BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL)
Abstract
1. Introduction
2. Results
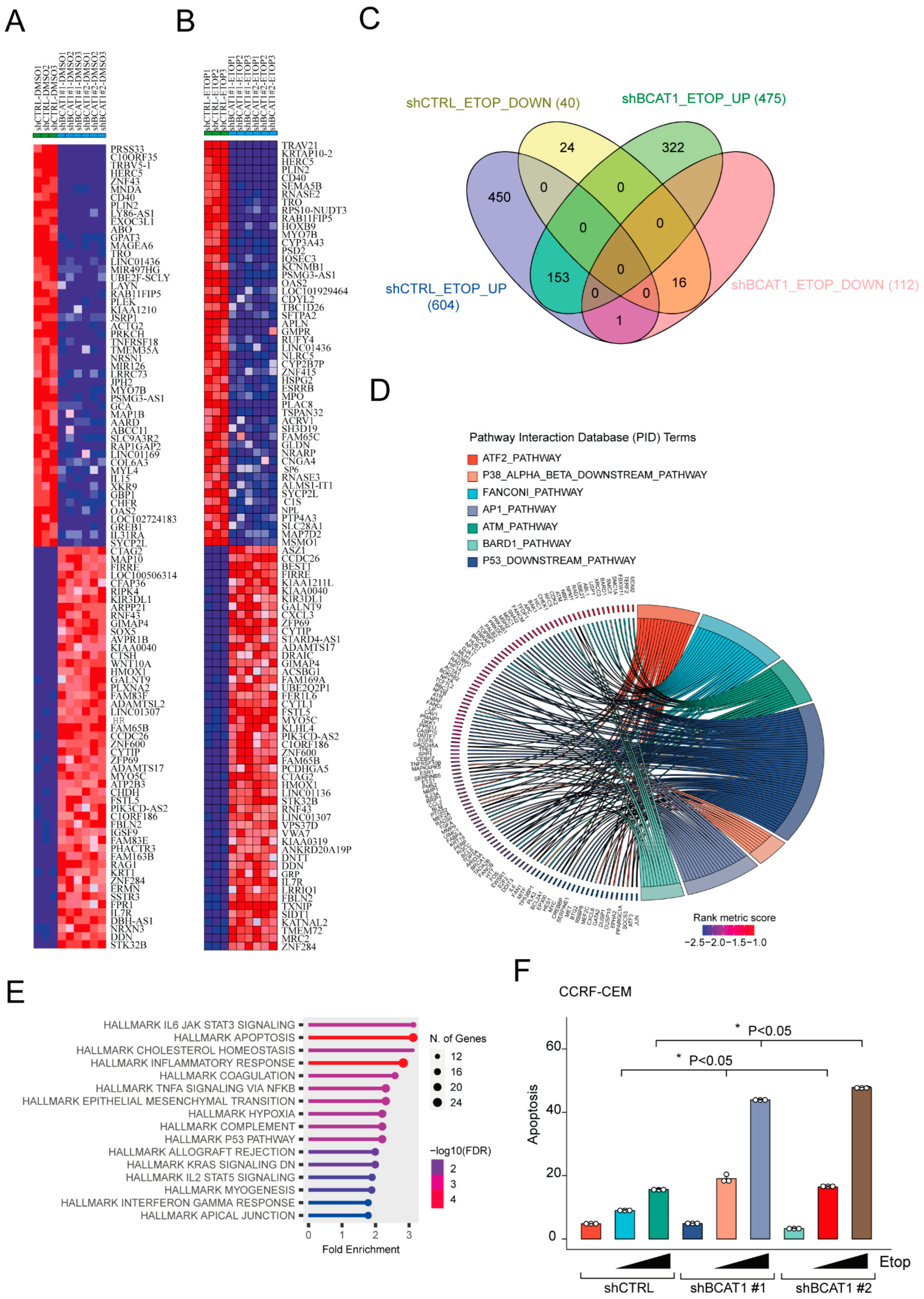
2.1. BCAT1 Depletion Activates an Ineffective DNA Damage Transcriptional Response
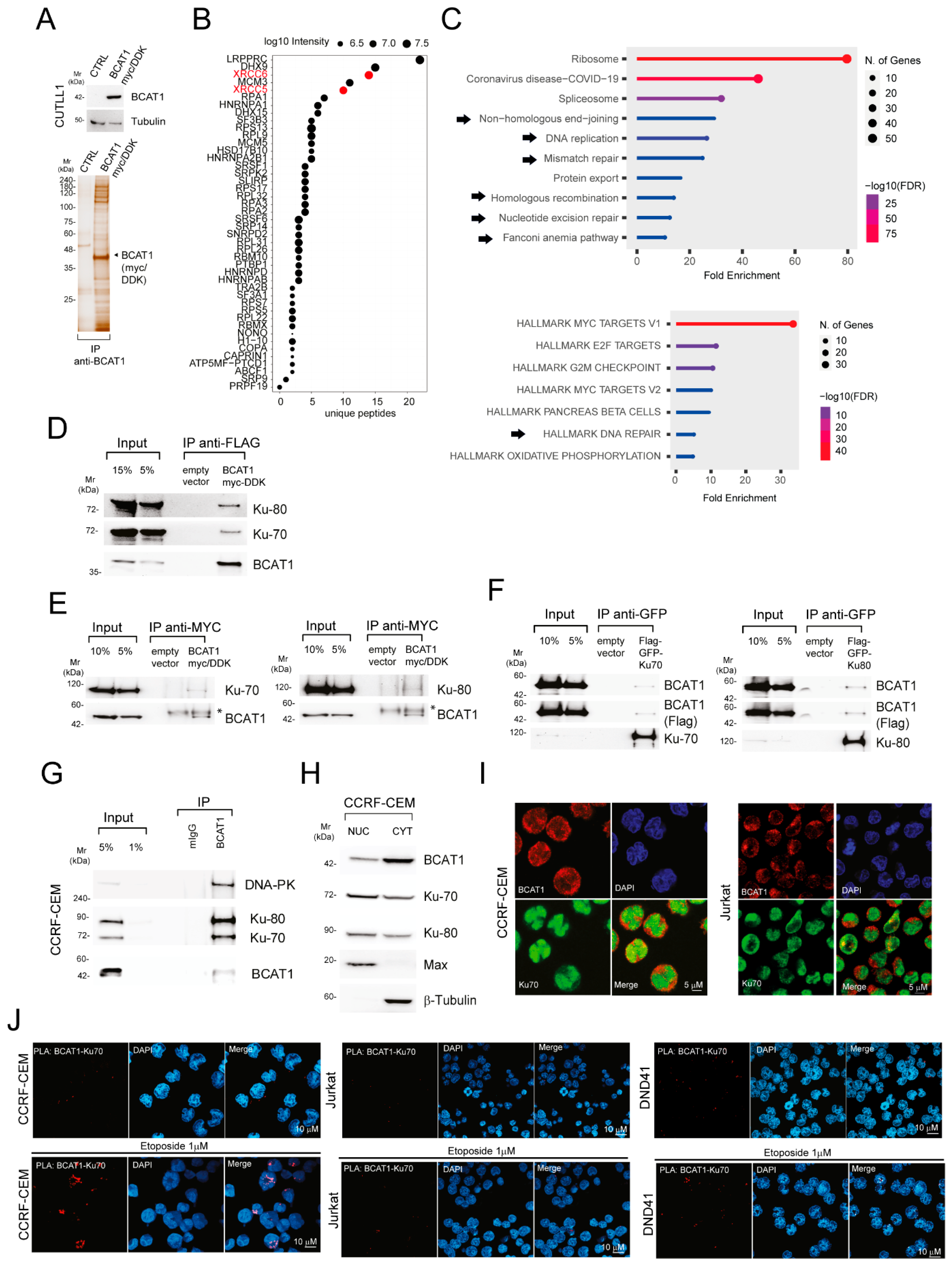
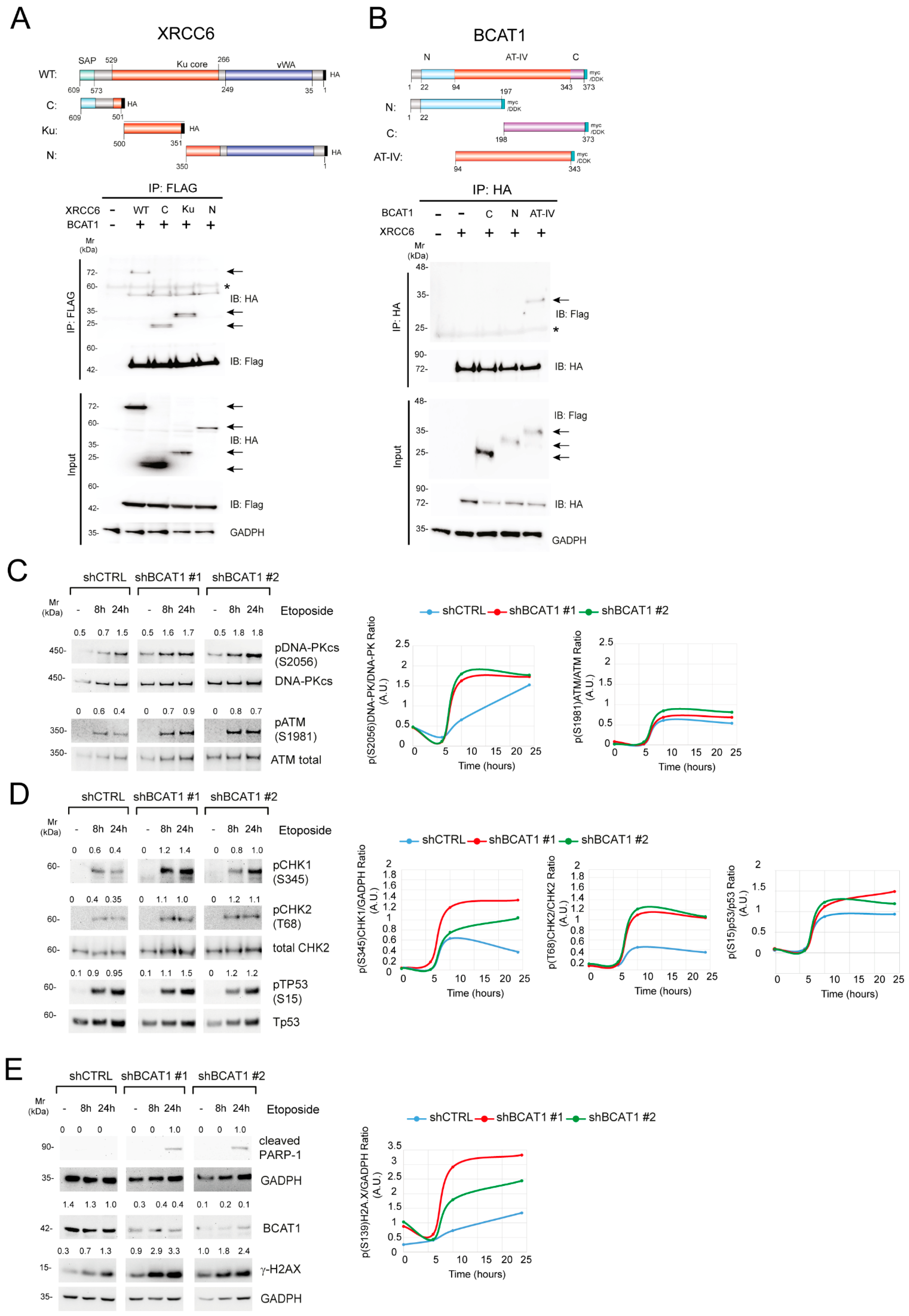
2.2. BCAT1 Binds with the KU Complex and Modulates DNA Damage Repair Signaling Pathways
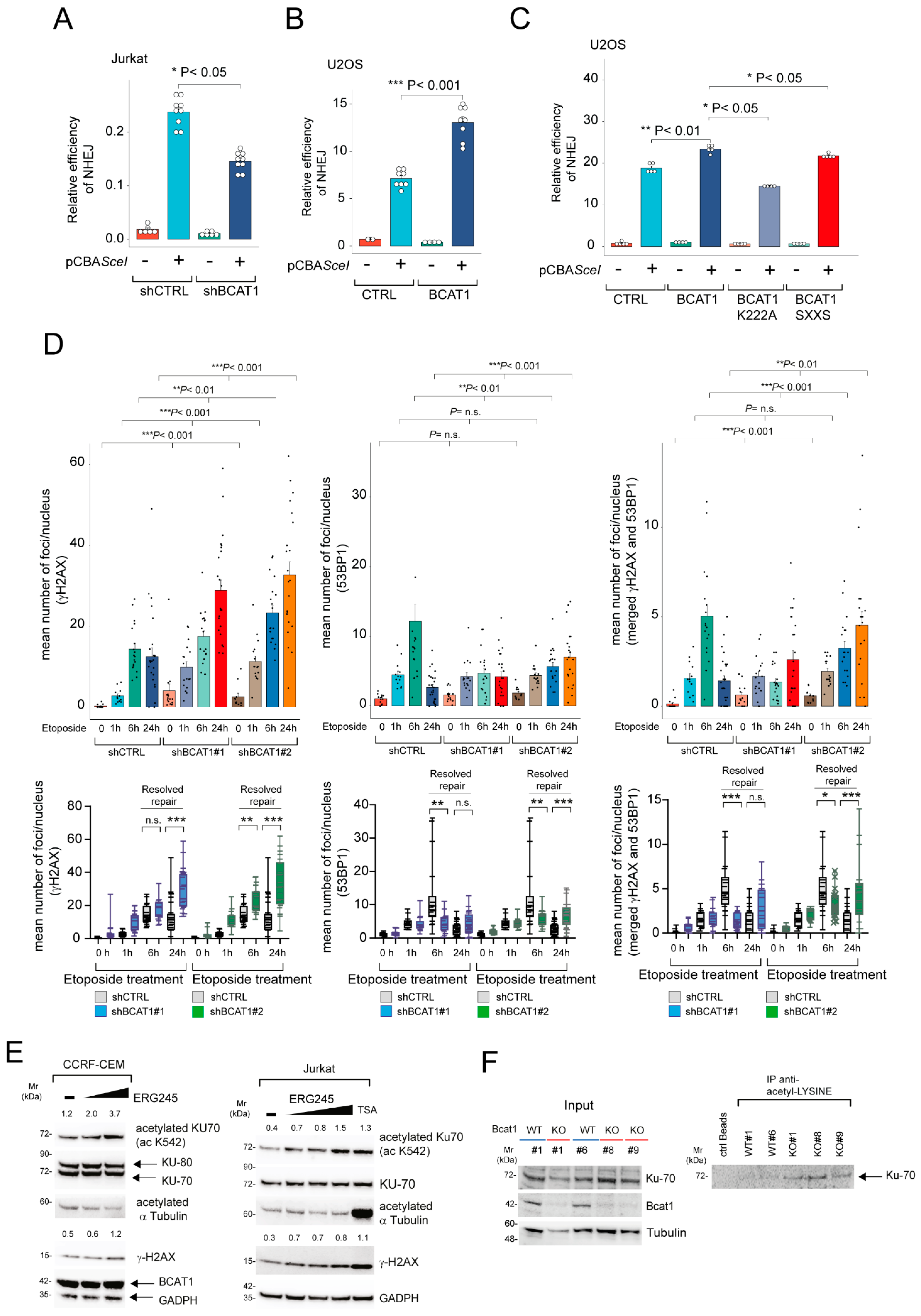
2.3. BCAT1 Depletion in Human T-ALL Leads to Reduced c-NHEJ Repair with Increased KU70/KU80 Acetylation
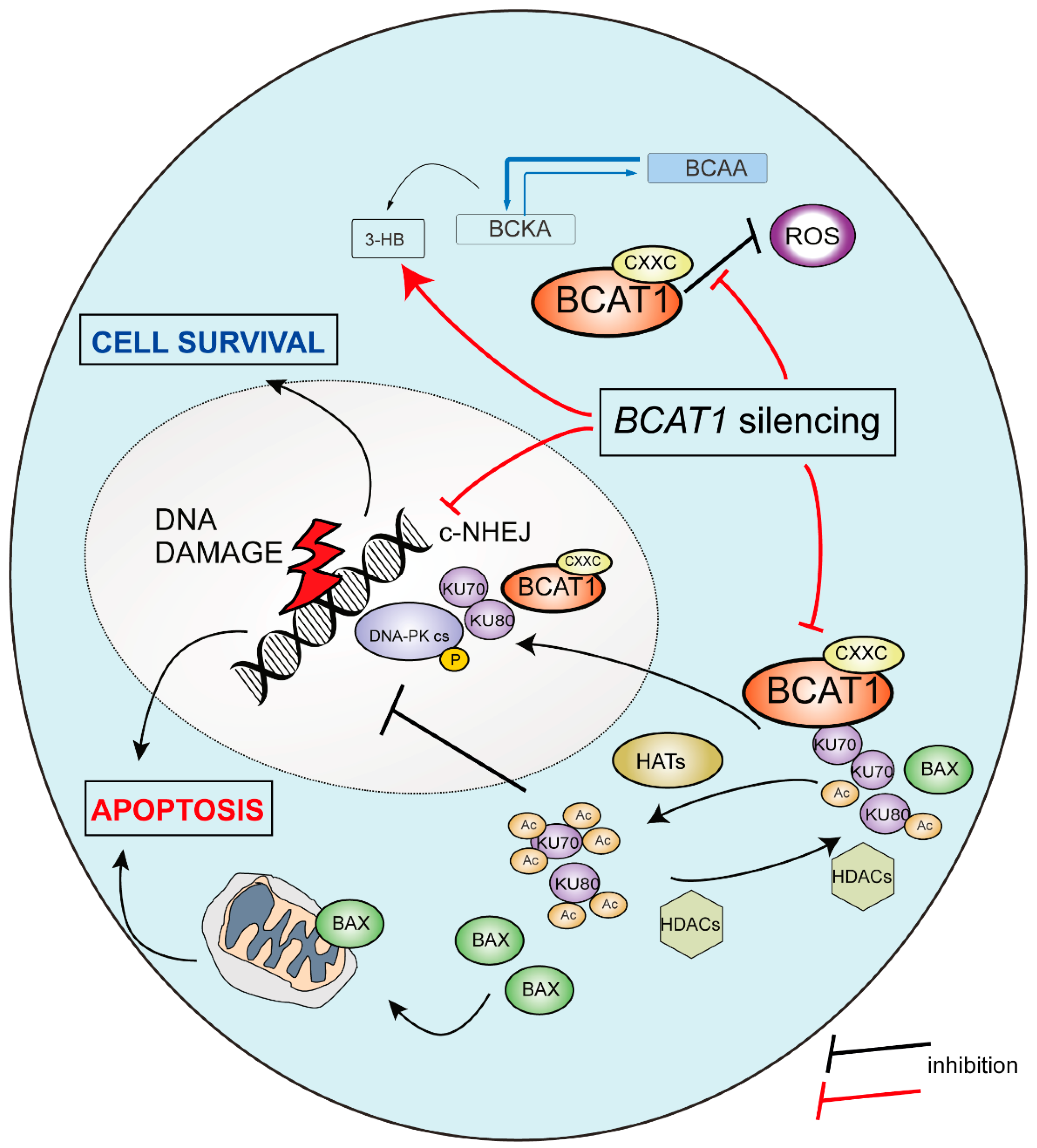
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Primary Leukemia Samples
4.2. RNA-Sequencing and Gene-Set Enrichment Analysis
4.3. Affinity Purification Mass Spectrometry and Bioinformatics
4.4. Apoptosis Analysis by Flow Cytometry
4.5. Western Blotting and Immunoprecipitation
4.6. Plasmids, Lentiviral Constructs, and Viral Production
4.7. Analysis of Double Strand Breaks (DSB) Repair by Reporter Assays
4.8. Cellular Fractionation
4.9. Immunoprecipitation of Acetylated Proteins
4.10. Immunofluorescence Analysis
4.11. Analysis of 53BP1 and γH2AX Foci by Immunofluorescence
4.12. Proximity Ligation Assay (PLA)
4.13. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Terwilliger, T.; Abdul-Hay, M. Acute lymphoblastic leukemia: A comprehensive review and 2017 update. Blood Cancer J. 2017, 7, e577. [Google Scholar] [CrossRef] [PubMed]
- Pui, C.H.; Robison, L.L.; Look, A.T. Acute lymphoblastic leukaemia. Lancet 2008, 371, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Belver, L.; Ferrando, A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nat. Rev. Cancer 2016, 16, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Bongiovanni, D.; Saccomani, V.; Piovan, E. Aberrant Signaling Pathways in T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2017, 18, 1904. [Google Scholar] [CrossRef]
- Weng, A.P.; Ferrando, A.A.; Lee, W.; Morris, J.P.t.; Silverman, L.B.; Sanchez-Irizarry, C.; Blacklow, S.C.; Look, A.T.; Aster, J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science 2004, 306, 269–271. [Google Scholar] [CrossRef]
- Palomero, T.; Lim, W.K.; Odom, D.T.; Sulis, M.L.; Real, P.J.; Margolin, A.; Barnes, K.C.; O’Neil, J.; Neuberg, D.; Weng, A.P.; et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA 2006, 103, 18261–18266. [Google Scholar] [CrossRef]
- Weng, A.P.; Millholland, J.M.; Yashiro-Ohtani, Y.; Arcangeli, M.L.; Lau, A.; Wai, C.; Del Bianco, C.; Rodriguez, C.G.; Sai, H.; Tobias, J.; et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006, 20, 2096–2109. [Google Scholar] [CrossRef]
- Vermezovic, J.; Adamowicz, M.; Santarpia, L.; Rustighi, A.; Forcato, M.; Lucano, C.; Massimiliano, L.; Costanzo, V.; Bicciato, S.; Del Sal, G.; et al. Notch is a direct negative regulator of the DNA-damage response. Nat. Struct. Mol. Biol. 2015, 22, 417–424. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Powell, J.D.; Hutson, S.M. Leucine Metabolism in T Cell Activation: mTOR Signaling and Beyond. Adv. Nutr. 2016, 7, 798S–805S. [Google Scholar] [CrossRef]
- Tosello, V.; Di Martino, L.; Papathanassiu, A.E.; Santa, S.D.; Pizzi, M.; Mussolin, L.; Liu, J.; Van Vlierberghe, P.; Piovan, E. BCAT1 is a NOTCH1 target and sustains the oncogenic function of NOTCH1. Haematologica 2024. early view. [Google Scholar] [CrossRef]
- Francois, L.; Boskovic, P.; Knerr, J.; He, W.; Sigismondo, G.; Schwan, C.; More, T.H.; Schlotter, M.; Krijgsveld, J.; Hiller, K.; et al. BCAT1 redox function maintains mitotic fidelity. Cell Rep. 2022, 41, 111524. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Zhang, M.; Yu, Y.; Li, S.; Ma, C.; Zhang, J.; Jiang, Y.; Li, Y.; Zheng, X.; Zhang, L.; et al. BCAT1 binds the RNA-binding protein ZNF423 to activate autophagy via the IRE1-XBP-1-RIDD axis in hypoxic PASMCs. Cell Death Dis. 2020, 11, 764. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Nowsheen, S.; Yang, E.S. The intersection between DNA damage response and cell death pathways. Exp. Oncol. 2012, 34, 243–254. [Google Scholar]
- Zahid, S.; Seif El Dahan, M.; Iehl, F.; Fernandez-Varela, P.; Le Du, M.H.; Ropars, V.; Charbonnier, J.B. The Multifaceted Roles of Ku70/80. Int. J. Mol. Sci. 2021, 22, 4134. [Google Scholar] [CrossRef]
- Harris, M.; El Hindy, M.; Usmari-Moraes, M.; Hudd, F.; Shafei, M.; Dong, M.; Hezwani, M.; Clark, P.; House, M.; Forshaw, T.; et al. BCAT-induced autophagy regulates Abeta load through an interdependence of redox state and PKC phosphorylation-implications in Alzheimer’s disease. Free Radic. Biol. Med. 2020, 152, 755–766. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Pierce, A.J.; Johnson, R.D.; Thompson, L.H.; Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999, 13, 2633–2638. [Google Scholar] [CrossRef]
- Bennardo, N.; Cheng, A.; Huang, N.; Stark, J.M. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008, 4, e1000110. [Google Scholar] [CrossRef]
- De Feraudy, S.; Revet, I.; Bezrookove, V.; Feeney, L.; Cleaver, J.E. A minority of foci or pan-nuclear apoptotic staining of gammaH2AX in the S phase after UV damage contain DNA double-strand breaks. Proc. Natl. Acad. Sci. USA 2010, 107, 6870–6875. [Google Scholar] [CrossRef]
- Lobbardi, R.; Pinder, J.; Martinez-Pastor, B.; Theodorou, M.; Blackburn, J.S.; Abraham, B.J.; Namiki, Y.; Mansour, M.; Abdelfattah, N.S.; Molodtsov, A.; et al. TOX Regulates Growth, DNA Repair, and Genomic Instability in T-cell Acute Lymphoblastic Leukemia. Cancer Discov. 2017, 7, 1336–1353. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, C.; Hada, M.; Opipari, A.W., Jr.; Castle, V.P.; Kwok, R.P. CREB-binding protein regulates Ku70 acetylation in response to ionization radiation in neuroblastoma. Mol. Cancer Res. MCR 2013, 11, 173–181. [Google Scholar] [CrossRef]
- Subramanian, C.; Opipari, A.W., Jr.; Bian, X.; Castle, V.P.; Kwok, R.P. Ku70 acetylation mediates neuroblastoma cell death induced by histone deacetylase inhibitors. Proc. Natl. Acad. Sci. USA 2005, 102, 4842–4847. [Google Scholar] [CrossRef]
- Shimazu, T.; Hirschey, M.D.; Newman, J.; He, W.; Shirakawa, K.; Le Moan, N.; Grueter, C.A.; Lim, H.; Saunders, L.R.; Stevens, R.D.; et al. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science 2013, 339, 211–214. [Google Scholar] [CrossRef] [PubMed]
- Lodi, F.; Certo, M.; Elkafrawy, H.; Li, W.; Vu, H.A.; Gilbo, K.; Su, L.; Pegg, I.L.; Weiss, T.; Bühler, M.; et al. BCAT1 inhibition affects CD8+ T cell activation, exhaustion, and tumoral immunity by altering iron homeostasis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Wang, Y.; Fu, Z.; Li, X.; Liang, Y.; Pei, S.; Hao, S.; Zhu, Q.; Yu, T.; Pei, Y.; Yuan, J.; et al. Cytoplasmic DNA sensing by KU complex in aged CD4(+) T cell potentiates T cell activation and aging-related autoimmune inflammation. Immunity 2021, 54, 632–647.e639. [Google Scholar] [CrossRef]
- Armstrong, F.; Brunet de la Grange, P.; Gerby, B.; Rouyez, M.C.; Calvo, J.; Fontenay, M.; Boissel, N.; Dombret, H.; Baruchel, A.; Landman-Parker, J.; et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood 2009, 113, 1730–1740. [Google Scholar] [CrossRef]
- Tosello, V.; Di Martino, L.; Piovan, E. Targeted Metabolomics of Tissue and Plasma Identifies Biomarkers in Mice with NOTCH1-Dependent T-Cell Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2024, 25, 6543. [Google Scholar] [CrossRef]
- Chiang, M.Y.; Xu, L.; Shestova, O.; Histen, G.; L’Heureux, S.; Romany, C.; Childs, M.E.; Gimotty, P.A.; Aster, J.C.; Pear, W.S. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J. Clin. Investig. 2008, 118, 3181–3194. [Google Scholar] [CrossRef]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Lee, B.; Luo, Y.; Choi, Y.; Choi, E.H.; Kim, K.B.; Seo, S.B.; Kim, Y.H.; Lee, H.H.; Kim, K.P.; et al. FOXL2 directs DNA double-strand break repair pathways by differentially interacting with Ku. Nat. Commun. 2020, 11, 2010. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tosello, V.; Rompietti, C.; Papathanassiu, A.E.; Arrigoni, G.; Piovan, E. BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL). Int. J. Mol. Sci. 2024, 25, 13571. https://doi.org/10.3390/ijms252413571
Tosello V, Rompietti C, Papathanassiu AE, Arrigoni G, Piovan E. BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL). International Journal of Molecular Sciences. 2024; 25(24):13571. https://doi.org/10.3390/ijms252413571
Chicago/Turabian StyleTosello, Valeria, Chiara Rompietti, Adonia E. Papathanassiu, Giorgio Arrigoni, and Erich Piovan. 2024. "BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL)" International Journal of Molecular Sciences 25, no. 24: 13571. https://doi.org/10.3390/ijms252413571
APA StyleTosello, V., Rompietti, C., Papathanassiu, A. E., Arrigoni, G., & Piovan, E. (2024). BCAT1 Associates with DNA Repair Proteins KU70 and KU80 and Contributes to Regulate DNA Repair in T-Cell Acute Lymphoblastic Leukemia (T-ALL). International Journal of Molecular Sciences, 25(24), 13571. https://doi.org/10.3390/ijms252413571






