from 5.7 cm to 18.2 cm. For sampling, MS222 (100 mg/L, Tricaine methanesulfonate, Sigma-Aldrich Co. LLC., St. Louis, MO, USA) was used to anesthetize the fish. One side of each gonad was used for histological observations to validate the gonadal status, while the other side of the gonad was used for qPCR experiments after gonadal status classification.
4.2. Gonadal Histology
The fish gonads were fixed with 4% paraformaldehyde in PBS at 4 °C for 12 h and then dehydrated using 100% methanol. After that, the gonads were embedded in paraffin wax and then cut into 5 μm thick sections. Finally, the gonadal sections were rehydrated in PBS and then stained with hematoxylin and eosin (H&E). The stages of gonadal development of all fish species in this study were examined histologically.
4.3. Total RNA Extraction and cDNA Synthesis
After the gonadal status of all fish had been determined via histology, six different stages of gonads were used to ascertain gene expression patterns. Extraction of total RNA followed the TransZol Up Kit (Transgene, ET111) manufacturer’s protocol. The quantity of total RNA was determined using a NanoDrop™ 1000 spectrometer (Thermo Fisher Scientific, Waltham, MA, USA). The first-strand cDNA was synthesized from 1 μg total RNA with HiFiScript gDNA Removal RT MasterMix Kit (CWBIO, Beijing, China). cDNA synthesis was performed using the manufacturer’s protocol.
4.4. Cloning of cyp19a1a and cyp19a1b cDNA
Full-length cDNA sequences of the P. argenteus cyp19a1a and cyp19a1b genes were amplified using PCR. Degenerate primers were designed based on conserved regions of the gene from other fish species. The primers used for amplification are listed in Table 1.

Table 1.
Oligonucleotides for specific primers.
4.5. Sequence Alignment and Phylogenetic Analysis
For phylogenic analysis, a set of Cyap19a1a and Cyp19a1b sequences in different fish species were retrieved from GenBank. The deduced amino acid sequences of P. argenteus Cyp19a1a and Cyp19a1b were subjected to multiple sequence alignment using Clustal W. The phylogenetic tree was constructed utilizing the Neighbor-Joining method within Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0. The evolutionary distances were computed using the Jones–Taylor–Thornton (JTT) matrix-based method and are in the units of the number of amino acid substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.7). The number displayed at each node signifies the bootstrap probability (expressed as a percentage from 1000 replicates). Branches corresponding to partitions reproduced in less than 30% bootstrap replicates are collapsed. The accession numbers for the sequences obtained in the analysis can be found in the legend of Figure 1.
4.6. Quantitative Real-Time PCR
Gene quantification of standards, samples, and controls were conducted simultaneously using quantitative real-time PCR (qPCR) (Quantagene q225 Real-Time PCR System) with the MagicSYBR mixture (CWBIO, Beijing, China). beta-actin (β-actin; GenBank accession no. KF982333) was used as an internal control to normalize the gene expression level. Specific primers for cyp19a1a, cyp19a1b, esr1, esr2a, arα, figla, and hsd17β1 are listed in Table 1. The specificity of PCR was confirmed through a single melting curve of unknown samples and standards. No signal was detected in non-template controls via qPCR. The data were calibrated according to the 2−ΔΔCt method. The reaction efficiency of different genes was assessed through the analysis of serially diluted cDNA templates. Based on linear regression analysis, the accuracy of real-time PCR assays was high (R2 > 0.99) across all dilutions. The relative expression values of target genes in all samples were normalized to β-actin, and the highest value of the target genes was set as 100%.
4.7. Recombinant Cyp19a1a Protein Production
The open reading frame (ORF) of cyp19a1a was introduced into the pcDNA 3.1 (+) Mammalian Expression Vector (Thermo Fisher Scientific, Waltham, MA, USA). The PCR primers 5′-CTAGCTAGCATGACTCCTGTCGGTTTGGACAC-3′ and 5′-CCCAAGCTTTTACCAGCTGCCTCTCTTTCTTG-3′ were used to amplify the ORF of cyp19a1a. The 293T (human embryonic kidney) cell line was used as a host for the recombinant construct. The recombinant Cyp19a1a (rCyp19a1a) was produced in 293T cells as in previous studies [21,63], and was used to check the specificity of the anti-Cyp19a1a antibody via Western blot analysis. Also, rCyp19a1a was used to elucidate the function of Cyp19a1a through the in vitro testicular and ovarian tissue culture experiments.
4.8. Western Blot Analysis
The rat polyclonal antibody was generated against the full-length amino acid of P. argenteus Cyp19a1a. The antisera were prepared by Ai Ting Biotechnology, Inc., Hangzhou, China. The specificity of anti-Cyp19a1a antiserum was confirmed via Western blotting analysis and used for immunohistochemical (IHC) staining in this study. Gonadal protein was extracted with RIPA buffer (R0010, Solarbio, Beijing, China). β-Actin was detected using β-Actin Rabbit mAb (AC026, ABclonal, Wuhan, China) (diluted 1:5000 with 1.5% nonfat milk powder) and alkaline phosphatase-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (SA00002-2, Proteintech, Wuhan, China) (diluted 1:2000 with 1.5% nonfat milk powder), which functioned as primary antibody and secondary antibody. Anti-Cyp19a1a antiserum, which was diluted 1:1000 with 1.5% nonfat milk powder, was used to detect rCyp19a1a and Cyp19a1a. Alkaline phosphatase-conjugated affinipure goat anti-rat IgG (H+L) antibody (SA00002-9, Proteintech, Wuhan, China), which was diluted 1:2000 with 1.5% nonfat milk powder, was used for the secondary reaction. After that, the NBT/BCIP Detection System (PR1100, Solarbio, Beijing, China) was used to visualize the immunoreactivity.
4.9. IHC Analyses
For IHC staining, we followed the methodology described previously [63]. The gonadal sections were cut into 5 μm thick sections, and the rehydrated sections were treated with sodium citrate antigen retrieval buffer (PR30001, Proteintech, Wuhan, China) to expose the antigens of the target protein. Primary antibodies against Cyp19a1a, which were diluted 1:100 with nonfat milk, were used to detect Cyp19a1a. For the secondary antibody reactions, biotin-SP-conjugated affinipure goat anti-rat IgG (H+L) antibody (SA00004-8, Proteintech, Wuhan, China diluted 1:200) was used to detect the Cyp19a1a antibody. Also, anti-bromodeoxyuridine (BrdU) polyclonal antibody (K010024P, ABclonal, Wuhan, China) (diluted 1:100 with nonfat milk) and HRP goat anti-rabbit IgG (H+L) (AS014, ABclonal, Wuhan, China) (diluted 1:200 with nonfat milk) were used to detect cell proliferation. Immunoreactivity was amplified with an SABC-HRP kit (P0603, Beyotime, Nanjing, China). After that, 3, 3′-diaminobenzidine (DAB, P0202, Beyotime, Nanjing, China) was used to visualize the immunoreactivity.
4.10. rCyp19a1a Treatment
Gonadal tissue culture followed previous studies [21,63]. In brief, the ovary or testis was dissected into less than 1 mm thick pieces. The gonadal tissues, which involved six replicates in each group, were incubated in the presence or absence of rCyp19a1a for 7 days at 26 °C. The basal culture medium consisted of Leibovitz’s L-15 Medium (11415064, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum and 2% penicillin–streptomycin liquid (P1400, Solarbio, Beijing, China). The low dose of rCyp19a1a was 15 μg/mL and the high dose of rCyp19a1a was 90 μg/mL. BrdU was used to analyze the effects of rCyp19a1a on the proliferation of mitotically active germ cells. Gonadal tissues were collected after 48 h of BrdU (60 μg/mL) treatment for immunohistology and gene expression analysis.
4.11. Sex Hormone Treatment
For sex hormone treatments, different concentrations of sex hormones including β-estradiol (GC11282, GLPBIO, Montclair, CA, USA) (E2, 0, 25, 50, 100, 200 ng/mL), methyltestosterone solution (GC19961, GLPBIO, Montclair, CA, USA) (MT, 0,15, 30, 60, 120 ng/mL), and anastrozole (GC10256, GLPBIO, Montclair, CA, USA) (aromatase inhibitor, AI, 0, 100, 1000, 10,000, 50,000 ng/mL) were used to culture gonadal tissue. BrdU was used to analyze the cell proliferation for each group. BrdU-incorporated cells were determined via IHC with anti-BrdU antibody. Sections of embedded gonadal tissues were collected from anterior, middle, and posterior parts for IHC staining. Sections were used to count the BrdU-incorporated cells. After counting, the best concentration of each sex hormone treatment was used for gene expression analysis.
4.12. Data Analysis
The data are presented as the mean ± SD (standard deviation). The Shapiro–Wilk method was used to analyze the normality of data distribution. The values were subjected to analysis via one-way ANOVA, followed by Tukey’s test or Duncan’s test, with p < 0.05 indicating a significant difference.
5. Conclusions
In this study, we demonstrated that the aromatase gene in P. argenteus exhibits two forms, which are brain-type aromatase and ovarian-type aromatase. cyp19a1b might be involved in brain development and also participates in the regulation of gonadal development, and cyp19a1a may play an important role in ovarian differentiation and ovarian maturation. Cyp19a1a is also involved in the synthesis of E2 through co-regulation with esr2a in P. argenteus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25031583/s1.
Author Contributions
C.G. designed the research; C.G., K.Z., C.L. and R.X. conducted the research; C.G. and K.Z. analyzed the data; C.G. participated in compiling the main manuscript. X.W., D.W. and S.X. gave comments and participated in compiling the manuscript. All authors reviewed and approved the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from Ningbo University Talent Introduction Research Initiation Fund Project (Natural Science Category) and KC Wong Magna Fund in Ningbo University.
Institutional Review Board Statement
All fish experiments were conducted in accordance with the recommendations of the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The Animal Care and Use Committee of Ningbo University approved the protocols (permit no. NBU20220079).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Material.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zhao, F.; Dong, Y.H.; Zhuang, P.; Zhang, T.; Zhang, L.Z.; Shi, Z.H. Genetic diversity of silver pomfret (Pampus argenteus) in the Southern Yellow and East China Seas. Biochem. Syst. Ecol. 2011, 39, 145–150. [Google Scholar] [CrossRef]
- Archangi, B.; Bazrafshan, K.H.; Ronagh, M.T.; Savari, A.; Abadi, M.S.A. Population genetic structure of silver pomfret (Pampus argenteus) in Persian Gulf and Oman Sea, inferred from 11 microsatellite loci. World J. Fish Mar. Sci. 2013, 5, 227–232. [Google Scholar]
- Mohitha, C.; Joy, L.; Divya, P.R.; Gopalakrishnan, A.; Basheer, V.S.; Koya, M.; Jena, J.K. Characterization of microsatellite markers in silver pomfret, Pampus argenteus (Perciformes: Stromateidae) through cross-species amplification and population genetic applications. J. Genet. 2014, 93, e89–e93. [Google Scholar] [CrossRef]
- Al-Abdul-Elah, K.; Hossain, M.A.; Akatsu, S. Recent advances in artificial breeding and larval rearing of silver pomfret Pampus argenteus (Euphrasen 1788) in Kuwait. Saudi. J. Biol. Sci. 2021, 28, 5808–5815. [Google Scholar] [CrossRef]
- Zhang, C.; Jacques, K.J.; Zhang, S.; Xu, S.L.; Wang, Y.J.; Wang, D.L. Analyses of growth performance and realized heritability of Pampus argenteus in a breeding program in China. Front. Mar. Sci. 2022, 9, 935924. [Google Scholar] [CrossRef]
- Conley, A.; Hinshelwood, M. Mammalian aromatases. Reproduction 2001, 121, 685–695. [Google Scholar] [CrossRef]
- Simpson, E.R.; Clyne, C.; Rubin, G.; Boon, W.C.; Robertson, K.; Britt, K.; Speed, C.; Jones, M. Aromatase-a brief overview. Annu. Rev. Physiol. 2002, 64, 93–127. [Google Scholar] [CrossRef]
- Chang, X.T.; Kobayashi, T.; Senthilkumaran, B.; Kobayashi-Kajura, H.; Sudhakumari, C.C.; Nagahama, Y. Two types of distribution aromatase with different encoding genes, tissue and developmental expression in Nile tilapia (Oreochromis niloticus). Gen. Comp. Endocrinol. 2005, 141, 101–115. [Google Scholar] [CrossRef]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Liu, Z.; Zhang, L.; Zhang, W. Epigenetic modifications during sex change repress gonadotropin stimulation of cyp19a1a in a teleost ricefield eel (Monopterus albus). Endocrinology 2013, 154, 2881–2890. [Google Scholar] [CrossRef]
- Lu, H.; Zhang, S.; Liu, Q.; Zhang, L.; Zhang, W. Cytoplasmic localization of Lrh-1 down-regulates ovarian follicular cyp19a1a expression in a teleost, the orange-spotted grouper Epinephelus coioides. Biol. Reprod. 2014, 91, 29. [Google Scholar] [CrossRef]
- Thomas, J.T.; Todd, E.V.; Muncaster, S.; Lokman, P.M.; Damsteegt, E.L.; Liu, H.; Soyano, K.; Gléonnec, F.; Lamm, M.S.; Godwin, J.R.; et al. Conservation and diversity in expression of candidate genes regulating socially-induced female-male sex change in wrasses. PeerJ 2019, 7, e7032. [Google Scholar] [CrossRef]
- Hou, M.; Feng, K.; Luo, H.; Jiang, Y.; Xu, W.; Li, Y.; Tao, B.; Chen, J.; Zhu, Z.; Song, Y.; et al. Multi-locus gene editing effectively knocked out cyp19a1a and foxl2 in Monopterus albus, a hermaphroditic fish. Aquaculture 2023, 565, 739130. [Google Scholar] [CrossRef]
- García-López, A.; Sánchez-Amaya, M.I.; Prat, F. Targeted gene expression profiling in European sea bass (Dicentrarchus labrax, L.) follicles from primary growth to late vitellogenesis. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2011, 160, 374–380. [Google Scholar] [CrossRef]
- Kwon, J.Y.; Kim, J. Differential expression of two distinct aromatase genes (cyp19a1a and cyp19a1b) during vitellogenesis and gestation in the viviparous black rockfish Sebastes schlegelii. Animal. Cell Syst. 2013, 17, 88–98. [Google Scholar] [CrossRef]
- Yan, T.; Cai, Y.; He, J.; Zhang, Q.; Wang, X.; Zhang, S.; He, L.; He, Z. Characterization and expression profiles of cyp19a1a in the schizothoracine fish Schizothorax prenanti. Tissue Cell 2019, 58, 70–75. [Google Scholar] [CrossRef]
- Mouriec, K.; Gueguen, M.M.; Manuel, C.; Percevault, F.; Thieulant, M.L.; Pakdel, F.; Kah, O. Androgens upregulate cyp19a1b (aromatase B) gene expression in the brain of zebrafish (Danio rerio) through estrogen receptors. Biol. Reprod. 2009, 80, 889–896. [Google Scholar] [CrossRef]
- Trubiroha, A.; Kroupova, H.; Wuertz, S.; Kloas, W. Up-regulation of gonadotropin mRNA-expression at the onset of gametogenesis in the roach (Rutilus rutilus): Evidence for an important role of brain-type aromatase (cyp19a1b) in the pituitary. Gen. Comp. Endocrinol. 2012, 178, 529–538. [Google Scholar] [CrossRef]
- Chaube, R.; Rawat, A.; Joy, K.P. Molecular cloning and characterization of brain and ovarian cytochrome P450 aromatase genes in the catfish Heteropneustes fossilis: Sex, tissue and seasonal variation in, and effects of gonadotropin on gene expression. Gen. Comp. Endocrinol. 2015, 221, 120–133. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Tsunematsu, T.; Motojima, Y.; Toriyama, K.; Horinouchi, A.; Ishii, Y.; Murata, H.; Yoshikawa, S.; Nyuji, M.; Shimizu, A. Pituitary luteinizing hormone synthesis starts in aromatase (cyp19a1b)-positive cells expressing esr1 and esr2b at the onset of puberty in Takifugu rubripes (fugu). Cell Tissue Res. 2022, 389, 259–287. [Google Scholar] [CrossRef]
- Wu, G.C.; Li, H.W.; Tey, W.G.; Lin, C.J.; Chang, C.F. Expression profile of amh/Amh during bi-directional sex change in the protogynous orange-spotted grouper Epinephelus coioides. PLoS ONE 2017, 12, e0185864. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Telecky, T.M.; Fukada, S.; Adachi, S.; Chen, S.; Nagahama, Y. Cloning and sequence analysis of the cDNA encoding P-450 aromatase (P450arom) from a rainbow trout (Oncorhynchus mykiss) ovary; relationship between the amount of P450arom mRNA and the production of oestradiol-17 beta in the ovary. J. Mol. Endocrinol. 1992, 8, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Valle, L.; Ramina, A.; Vianello, S.; Belvedere, P.; Colombo, L. Cloning of two mRNA variants of brain aromatase cytochrome P450 in rainbow trout (Oncorhynchus mykiss Walbaum). J. Steroid. Biochem. Mol. Biol. 2002, 82, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Kishida, M.; Callard, G.V. Distinct cytochrome P450 aromatase isoforms in zebrafish (Danio rerio) brain and ovary are differentially programmed and estrogen regulated during early development. Endocrinology 2001, 142, 740–750. [Google Scholar] [CrossRef]
- Kwon, J.Y.; McAndrew, B.J.; Penman, D.J. Cloning of brain aromatase gene and expression of brain and ovarian aromatase genes during sexual differentiation in genetic male and female Nile tilapia Oreochromis niloticus. Mol. Reprod. Dev. 2001, 59, 359–370. [Google Scholar] [CrossRef]
- Blázquez, M.; Piferrer, F. Cloning, sequence analysis, tissue distribution, and sex-specific expression of the neural form of P450 aromatase in juvenile sea bass (Dicentrarchus labrax). Mol. Cell. Endocrinol. 2004, 219, 83–94. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, W.; Zhang, L.; Zhu, T.; Tian, J.; Li, X.; Lin, H. Two distinct cytochrome P450 aromatases in the orange-spotted grouper (Epinephelus coioides): cDNA cloning and differential mRNA expression. J. Steroid. Biochem. Mol. Biol. 2004, 92, 39–50. [Google Scholar] [CrossRef]
- Choi, J.Y.; Park, J.G.; Jeong, H.B.; Lee, Y.D.; Takemura, A.; Kim, S.J. Molecular cloning of cytochrome P450 aromatases in the protogynous wrasse, Halichoeres tenuispinis. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2005, 141, 49–59. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, W.; Yang, H.; Zhou, W.; Hu, C.; Zhang, L. Two cytochrome P450 aromatase genes in the hermaphrodite ricefield eel Monopterus albus: mRNA expression during ovarian development and sex change. J. Endocrinol. 2008, 199, 317–331. [Google Scholar] [CrossRef]
- Steinke, D.; Hoegg, S.; Brinkmann, H.; Meyer, A. Three rounds (1R/2R/3R) of genome duplications and the evolution of the glycolytic pathway in vertebrates. BMC Biol. 2006, 4, 16. [Google Scholar] [CrossRef]
- Lin, C.J.; Maugars, G.; Lafont, A.G.; Jeng, S.R.; Wu, G.C.; Dufour, S.; Chang, C.F. Basal teleosts provide new insights into the evolutionary history of teleost-duplicated aromatase. Gen. Comp. Endocrinol. 2020, 291, 113395. [Google Scholar] [CrossRef]
- Gohin, M.; Bodinier, P.; Fostier, A.; Chesnel, F.; Bobe, J. Aromatase is expressed and active in the rainbow trout oocyte during final oocyte maturation. Mol. Reprod. Dev. 2011, 78, 510–518. [Google Scholar] [CrossRef]
- Rodríguez-Marí, A.; Yan, Y.L.; Bremiller, R.A.; Wilson, C.; Cañestro, C.; Postlethwait, J.H. Characterization and expression pattern of zebrafish Anti-Müllerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 2005, 5, 655–667. [Google Scholar] [CrossRef]
- Liu, H.; Mu, X.; Gui, L.; Su, M.; Li, H.; Zhang, G.; Liu, Z.; Zhang, J. Characterization and gonadal expression of FOXL2 relative to Cyp19a genes in spotted scat Scatophagus argus. Gene 2015, 561, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Gen, K.; Okuzawa, K.; Kumakura, N.; Yamaguchi, S.; Kagawa, H. Correlation between messenger RNA expression of cytochrome P450 aromatase and its enzyme activity during oocyte development in the red seabream (Pagrus major). Biol. Reprod. 2001, 65, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C.; Kwon, J.Y.; Penman, D.J. Physical mapping of the brain and ovarian aromatase genes in the Nile Tilapia, Oreochromis niloticus, by fluorescence in situ hybridization. Anim. Genet. 2003, 34, 62–64. [Google Scholar] [CrossRef] [PubMed]
- Paixão, R.V.; Silva, G.F.; Caetano, A.R.; Cintra, L.C.; Varela, E.S.; O’Sullivan, F.L.A. Phylogenomic and expression analysis of Colossoma macropomum cyp19a1a and cyp19a1b and their non-classical role in tambaqui sex differentiation. Gene 2022, 843, 146795. [Google Scholar] [CrossRef]
- Li, C.; Li, Y.Y.; Qin, C.L.; Yu, C.H.; Hu, J.B.; Wang, Y.J.; Guo, C.Y. Determination of the timing of early gonadal differentiation in silver pomfret, Pampus argenteus. Anim. Reprod. Sci. 2023, 107373. [Google Scholar] [CrossRef]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile Tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef]
- Nakamoto, M.; Muramatsu, S.; Yoshida, S.; Matsuda, M.; Nagahama, Y.; Shibata, N. Gonadal sex differentiation and expression of Sox9a2, Dmrt1, and Foxl2 in Oryzias luzonensis. Genesis 2009, 47, 289–299. [Google Scholar] [CrossRef]
- Jiang, M.; Jia, S.; Chen, J.; Chen, K.; Ma, W.; Wu, X.; Luo, H.; Li, Y.; Zhu, Z.; Hu, W. Timing of gonadal development and dimorphic expression of sex-related genes in gonads during early sex differentiation in the Yellow River carp. Aquaculture 2020, 518, 734825. [Google Scholar] [CrossRef]
- Lubzens, E.; Young, G.; Bobe, J.; Cerda, J. Oogenesis inteleosts: How eggs are formed. Gen. Comp. Endocrinol. 2010, 165, 367–389. [Google Scholar] [CrossRef]
- Nagahama, Y.; Yamashita, M. Regulation of oocyte maturation in fish. Dev. Growth Differ. 2008, 50, S195–S219. [Google Scholar] [CrossRef]
- Callard, G.V.; Tchoudakova, A.V.; Kishida, M.; Wood, E. Differential tissue distribution, developmental programming, estrogen regulation and promoter characteristics of cyp19 genes in teleost fish. J. Steroid Biochem. Mol. Biol. 2001, 79, 305–314. [Google Scholar] [CrossRef]
- Godwin, J. Neuroendocrinology of sexual plasticity in teleost fishes. Front. Neuroendocrinol. 2010, 31, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Diotel, N.; Vaillant, C.; Kah, O.; Pellegrini, E. Mapping of brain lipid binding protein (Blbp) in the brain of adult zebrafish, co-expression with aromatase B and links with proliferation. Gene. Expr. Patterns 2016, 20, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Ulhaq, Z.S.; Kishida, M. Brain aromatase modulates serotonergic neuron by regulating serotonin levels in Zebrafish embryos and larvae. Front. Endocrinol. 2018, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.; Therrien, M.; Lu, C.; Liu, X.; Trudeau, V.L. Mutation of brain aromatase disrupts spawning behavior and reproductive health in female zebrafish. Front. Endocrinol. 2023, 14, 1225199. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.J.; Wu, G.C.; Dufour, S.; Chang, C.F. Activation of the brain-pituitary-gonadotropic axis in the black porgy Acanthopagrus schlegelii during gonadal differentiation and testis development and effect of estradiol treatment. Gen. Comp. Endocrinol. 2019, 281, 17–29. [Google Scholar] [CrossRef]
- Jagarlamudi, K.; Rajkovic, A. Oogenesis: Transcriptional regulators and mouse models. Mol. Cell. Endocrinol. 2012, 356, 31–39. [Google Scholar] [CrossRef]
- Fang, D.A.; Yang, X.J.; Feng, X.; Zhou, Y.F.; Xu, D.P.; Zhang, M.Y.; Liu, K. FoxL2 combined with Cyp19a1a regulate the spawning upstream migration in Coilia nasus. Gene 2019, 10, 307–315. [Google Scholar] [CrossRef]
- Zhang, X.; Li, M.; Ma, H.; Liu, X.; Shi, H.; Li, M.; Wang, D. Mutation of foxl2 or cyp19a1a results in female to male sex reversal in XX Nile tilapia. Endocrinology 2017, 158, 2634–2647. [Google Scholar] [CrossRef]
- Lau, E.S.; Zhang, Z.; Qin, M.; Ge, W. Knockout of zebrafish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci. Rep. 2016, 6, 37357. [Google Scholar] [CrossRef]
- Nakamoto, M.; Shibata, Y.; Ohno, K.; Usami, T.; Kamei, Y.; Taniguchi, Y.; Todo, T.; Sakamoto, T.; Young, G.; Swanson, P.; et al. Ovarian aromatase loss-of-function mutant medaka undergo ovary degeneration and partial female-to-male sex reversal after puberty. Mol. Cell. Endocrinol. 2018, 460, 104–122. [Google Scholar] [CrossRef] [PubMed]
- Piferrer, F. Endocrine sex control strategies for the feminization of teleost fish. Auaculture 2001, 197, 229–281. [Google Scholar] [CrossRef]
- Kobayashi, T.; Kajiura-Kobayashi, H.; Nagahama, Y. Induction of XY sex reversal by estrogen involves altered gene expression in a teleost, tilapia. Cytogenet. Genome. Res. 2003, 101, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Gárriz, Á.; Del Fresno, P.S.; Miranda, L.A. Exposure to E2 and EE2 environmental concentrations affect different components of the Brain-Pituitary-Gonadal axis in pejerrey fish (Odontesthes bonariensis). Ecotoxicol. Environ. Saf. 2017, 144, 45–53. [Google Scholar] [CrossRef]
- Rodrigues-Filho, J.A.; Garcia, C.E.O.; Chehade, C.G.; Sanches, E.G.; Borella, M.I.; Nostro, F.L.L.; Araújo, B.C.; Branco, G.S.; Moreira, R.G. Gonadal remodeling and hormonal regulation during sex change of juvenile dusky grouper Epinephelus marginatus (Teleostei, Serranidae), an endangered protogynous hermaphrodite fish. Fish. Physiol. Biochem. 2020, 46, 1809–1824. [Google Scholar] [CrossRef]
- Jiang, Y.; Luo, H.; Hou, M.; Chen, J.; Tao, B.; Zhu, Z.; Song, Y.; Hu, W. Aromatase inhibitor induces sex reversal in the protogynous hermaphroditic rice field eel (Monopterus albus). Aquaculture 2022, 551, 737960. [Google Scholar] [CrossRef]
- Pinto, P.I.S.; Passos, A.L.; Martins, R.S.; Power, D.M.; Canário, A.V.M. Characterization of estrogen receptor βb in sea bream (Sparus auratus): Phylogeny, ligand-binding, and comparative analysis of expression. Gen. Comp. Endocrinol. 2006, 145, 197–207. [Google Scholar] [CrossRef]
- Choi, C.Y.; Habibi, H.R. Molecular cloning of estrogen receptor α and expression pattern of estrogen receptor subtypes in male and female goldfish. Mol. Cell. Endocrinol. 2003, 204, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, M.B.; Thomas, P. The unusual binding properties of the third distinct teleost estrogen receptor subtype ERβa are accompanied by highly conserved amino acid changes in the ligand binding domain. Endocrinology 2004, 145, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.Y.; Tseng, P.W.; Hwang, J.S.; Wu, G.C.; Chang, C.F. The characteristics and expression profile of gonadal soma-derived factor (gsdf/Gsdf) during androgen-induced reversible sex change in the protogynous orange-spotted grouper, Epinephelus coioides. Aquaculture 2021, 542, 736832. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).