Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation
Abstract
1. Introduction
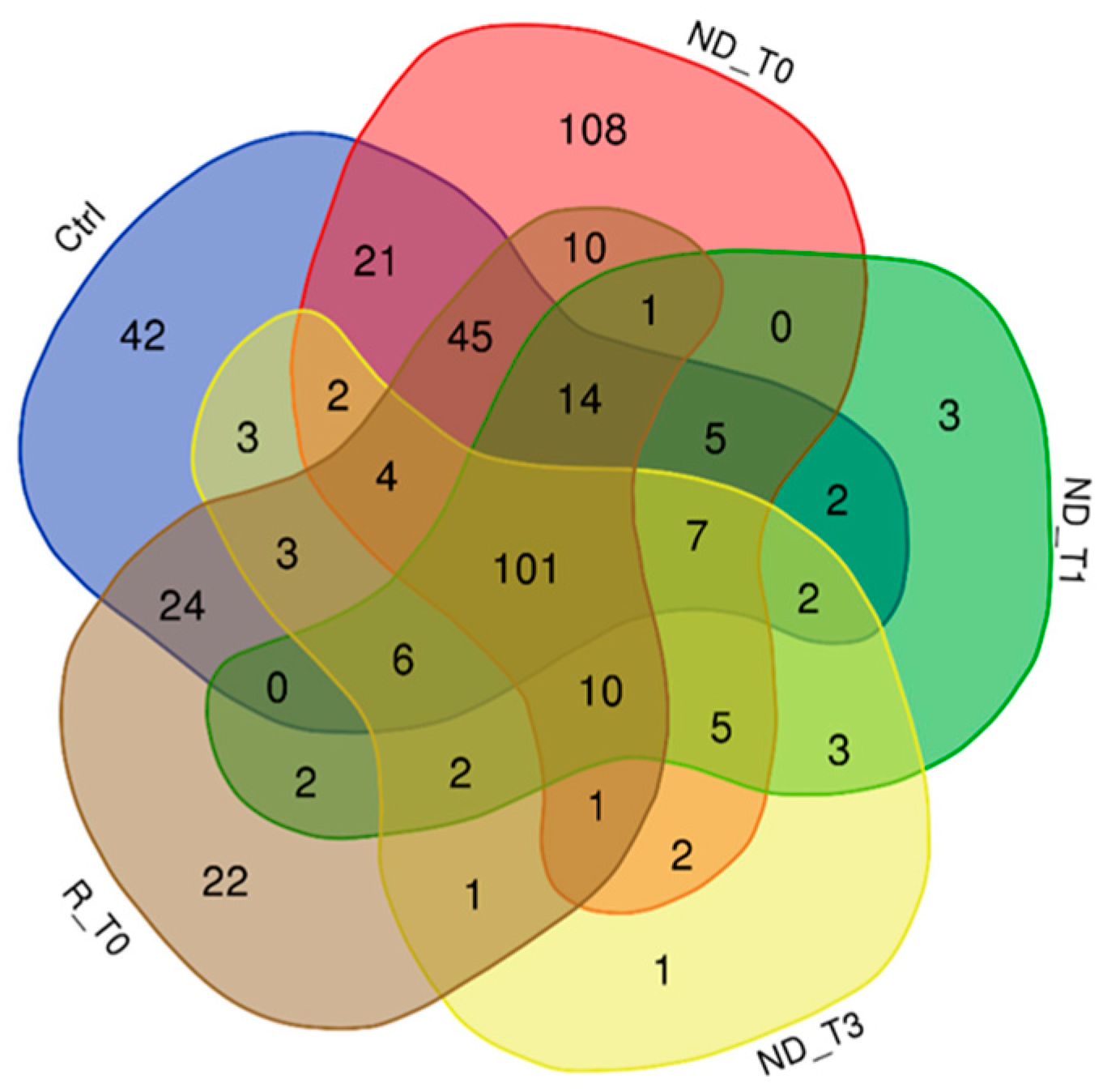
2. Results and Discussion
2.1. Proteins Exclusive of ND_ and R_T0 Saliva
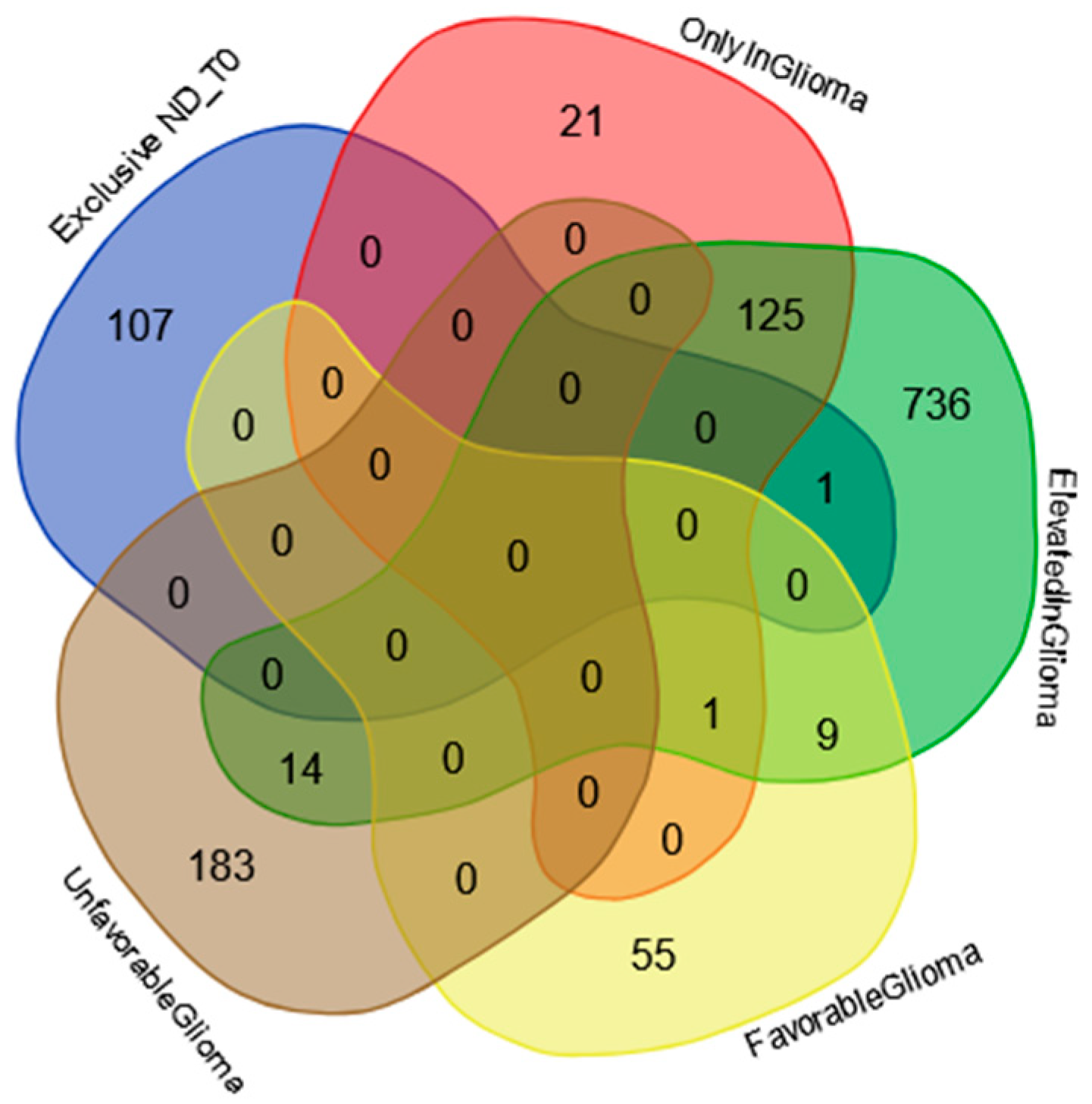
2.2. Proteins Exclusive of ND_T0 Saliva
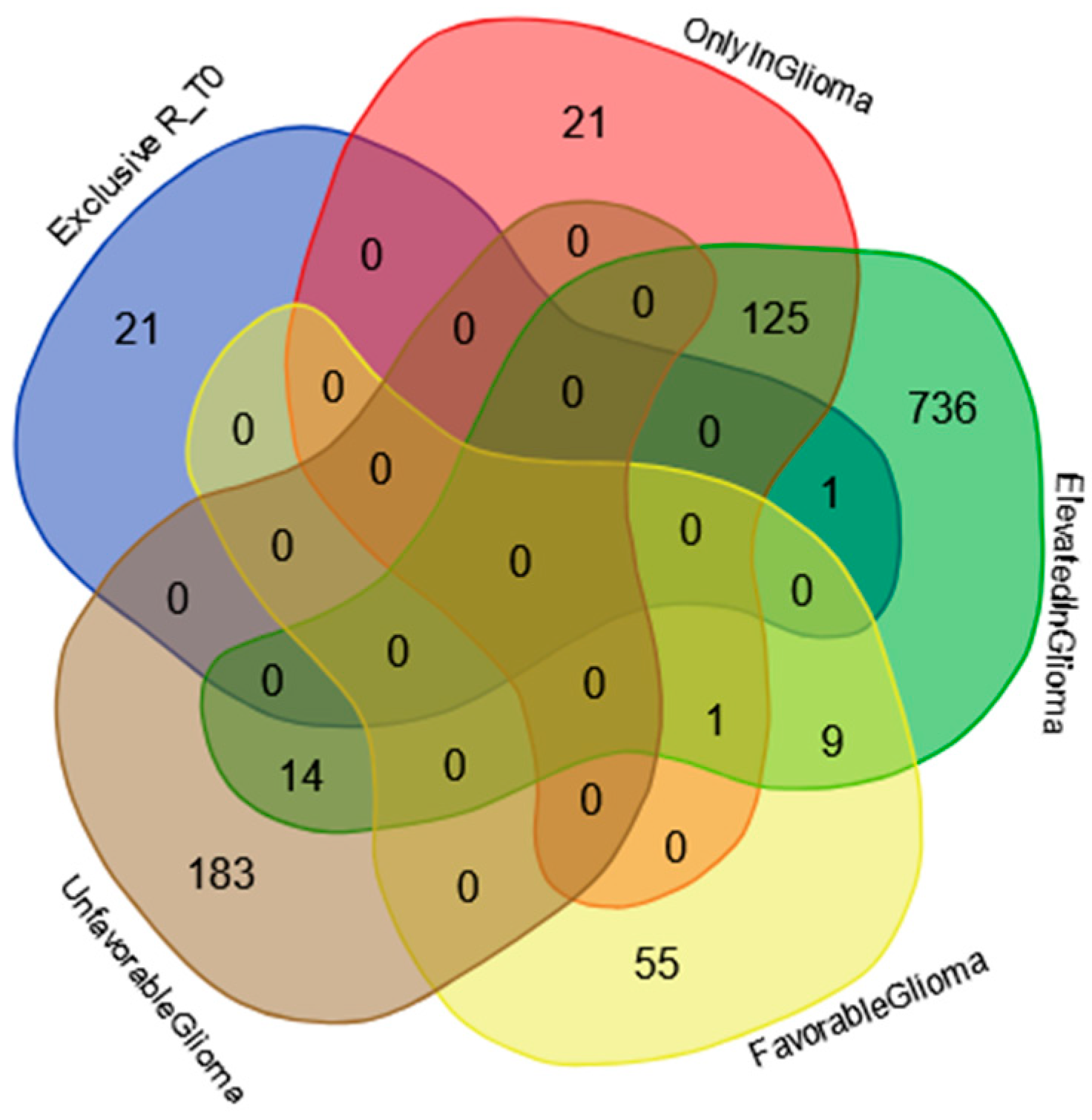
2.3. Proteins Exclusive of R_T0 Saliva
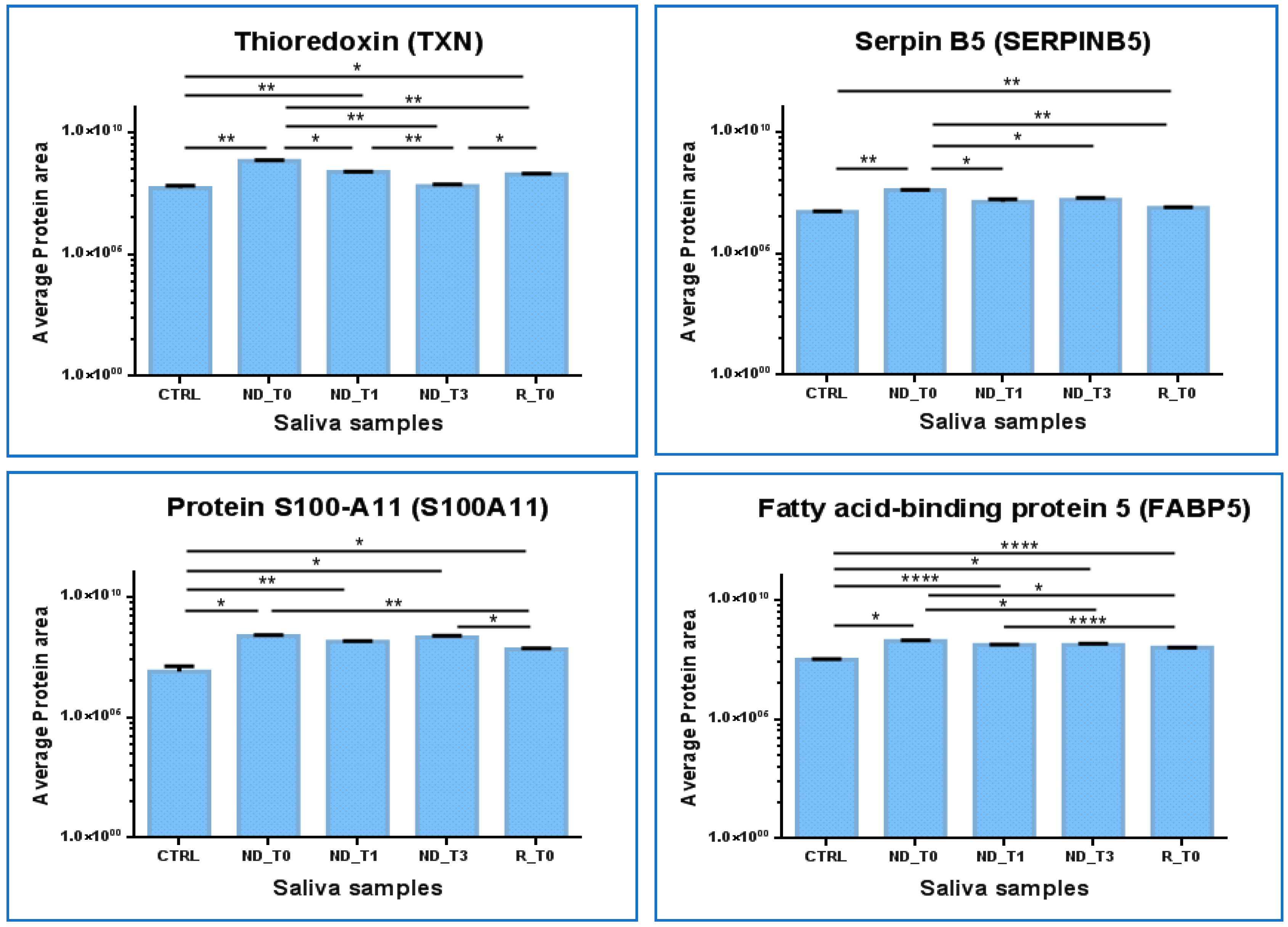
2.4. Label-Free Relative Quantitation of the Proteins Identified in All Saliva Pools
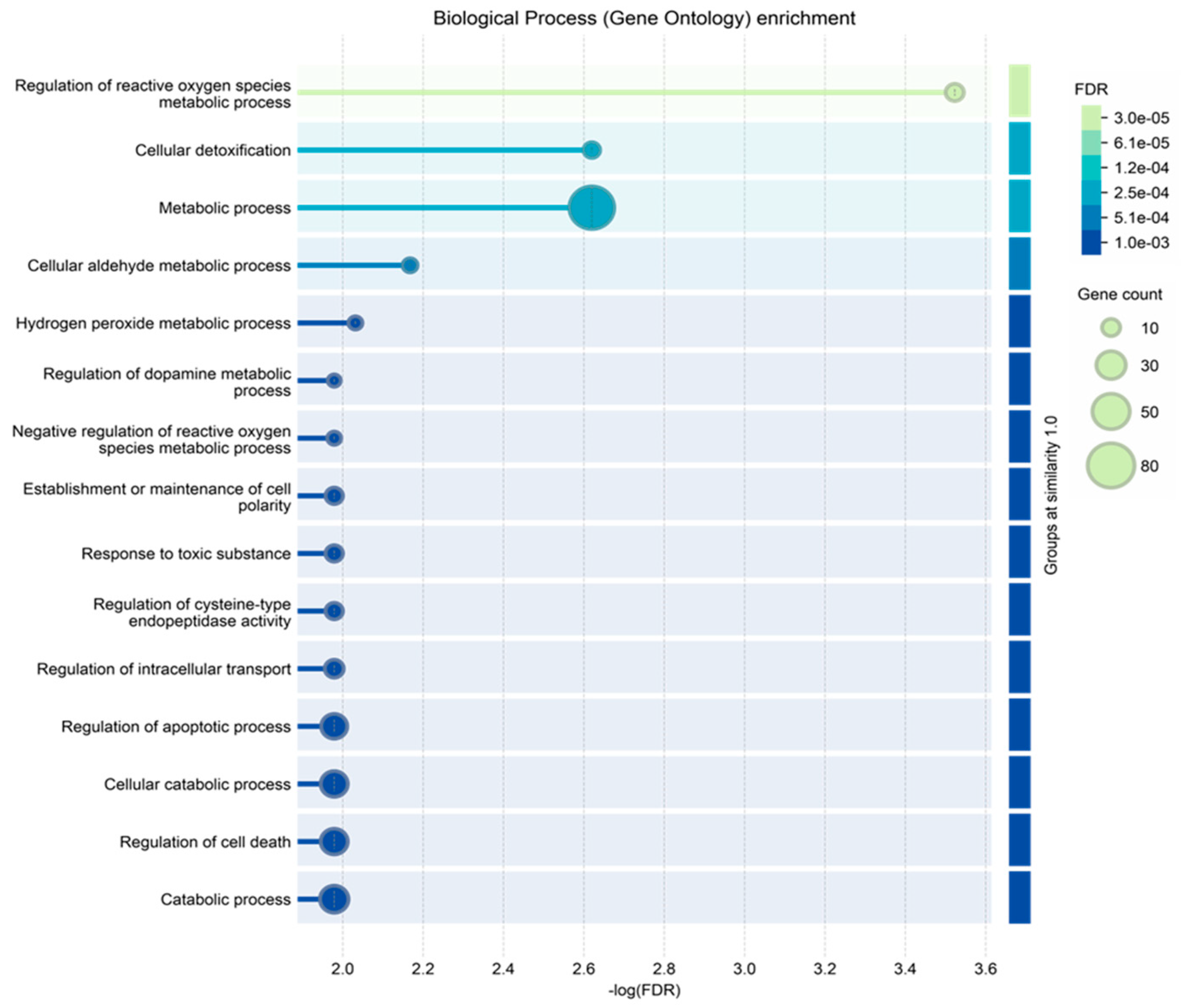
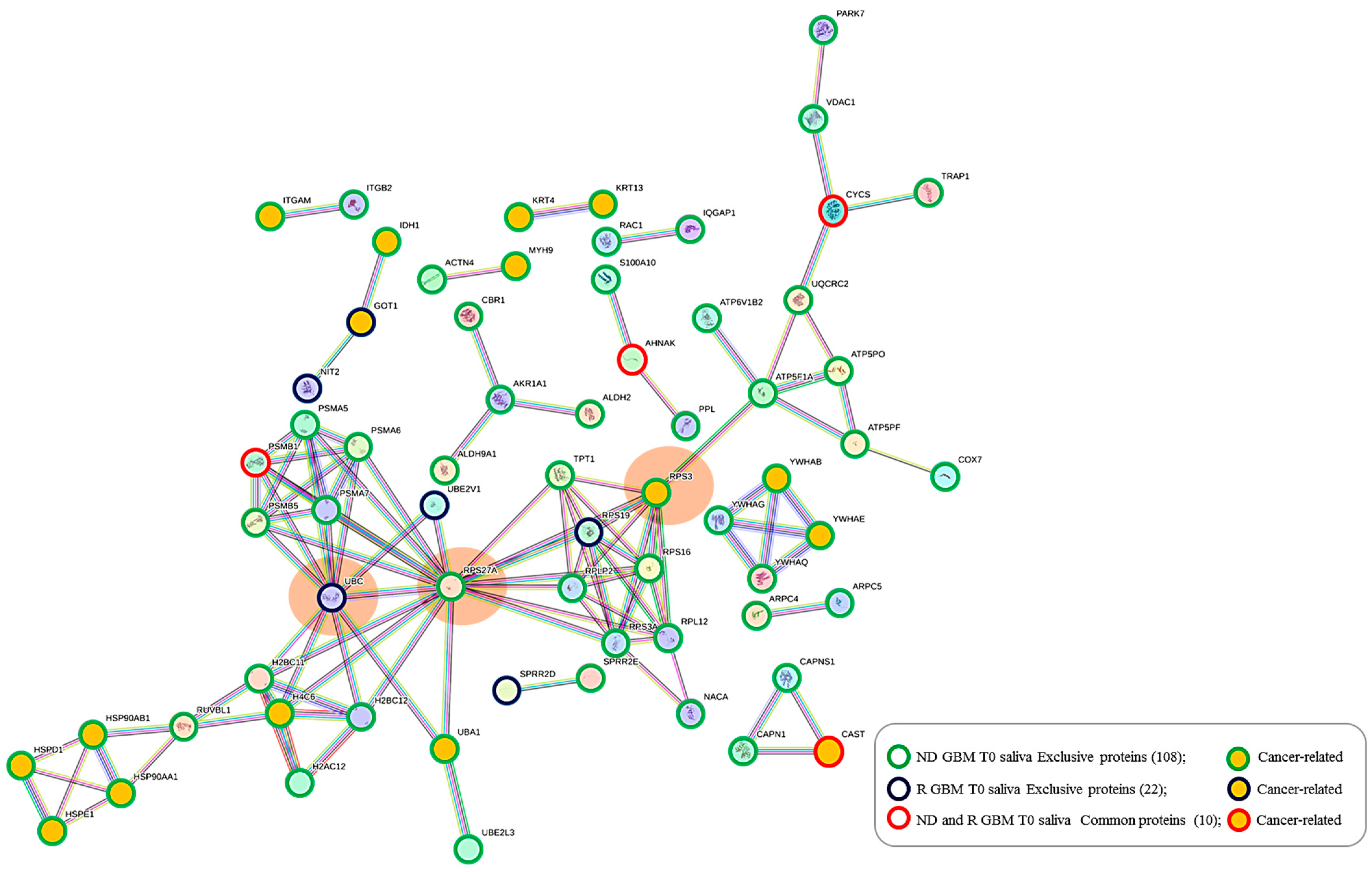
2.5. Protein–Protein Functional Interaction Network of GBM Tumor Salivary Proteins
3. Materials and Methods
3.1. Sample Collection
3.2. Chemicals
3.3. Sample Preparation
3.4. LC-MS Proteomic Analysis
3.5. LC-MS Data Elaboration and Bioinformatic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.; Dang, R.; Liu, H.; Dai, L.; Liu, H.; Adegboro, A.A.; Zhang, Y.; Li, W.; Peng, K.; Hong, J.; et al. Machine learning-based investigation of regulated cell death for predicting prognosis and immunotherapy response in glioma patients. Sci. Rep. 2024, 14, 4173. [Google Scholar] [CrossRef]
- Wu, W.; Klockow, J.L.; Zhang, M.; Lafortune, F.; Chang, E.; Jin, L.; Wu, Y.; Daldrup-Link, H.E. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacol. Res. 2021, 171, 105780. [Google Scholar] [CrossRef] [PubMed]
- Senhaji, N.; Squalli Houssaini, A.; Lamrabet, S.; Louati, S.; Bennis, S. Molecular and Circulating Biomarkers in Patients with Glioblastoma. Int. J. Mol. Sci. 2022, 23, 7474. [Google Scholar] [CrossRef] [PubMed]
- Ius, T.; Pignotti, F.; Della Pepa, G.M.; Bagatto, D.; Isola, M.; Battistella, C.; Gaudino, S.; Pegolo, E.; Chiesa, S.; Arcicasa, M.; et al. Glioblastoma: From volumetric analysis to molecular predictors. J. Neurosurg. Sci. 2022, 66, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, T.; Jiang, Y.; Xu, H.; Yan, Y.; Fu, D.; Chen, J. The challenges and the promise of molecular targeted therapy in malignant gliomas. Neoplasia 2015, 17, 239–255. [Google Scholar] [CrossRef]
- Rodríguez-Camacho, A.; Flores-Vázquez, J.G.; Moscardini-Martelli, J.; Torres-Ríos, J.A.; Olmos-Guzmán, A.; Ortiz-Arce, C.S.; Cid-Sánchez, D.R.; Pérez, S.R.; Macías-González, M.D.S.; Hernández-Sánchez, L.C.; et al. Glioblastoma Treatment: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2022, 29, 7207. [Google Scholar] [CrossRef]
- Boroumand, M.; Olianas, A.; Cabras, T.; Manconi, B.; Fanni, D.; Faa, G.; Desiderio, C.; Messana, I.; Castagnola, M. Saliva, a bodily fluid with recognized and potential diagnostic applications. J. Sep. Sci. 2021, 44, 3677–3690. [Google Scholar] [CrossRef]
- Shakeeb, N.; Varkey, P.; Ajit, A. Human Saliva as a Diagnostic Specimen for Early Detection of Inflammatory Biomarkers by Real-Time RT-PCR. Inflammation 2021, 44, 1713–1723. [Google Scholar] [CrossRef]
- Messana, I.; Cabras, T.; Iavarone, F.; Manconi, B.; Huang, L.; Martelli, C.; Olianas, A.; Sanna, M.T.; Pisano, E.; Sanna, M.; et al. Chrono-proteomics of human saliva: Variations of the salivary proteome during human development. J. Proteome Res. 2015, 14, 1666–1677. [Google Scholar] [CrossRef]
- Cabras, T.; Pisano, E.; Boi, R.; Olianas, A.; Manconi, B.; Inzitari, R.; Fanali, C.; Giardina, B.; Castagnola, M.; Messana, I. Age-dependent modifications of the human salivary secretory protein complex. J. Proteome Res. 2009, 8, 4126–4134. [Google Scholar] [CrossRef]
- Esteves, C.V.; Campos, W.G.; Souza, M.M.; Lourenço, S.V.; Siqueira, W.L.; Lemos-Júnior, C.A. Diagnostic potential of saliva proteome analysis: A review and guide to clinical practice. Braz. Oral Res. 2019, 33, 43. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Scarano, E.; Passali, G.C.; Messana, I.; Cabras, T.; Iavarone, F.; Di Cintio, G.; Fiorita, A.; De Corso, E.; Paludetti, G. Salivary biomarkers and proteomics: Future diagnostic and clinical utilities. Acta Otorhinolaryngol. Ital. 2017, 37, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wulfkuhle, J.D.; Paweletz, C.P.; Steeg, P.S.; Petricoin, E.F., III; Liotta, L. Approaches to the diagnosis, treatment, and monitoring of cancer. Adv. Exp. Med. Biol. 2003, 532, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Streckfus, C.F.; Brown, R.E.; Bull, J.M. Proteomics, morphoproteomics, saliva and breast cancer: An emerging approach to guide the delivery of individualised thermal therapy, thermochemotherapy and monitor therapy response. Int. J. Hyperth. 2010, 26, 649–661. [Google Scholar] [CrossRef] [PubMed]
- Skallevold, H.E.; Vallenari, E.M.; Sapkota, D. Salivary Biomarkers in Lung Cancer. Mediat. Inflamm. 2021, 6019791. [Google Scholar] [CrossRef]
- Wang, X.; Kaczor-Urbanowicz, K.E.; Wong, D.T. Salivary biomarkers in cancer detection. Med. Oncol. 2017, 34, 7. [Google Scholar] [CrossRef]
- Koopaie, M.; Kolahdooz, S.; Fatahzadeh, M.; Manifar, S. Salivary biomarkers in breast cancer diagnosis: A systematic review and diagnostic meta-analysis. Cancer Med. 2022, 11, 2644–2661. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Lane, L.; Overall, C.M.; Bandeira, N.; Baker, M.S.; Pineau, C.; Moritz, R.L.; Corrales, F.; Orchard, S.; Van Eyk, J.E.; et al. Human proteome project mass spectrometry data interpretation guidelines 3.0. J. Proteome Res. 2019, 18, 4108–4116. [Google Scholar] [CrossRef]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [PubMed]
- La Rocca, G.; Simboli, G.A.; Vincenzoni, F.; Rossetti, D.V.; Urbani, A.; Ius, T.; Della Pepa, G.M.; Olivi, A.; Sabatino, G.; Desiderio, C. Glioblastoma CUSA Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers 2020, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Drucker, K.L.; Gianinni, C.; Decker, P.A.; Diamandis, E.P.; Scarisbrick, I.A. Prognostic significance of multiple kallikreins in high-grade astrocytoma. BMC Cancer 2015, 15, 565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z.; Yang, L.; Xu, B.; Li, W.; Tang, P.; Zhang, Z.; Han, N.; Gao, Y.; Cheng, S.; et al. Identification of human tissue kallikrein 6 as a potential marker of laryngeal cancer based on the relevant secretory/releasing protein database. Dis Markers 2014, 2014, 594093. [Google Scholar] [CrossRef] [PubMed]
- Talieri, M.; Zoma, M.; Devetzi, M.; Scorilas, A.; Ardavanis, A. Kallikrein-related peptidase 6 (KLK6) gene expression in intracranial tumors. Tumor Biol. 2012, 33, 1375–1383. [Google Scholar] [CrossRef]
- Ghosh, M.C.; Grass, L.; Soosaipillai, A.; Sotiropoulou, G.; Diamandis, E.P. Human kallikrein 6 degrades extracellular matrix proteins and may enhance the metastatic potential of tumour cells. Tumour Biol. 2004, 25, 193–199. [Google Scholar] [CrossRef]
- Lee, N.H.; Park, S.R.; Lee, J.W.; Lim, S.; Lee, S.H.; Nam, S.; Kim, D.Y.; Hah, S.Y.; Hong, I.S.; Lee, H.Y. SERPINB2 Is a Novel Indicator of Cancer Stem Cell Tumorigenicity in Multiple Cancer Types. Cancers 2019, 11, 499. [Google Scholar] [CrossRef]
- Murphy, P.G.; Lenz, S.P.; Dobson, M.; Arndt, A.D.; Hart, D.A. Purification and characterization of the plasminogen activator inhibitors PAI-1, PAI-2, and PN-1 from the human glioblastoma U138. Biochem. Cell Biol. 1993, 71, 248–254. [Google Scholar] [CrossRef]
- Al-Khatib, S.M.; Al-Bzour, A.N.; Al-Majali, M.N.; Sa’d, L.M.; Alramadneh, J.A.; Othman, N.R.; Al-Mistarehi, A.H.; Alomari, S. Exploring Genetic Determinants: A Comprehensive Analysis of Serpin B Family SNPs and Prognosis in Glioblastoma Multiforme Patients. Cancers 2024, 10, 1112. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, P.; Hu, W.; Chen, D. New insights into the roles of peroxiredoxins in cancer. Biomed. Pharmacother. 2023, 164, 114896. [Google Scholar] [CrossRef]
- Lamprou, I.; Kakouratos, C.; Tsolou, A.; Pavlidis, P.; Xanthopoulou, E.T.; Nanos, C.; Tsaroucha, A.; Sivridis, E.; Giatromanolaki, A.; Koukourakis, M.I. Lipophagy-Related Protein Perilipin-3 and Resistance of Prostate Cancer to Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Chung, Y.M.; Lee, Y.S.; Kim, H.J.; Kim, J.S.; Chae, H.Z.; Yoo, Y.D. Antisense of human peroxiredoxin II enhances radiation-induced cell death. Clin. Cancer Res. 2000, 6, 4915–4920. [Google Scholar] [PubMed]
- Müller Bark, J.; Trevisan França de Lima, L.; Zhang, X.; Broszczak, D.; Leo, P.J.; Jeffree, R.L.; Chua, B.; Day, B.W.; Punyadeera, C. Proteome profiling of salivary small extrace llular vesicles in glioblastoma patients. Cancer 2023, 129, 2836–2847. [Google Scholar] [CrossRef] [PubMed]
- Cumba Garcia, L.M.; Peterson, T.E.; Cepeda, M.A.; Johnson, A.J.; Parney, I.F. Isolation and analysis of plasma-derived exosomes in patients with glioma. Front. Oncol. 2019, 9, 651. [Google Scholar] [CrossRef]
- García-Romero, N.; Carrión-Navarro, J.; Esteban-Rubio, S.; Lazaro-Ibanez, E.; Peris-Celda, M.; Alonso, M.M.; Guzman-De-Villoria, J.; Fernandez-Carballal, C.; de Mendivil, A.O.; Garcia-Duque, S.; et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 2017, 8, 1416–1428. [Google Scholar] [CrossRef]
- Cai, L.; Li, Q.; Li, W.; Wang, C.; Tu, M.; Zhu, Z.; Su, Z.; Lu, X. Calpain suppresses cell growth and invasion of glioblastoma multiforme by producing the cleavage of filamin A. Int. J. Clin. Oncol. 2020, 25, 1055–1066. [Google Scholar] [CrossRef]
- Spinelli, S.; Barbieri, F.; Averna, M.; Florio, T.; Pedrazzi, M.; Tremonti, B.F.; Capraro, M.; De Tullio, R. Expression of calpastatin hcast 3-25 and activity of the calpain/calpastatin system in human glioblastoma stem cells: Possible involvement of hcast 3-25 in cell differentiation. Front. Mol. Biosci. 2024, 11, 1359956. [Google Scholar] [CrossRef]
- Bassett, E.A.; Palanichamy, K.; Pearson, M.; McElroy, J.P.; Haque, S.J.; Bell, E.H.; Chakravarti, A. Calpastatin phosphorylation regulates radiation-induced calpain activity in glioblastoma. Oncotarget 2018, 9, 14597–14607. [Google Scholar] [CrossRef]
- Suárez-Arroyo, I.J.; Feliz-Mosquea, Y.R.; Pérez-Laspiur, J.; Arju, R.; Giashuddin, S.; Maldonado-Martínez, G.; Cubano, L.A.; Schneider, R.J.; Martínez-Montemayor, M.M. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. Am. J. Cancer Res. 2016, 6, 1720–1740. [Google Scholar]
- Wheeler, D.B.; Zoncu, R.; Root, D.E.; Sabatini, D.M.; Sawyers, C.L. Identification of an oncogenic RAB protein. Science 2015, 350, 211–217. [Google Scholar] [CrossRef]
- Homma, Y.; Hiragi, S.; Fukuda, M. Rab family of small GTPases: An updated view on their regulation and functions. FEBS J. 2021, 288, 36–55. [Google Scholar] [CrossRef] [PubMed]
- Villagomez, F.R.; Medina-Contreras, O.; Cerna-Cortes, J.F.; Patino-Lopez, G. The role of the oncogenic Rab35 in cancer invasion, metastasis, and immune evasion, especially in leukemia. Small GTPases 2020, 11, 334–345. [Google Scholar] [CrossRef]
- Kulasekaran, G.; Chaineau, M.; Piscopo, V.E.C.; Verginelli, F.; Fotouhi, M.; Girard, M.; Tang, Y.; Dali, R.; Lo, R.; Stifani, S.; et al. An Arf/Rab cascade controls the growth and invasiveness of glioblastoma. J. Cell Biol. 2021, 220, e202004229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cai, Z.; Li, H. AHNAKs roles in physiology and malignant tumors. Front. Oncol. 2023, 13, 1258951. [Google Scholar] [CrossRef] [PubMed]
- Azimi, P.; Karimpour, M.; Yazdanian, T.; Totonchi, M.; Ahmadiani, A. Comprehensive somatic mutational analysis in glioblastoma: Implications for precision medicine approaches. PLoS ONE 2024, 19, e0295698. [Google Scholar] [CrossRef] [PubMed]
- Rana, R.; Huirem, R.S.; Kant, R.; Chauhan, K.; Sharma, S.; Yashavarddhan, M.H.; Chhabra, S.S.; Acharya, R.; Kalra, S.K.; Gupta, A.; et al. Cytochrome C as a potential clinical marker for diagnosis and treatment of glioma. Front. Oncol. 2022, 12, 960787. [Google Scholar] [CrossRef]
- Kopeć, M.; Borek-Dorosz, A.; Jarczewska, K.; Barańska, M.; Abramczyk, H. The role of cardiolipin and cytochrome c in mitochondrial metabolism of cancer cells determined by Raman imaging: In vitro study on the brain glioblastoma U-87 MG cell line. Analyst 2024, 149, 2697–2708. [Google Scholar] [CrossRef]
- Shang, E.; Nguyen, T.T.T.; Westhoff, M.A.; Karpel-Massler, G.; Siegelin, M.D. Targeting cellular respiration as a therapeutic strategy in glioblastoma. Oncotarget 2023, 14, 419–425. [Google Scholar] [CrossRef]
- Jia, Z.; Ai, X.; Sun, F.; Zang, T.; Guan, Y.; Gao, F. Identification of new hub genes associated with bladder carcinoma via bioinformatics analysis. Tumor 2015, 101, 117–122. [Google Scholar] [CrossRef]
- Guo, J.Y.; Jing, Z.Q.; Li, X.J.; Liu, L.Y. Bioinformatic Analysis Identifying PSMB 1/2/3/4/6/8/9/10 as Prognostic Indicators in Clear Cell Renal Cell Carcinoma. Int. J. Med. Sci. 2022, 19, 796–812. [Google Scholar] [CrossRef]
- Thaker, N.G.; Zhang, F.; McDonald, P.R.; Shun, T.Y.; Lewen, M.D.; Pollack, I.F.; Lazo, J.S. Identification of survival genes in human glioblastoma cells by small interfering RNA screening. Mol. Pharmacol. 2009, 76, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, Z.; Tan, Z.; Liu, X.; Wang, Z.; Xiong, B.; Shen, X.; Zhu, X. PSMB2 plays an oncogenic role in glioma and correlates to the immune microenvironment. Sci. Rep. 2024, 14, 5861. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.C.; Tsai, W.C.; Sung, Y.C.; Chang, H.H.; Chen, Y. Interference with PSMB4 Expression Exerts an Anti-Tumor Effect by Decreasing the Invasion and Proliferation of Human Glioblastoma Cells. Cell. Physiol. Biochem. 2018, 45, 819–831. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Zhou, G.; Chen, L.; Zhang, Y.; Ma, W.; Gao, L.; Gao, G. The prognostic and immune significance of PLBD1 in pan-cancer and its roles in proliferation and invasion of glioma. J. Cancer 2024, 15, 3857–3872. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, S.; Gokhin, D.S.; Sui, Z.; Bergeron, S.E.; Rubenstein, P.A.; Fowler, V.M. Differential actin-regulatory activities of Tropomodulin1 and Tropomodulin3 with diverse tropomyosin and actin isoforms. J. Biol. Chem. 2014, 289, 11616–11629. [Google Scholar] [CrossRef]
- Suresh, R.; Diaz, R.J. The remodelling of actin composition as a hallmark of cancer. Transl. Oncol. 2021, 14, 101051. [Google Scholar] [CrossRef]
- Chen, B.; Wang, M.; Qiu, J.; Liao, K.; Zhang, W.; Lv, Q.; Ma, C.; Qian, Z.; Shi, Z.; Liang, R.; et al. Cleavage of tropomodulin-3 by asparagine endopeptidase promotes cancer malignancy by actin remodeling and SND1/RhoA signaling. J. Exp. Clin. Cancer Res. 2022, 41, 209. [Google Scholar] [CrossRef]
- Lu, S.Z.; Guo, Y.S.; Liang, P.Z.; Zhang, S.Z.; Yin, S.; Yin, Y.Q.; Wang, X.M.; Ding, F.; Gu, X.S.; Zhou, J.W. Suppression of astrocytic autophagy by αB-crystallin contributes to α-synuclein inclusion formation. Transl. Neurodegener. 2019, 8, 3. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Wu, J.; Li, W.; Chen, Z.; Yang, L. Progression of the role of CRYAB in signaling pathways and cancers. OncoTargets Ther. 2019, 12, 4129–4139. [Google Scholar] [CrossRef]
- Cai, H.B.; Zhao, M.Y.; Li, X.H.; Li, Y.Q.; Yu, T.H.; Wang, C.Z.; Wang, L.N.; Xu, W.Y.; Liang, B.; Cai, Y.P.; et al. Single cell sequencing revealed the mechanism of CRYAB in glioma and its diagnostic and prognostic value. Front. Immunol. 2024, 14, 1336187. [Google Scholar] [CrossRef]
- Cheng, L.; Zou, X.; Wang, J.; Zhang, J.; Mo, Z.; Huang, H. The role of CRYAB in tumor prognosis and immune infiltration: A Pan-cancer analysis. Front. Surg. 2023, 9, 1117307. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, Y.; Tu, K.; Xu, Q.; Yang, Y.; Salman, S.; Le, N.; Lu, H.; Chen, C.; Zhu, Y.; et al. Histone citrullination by PADI4 is required for HIF-dependent transcriptional responses to hypoxia and tumor vascularization. Sci. Adv. 2021, 7, eabe3771. [Google Scholar] [CrossRef] [PubMed]
- Rossetti, D.V.; Muntiu, A.; Massimi, L.; Tamburrini, G.; Desiderio, C. Citrullination Post-Translational Modification: State of the Art of Brain Tumor Investigations and Future Perspectives. Diagnostics 2023, 13, 2872. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, Y.; Wang, W. The Role of Peptidyl Arginine Deiminase IV (PADI4) in Cancers. Anticancer Agents Med. Chem. 2023, 23, 256–265. [Google Scholar] [CrossRef]
- Zhu, D.; Lu, Y.; Wang, Y.; Wang, Y. PAD4 and Its Inhibitors in Cancer Progression and Prognosis. Pharmaceutics 2022, 14, 2414. [Google Scholar] [CrossRef]
- Okura, G.C.; Bharadwaj, A.G.; Waisman, D.M. Recent advances in molecular and cellular functions of S100A10. Biomolecules 2023, 13, 1450. [Google Scholar] [CrossRef]
- Ma, K.; Chen, S.; Chen, X.; Yang, C.; Yang, J. S100A10 Is a New Prognostic Biomarker Related to the Malignant Molecular Features and Immunosuppression Process of Adult Gliomas. World Neurosurg. 2022, 165, e650–e663. [Google Scholar] [CrossRef]
- Alarm Signal S100-Related Signature Is Correlated with Tumor Microenvironment and Predicts Prognosis in Glioma. Dis. Markers 2022, 22, 4968555. [CrossRef]
- Di Giuseppe, F.; Carluccio, M.; Zuccarini, M.; Giuliani, P.; Ricci-Vitiani, L.; Pallini, R.; De Sanctis, P.; Di Pietro, R.; Ciccarelli, R.; Angelucci, S. Proteomic Characterization of Two Extracellular Vesicle Subtypes Isolated from Human Glioblastoma Stem Cell Secretome by Sequential Centrifugal Ultrafiltration. Biomedicines 2021, 9, 146. [Google Scholar] [CrossRef]
- Broggi, G.; Angelico, G.; Farina, J.; Tinnirello, G.; Barresi, V.; Zanelli, M.; Palicelli, A.; Certo, F.; Barbagallo, G.; Magro, G.; et al. Tumor-associated microenvironment, PD-L1 expression and their relationship with immunotherapy in glioblastoma, IDH-wild type: A comprehensive review with emphasis on the implications for neuropathologists. Pathol. Res. Pract. 2024, 254, 155144. [Google Scholar] [CrossRef]
- Shu, C.; Li, Q. Current advances in PD-1/PD-L1 axis-related tumour-infiltrating immune cells and therapeutic regimens in glioblastoma. Crit. Rev. Oncol. Hematol. 2020, 151, 102965. [Google Scholar] [CrossRef] [PubMed]
- Litak, J.; Mazurek, M.; Grochowski, C.; Kamieniak, P.; Roliński, J. PD-L1/PD-1 Axis in Glioblastoma Multiforme. Int. J. Mol. Sci. 2019, 20, 5347. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Kong, Z.; Ma, W. PD-1/PD-L1 immune checkpoint inhibitors in glioblastoma: Clinical studies, challenges and potential. Hum. Vaccin. Immunother. 2021, 17, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ma, X.; Gao, P.; Han, X.; Zhao, P.; Xie, F.; Liu, M. Advancing glioblastoma treatment by targeting metabolism. Neoplasia 2024, 51, 100985. [Google Scholar] [CrossRef] [PubMed]
- Garcia, J.H.; Jain, S.; Aghi, M.K. Metabolic Drivers of Invasion in Glioblastoma. Front. Cell Dev. Biol. 2021, 9, 683276. [Google Scholar] [CrossRef]
- Dewdney, B.; Jenkins, M.R.; Best, S.A.; Freytag, S.; Prasad, K.; Holst, J.; Endersby, R.; Johns, T.G. From signalling pathways to targeted therapies: Unravelling glioblastoma’s secrets and harnessing two decades of progress. Signal Transduct. Target. Ther. 2023, 8, 400. [Google Scholar] [CrossRef]
- Castro-Gamero, A.M.; Pezuk, J.A.; Brassesco, M.S.; Tone, L.G. G2/M inhibitors as pharmacotherapeutic opportunities for glioblastoma: The old, the new, and the future. Cancer Biol. Med. 2018, 15, 354–374. [Google Scholar] [CrossRef]
- Vlatkovic, T.; Veldwijk, M.R.; Giordano, F.A.; Herskind, C. Targeting Cell Cycle Checkpoint Kinases to Overcome Intrinsic Radioresistance in Brain Tumor Cells. Cancers 2022, 14, 701. [Google Scholar] [CrossRef]
- Fisher, R.; Pusztai, L.; Swanton, C. Cancer heterogeneity: Implications for targeted therapeutics. Br. J. Cancer 2013, 108, 479–485. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Tennstedt, P.; Bölch, C.; Strobel, G.; Minner, S.; Burkhardt, L.; Grob, T.; Masser, S.; Sauter, G.; Schlomm, T.; Simon, R. Patterns of TPD52 overexpression in multiple human solid tumor types analyzed by quantitative PCR. Int. J. Oncol. 2014, 44, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Zahra, K.; Shabbir, M.; Badshah, Y.; Trembley, J.H.; Badar, Z.; Khan, K.; Razak, S. Determining KLF14 tertiary structure and diagnostic significance in brain cancer progression. Sci. Rep. 2022, 12, 8039. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Liu, Y.; Yang, M.; Tu, M.; Zhu, W.; Chen, M. Glutamate-Oxaloacetate Transaminase 1 Impairs Glycolysis by Interacting with Pyruvate Carboxylase and Further Inhibits the Malignant Phenotypes of Glioblastoma Cells. World Neurosurg. 2021, 154, e616–e626. [Google Scholar] [CrossRef] [PubMed]
- Roh, J.; Im, M.; Chae, Y.; Kang, J.; Kim, W. The Involvement of Long Non-Coding RNAs in Glutamine-Metabolic Reprogramming and Therapeutic Resistance in Cancer. Int. J. Mol. Sci. 2022, 23, 14808. [Google Scholar] [CrossRef] [PubMed]
- Yin, A.; Shang, Z.; Etcheverry, A.; He, Y.; Aubry, M.; Lu, N.; Dong, Y. Integrative analysis identifies an immune-relevant epigenetic signature for prognostication of non-G-CIMP glioblastomas. OncoImmunology 2021, 10, e1902071. [Google Scholar] [CrossRef]
- Sugino, H.; Sawada, Y. Influence of S100A2 in Human Diseases. Diagnostics 2022, 12, 1756. [Google Scholar] [CrossRef]
- Caspa Gokulan, R.; Yap, L.F.; Paterson, I.C. HOPX: A Unique Homeodomain Protein in Development and Tumor Suppression. Cancers 2022, 14, 2764. [Google Scholar] [CrossRef]
- Yap, L.F.; Lai, S.L.; Patmanathan, S.N.; Gokulan, R.; Robinson, C.M.; White, J.B.; Paterson, I.C. HOPX functions as a tumour suppressor in head and neck cancer. Sci. Rep. 2016, 6, 38758. [Google Scholar] [CrossRef]
- Yi, G.Z.; Xiang, W.; Feng, W.Y.; Chen, Z.Y.; Li, Y.M.; Deng, S.Z.; Liu, Y.W. Identification of Key Candidate Proteins and Pathways Associated with Temozolomide Resistance in Glioblastoma Based on Subcellular Proteomics and Bioinformatical Analysis. BioMed Res. Int. 2018, 5238760. [Google Scholar] [CrossRef]
- Di Francesco, A.M.; Verrecchia, E.; Manna, S.; Urbani, A.; Manna, R. The chitinases as biomarkers in immune-mediated diseases. Clin. Chem. Lab Med. 2023, 61, 1363–1381. [Google Scholar] [CrossRef]
- Bramatti, I.; Aschner, M.; Branco, V.; Carvalho, C. Exposure of human glioblastoma cells to thimerosal inhibits the thioredoxin system and decreases tumor growth-related factors. Toxicol. Appl. Pharmacol. 2024, 484, 116844. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Qian, Y.; Zhang, W.; Wang, Q.; Jia, M.; Yue, J.; Fan, Z.; Jiang, Y.; Wang, L.; Wang, Y.; et al. Dual targeting agent Thiotert inhibits the progression of glioblastoma by inducing ER stress-dependent autophagy. Biomed. Pharmacother. 2024, 170, 115867. [Google Scholar] [CrossRef] [PubMed]
- Lauko, A.; Volovetz, J.; Turaga, S.M.; Bayik, D.; Silver, D.J.; Mitchell, K.; Mulkearns-Hubert, E.E.; Watson, D.C.; Desai, K.; Midha, M.; et al. SerpinB3 drives cancer stem cell survival in glioblastoma. Cell Rep. 2022, 40, 111348. [Google Scholar] [CrossRef] [PubMed]
- George Warren, W.; Osborn, M.; Yates, A.; Wright, K.; O’Sullivan, S.E. The emerging role of fatty acid binding protein 5 (FABP5) in cancers. Drug Discov. Today 2023, 28, 103628. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mao, X.; Ye, L.; Cheng, H.; Dai, X. The Role of the S100 Protein Family in Glioma. J. Cancer 2022, 13, 3022–3030. [Google Scholar] [CrossRef]
- Ren, S.; Zhang, Z.; Li, M.; Wang, D.; Guo, R.; Fang, X.; Chen, F. Cancer testis antigen subfamilies: Attractive targets for therapeutic vaccine. Int. J. Oncol. 2023, 62, 71. [Google Scholar] [CrossRef]
- Uribe, M.L.; Marrocco, I.; Yarden, Y. EGFR in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance. Cancers 2021, 13, 2748. [Google Scholar] [CrossRef]
- Colella, B.; Faienza, F.; Carinci, M.; D’Alessandro, G.; Catalano, M.; Santoro, A.; Cecconi, F.; Limatola, C.; Di Bartolomeo, S. Autophagy induction impairs Wnt/β-catenin signalling through β-catenin relocalisation in glioblastoma cells. Cell. Signal. 2019, 53, 357–364. [Google Scholar] [CrossRef]
- Colella, B.; Faienza, F.; Di Bartolomeo, S. EMT Regulation by Autophagy: A New Perspective in Glioblastoma Biology. Cancers 2019, 11, 312. [Google Scholar] [CrossRef]
- Hallal, S.M.; Tűzesi, Á.; Sida, L.A.; Xian, E.; Madani, D.; Muralidharan, K.; Shivalingam, B.; Buckland, M.E.; Satgunaseelan, L.; Alexander, K.L. Glioblastoma biomarkers in urinary extracellular vesicles reveal the potential for a ‘liquid gold’ biopsy. Br. J. Cancer 2024, 130, 836–851. [Google Scholar] [CrossRef]
- Hallal, S.; Azimi, A.; Wei, H.; Ho, N.; Lee, M.Y.T.; Sim, H.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Alexander-Kaufman, K.L. A Comprehensive Proteomic SWATH-MS Workflow for Profiling Blood Extracellular Vesicles: A New Avenue for Glioma Tumour Surveillance. Int. J. Mol. Sci. 2020, 21, 4754. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Russell, B.P.; Wei, H.; Lee, M.Y.T.; Toon, C.W.; Sy, J.; Shivalingam, B.; Buckland, M.E.; Kaufman, K.L. Extracellular Vesicles from Neurosurgical Aspirates Identifies Chaperonin Containing TCP1 Subunit 6A as a Potential Glioblastoma Biomarker with Prognostic Significance. Proteomics 2019, 19, e1800157. [Google Scholar] [CrossRef] [PubMed]
- Mallawaaratchy, D.M.; Hallal, S.; Russell, B.; Ly, L.; Ebrahimkhani, S.; Wei, H.; Christopherson, R.I.; Buckland, M.E.; Kaufman, K.L. Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J. Neuro-Oncol. 2017, 131, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Moresi, F.; Rossetti, D.V.; Vincenzoni, F.; Simboli, G.A.; La Rocca, G.; Olivi, A.; Urbani, A.; Sabatino, G.; Desiderio, C. Investigating Glioblastoma Multiforme Sub-Proteomes: A Computational Study of CUSA Fluid Proteomic Data. Int. J. Mol. Sci. 2022, 23, 2058. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R. Filter Aided Sample Preparation—A tutorial. Anal. Chim. Acta 2019, 1090, 23–30. [Google Scholar] [CrossRef]
- Oliveros, J.C. Venny: An Interactive Tool for Comparing Lists with Venn’s Diagrams. 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 7 August 2024).
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]



| Accession * | Description #§ | Gene Name | kDa | CUSA ND CORE | CUSA ND A+ | CUSA ND A− | CUSA R CORE | CUSA R A+ | CUSA R A− |
|---|---|---|---|---|---|---|---|---|---|
| P99999 * | Cytochrome c | CYCS | 11.7 | - | x | - | - | x | - |
| P32119 * | Peroxiredoxin-2 | PRDX2 | 21.9 | x | x | x | x | x | x |
| Q92928 * | Putative Ras-related protein Rab-1C | RAB1C | 22.0 | - | x | - | - | - | - |
| P20618 | Proteasome subunit beta type-1 | PSMB1 | 26.5 | - | - | - | - | - | - |
| Q92876 | Kallikrein-6 | KLK6 | 26.8 | - | - | - | - | - | - |
| Q9NYL9 | Tropomodulin-3 | TMOD3 | 39.6 | - | - | - | - | - | - |
| P05120 | Plasminogen activator inhibitor 2 | (PAI2) | 46.6 | - | - | - | - | - | - |
| Q6P4A8 | Phospholipase B-like 1 | PLBD1 | 6.3 | - | - | - | - | - | - |
| P20810 * | Calpastatin | CAST | 76.5 | - | - | x | - | x | - |
| Q09666 * | Neuroblast differentiation-associated protein AHNAK | AHNAK | 62.9 | - | - | - | x | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muntiu, A.; Moresi, F.; Vincenzoni, F.; Rossetti, D.V.; Iavarone, F.; Messana, I.; Castagnola, M.; La Rocca, G.; Mazzucchi, E.; Olivi, A.; et al. Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation. Int. J. Mol. Sci. 2024, 25, 12984. https://doi.org/10.3390/ijms252312984
Muntiu A, Moresi F, Vincenzoni F, Rossetti DV, Iavarone F, Messana I, Castagnola M, La Rocca G, Mazzucchi E, Olivi A, et al. Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation. International Journal of Molecular Sciences. 2024; 25(23):12984. https://doi.org/10.3390/ijms252312984
Chicago/Turabian StyleMuntiu, Alexandra, Fabiana Moresi, Federica Vincenzoni, Diana Valeria Rossetti, Federica Iavarone, Irene Messana, Massimo Castagnola, Giuseppe La Rocca, Edoardo Mazzucchi, Alessandro Olivi, and et al. 2024. "Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation" International Journal of Molecular Sciences 25, no. 23: 12984. https://doi.org/10.3390/ijms252312984
APA StyleMuntiu, A., Moresi, F., Vincenzoni, F., Rossetti, D. V., Iavarone, F., Messana, I., Castagnola, M., La Rocca, G., Mazzucchi, E., Olivi, A., Urbani, A., Sabatino, G., & Desiderio, C. (2024). Proteomic Profiling of Pre- and Post-Surgery Saliva of Glioblastoma Patients: A Pilot Investigation. International Journal of Molecular Sciences, 25(23), 12984. https://doi.org/10.3390/ijms252312984








