Genetic Diversity Analysis and Identification of Candidate Genes for Growth Traits in Chengkou Mountain Chicken
Abstract
1. Introduction
2. Results
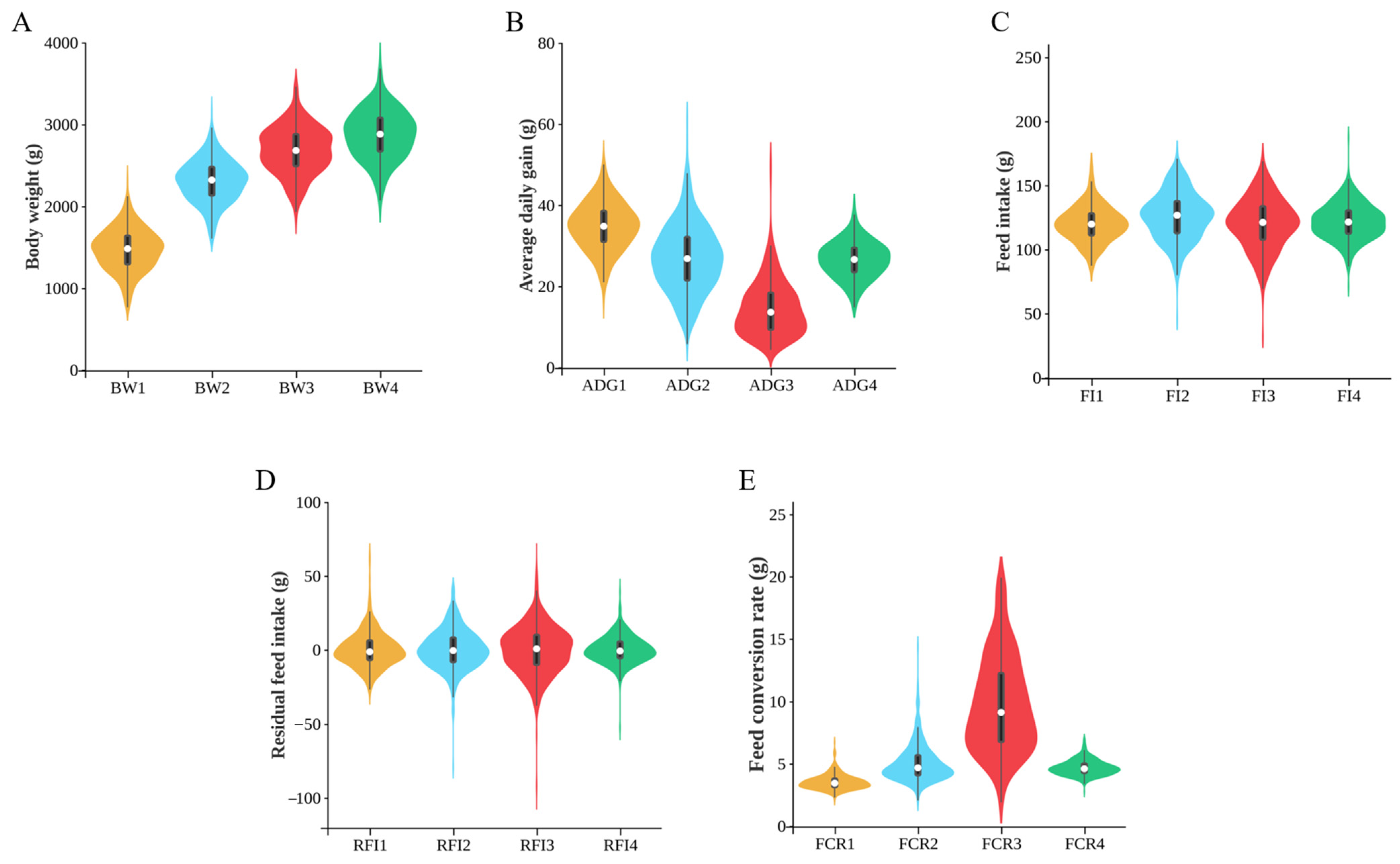
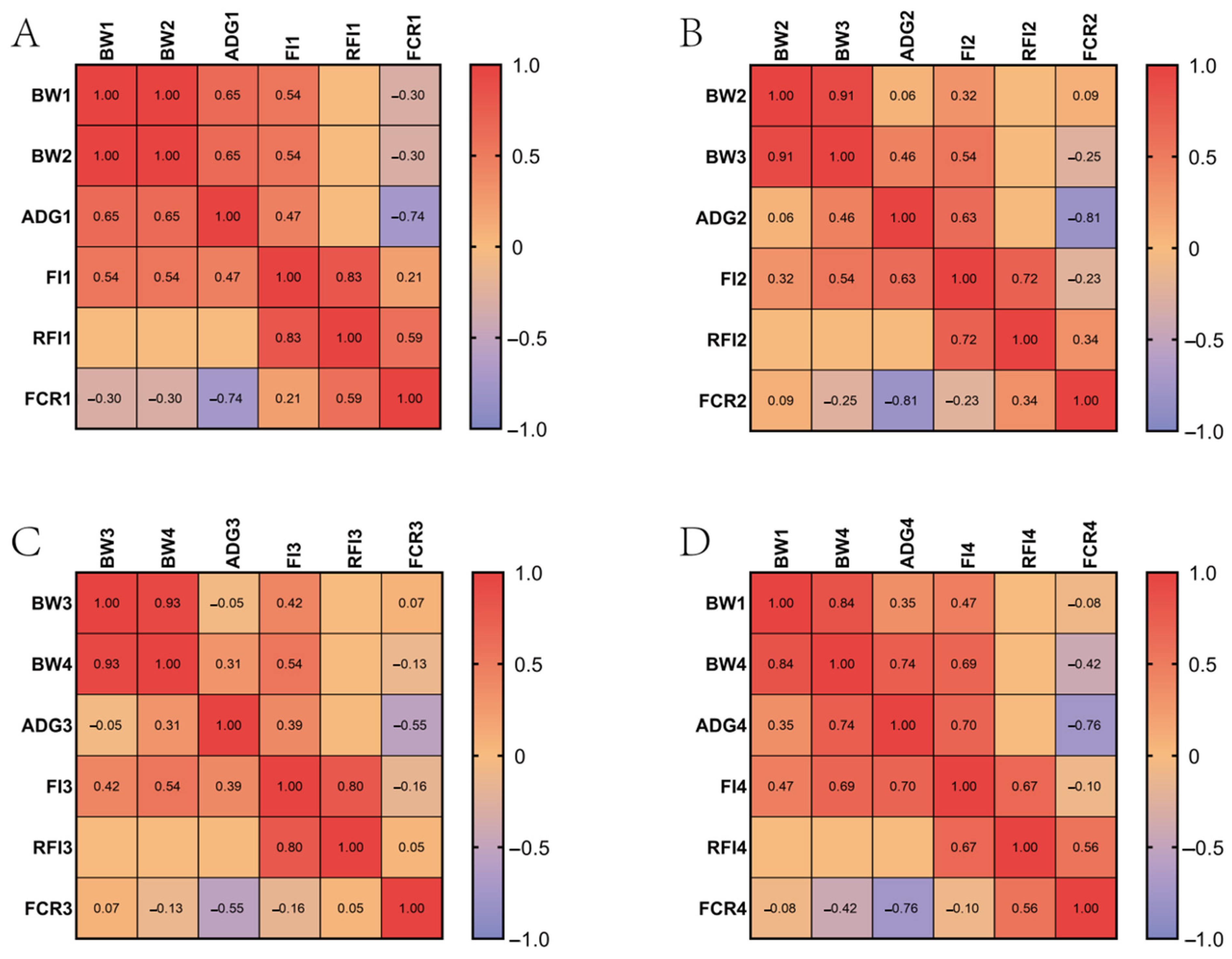
2.1. Phenotypic Data Analysis
2.2. Genotypic Data
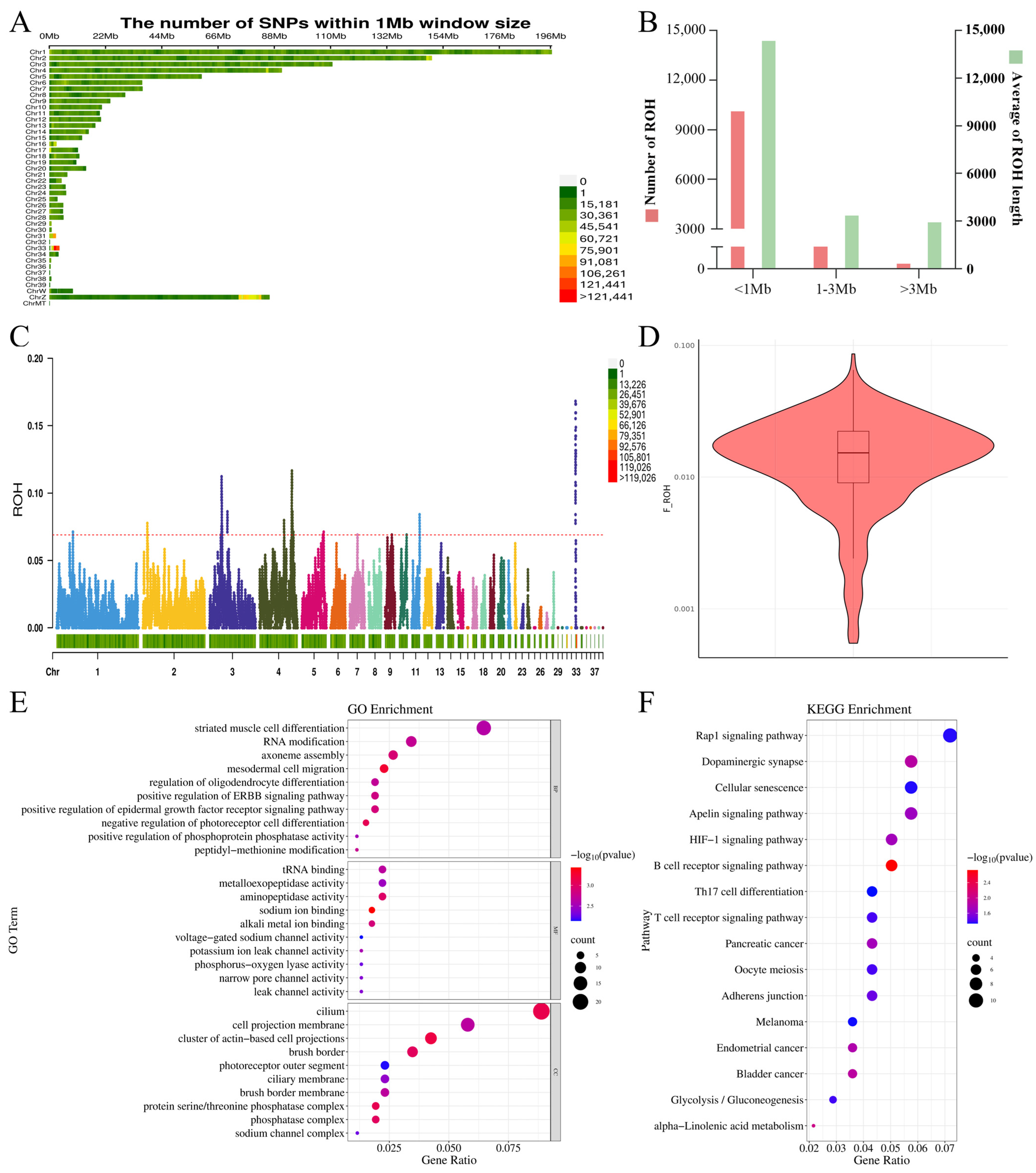
2.3. Genetic Diversity Analysis and Inbreeding Estimation
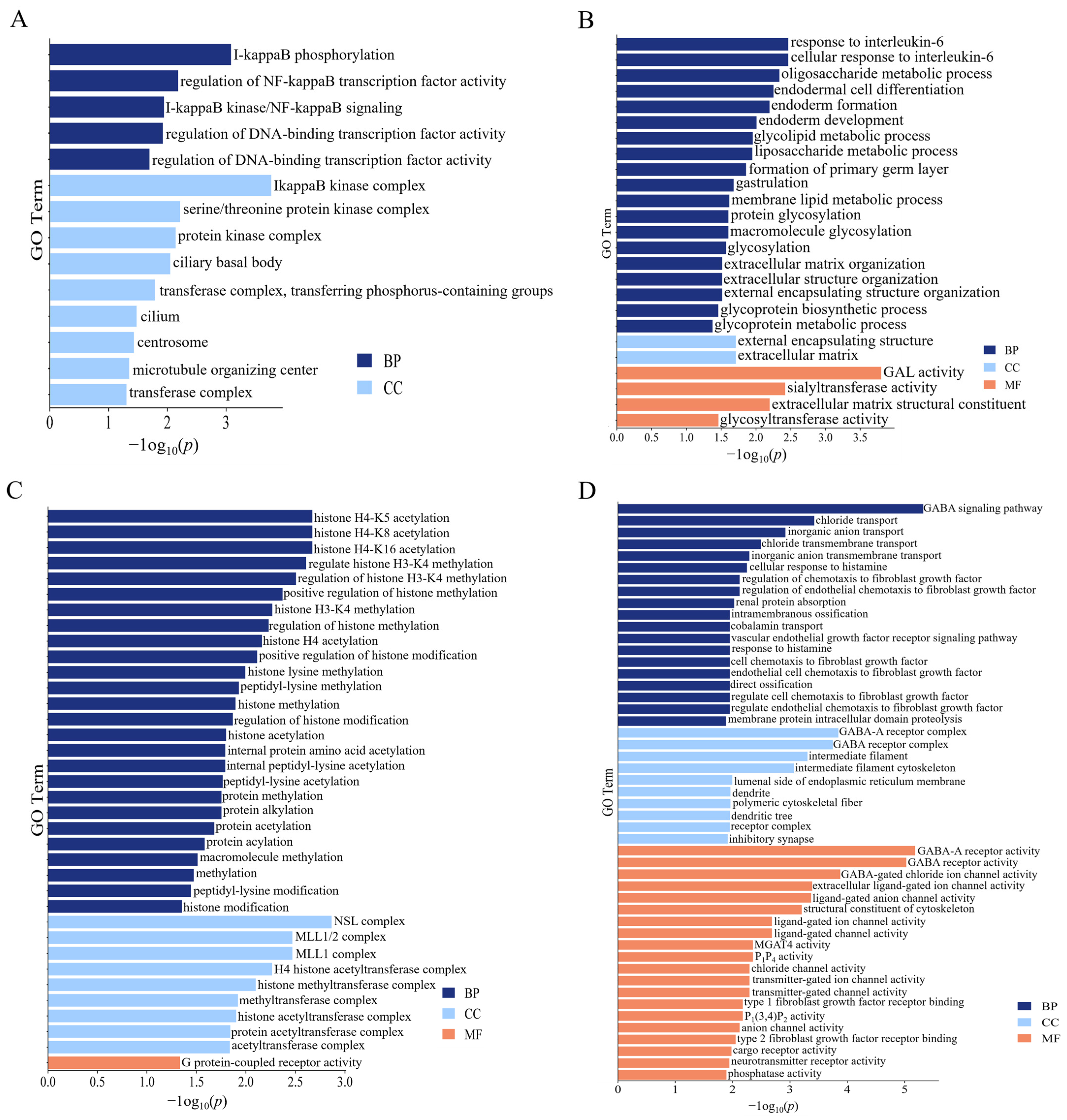
2.4. Identification of ROH Island Candidate Genes
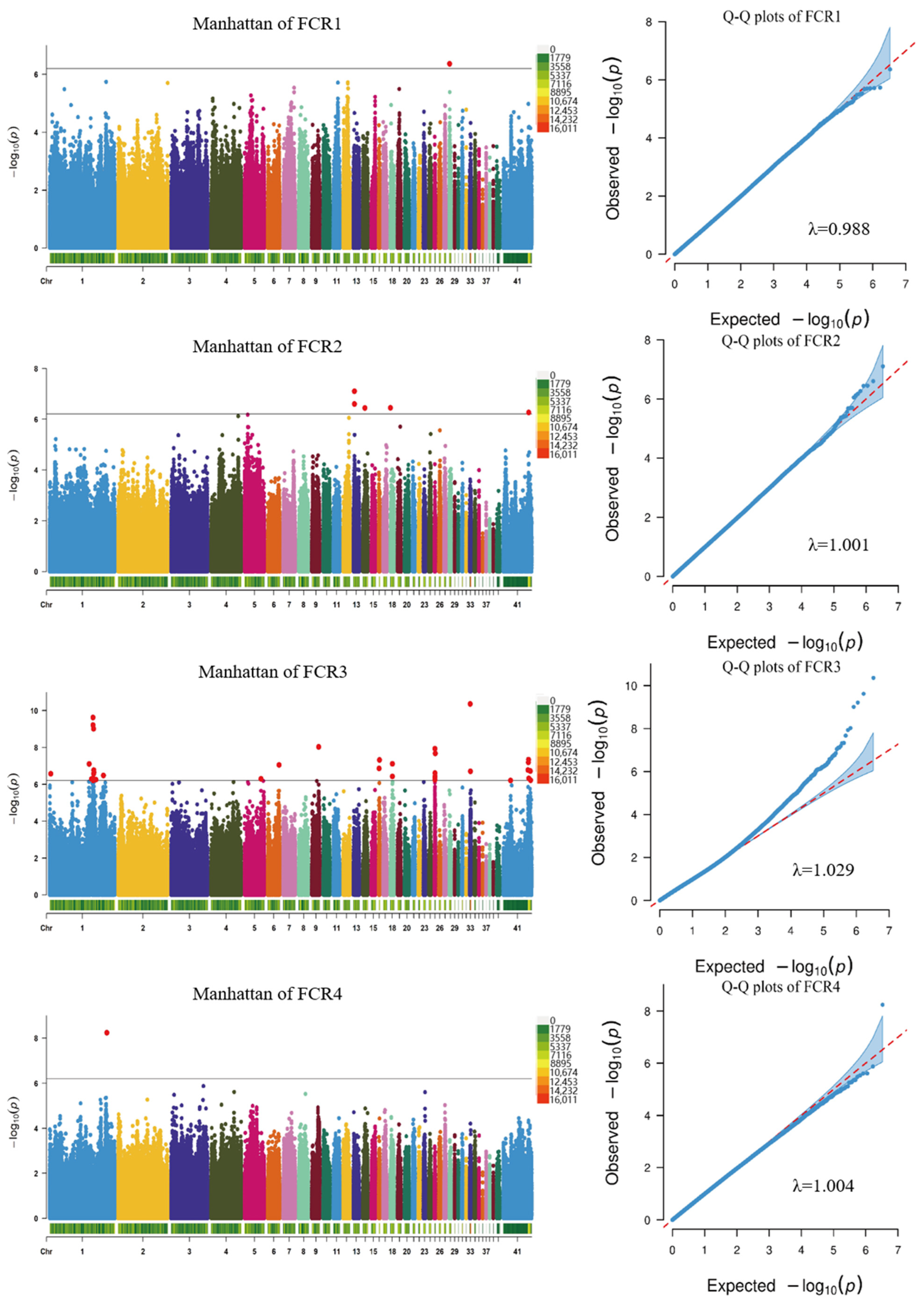
2.5. Whole-Genome Association Study and Identification of Candidate Genes
3. Discussion
3.1. Phenotypic Analysis
3.2. Genetic Diversity and Inbreeding Analysis
3.3. Candidate Genes Within ROH Islands
3.4. Genome-Wide Association Analysis
4. Materials and Methods
4.1. Animal Rearing and Phenotype Measuring
4.2. Sequencing and SNP Calling
4.3. ROH Island Identification and Significant SNP Identification
4.4. Genetic Diversity Analysis and Inbreeding Level Assessment
4.5. GWAS
4.6. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment of Candidate Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ren, L.; Liu, A.; Wang, Q.; Wang, H.; Dong, D.; Liu, L. Transcriptome analysis of embryonic muscle development in Chengkou Mountain Chicken. BMC Genom. 2021, 22, 431. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Shen, L.; Zhou, J.; Cao, Z.; Luan, P.; Li, Y.; Xiao, F.; Guo, H.; Li, H.; Zhang, H. Genome-wide association studies for growth traits in broilers. BMC Genom. 2022, 23, 1. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, F.C.; Joshi, P.K.; Clark, D.W.; Ramsay, M.; Wilson, J.F. Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 2018, 19, 220–234. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Nicolazzi, E.L.; Stella, A.; Boettcher, P.J.; Gandini, G. Challenges and opportunities in genetic improvement of local livestock breeds. Front. Genet. 2015, 6, 33. [Google Scholar] [CrossRef]
- Bosse, M.; Megens, H.J.; Madsen, O.; Crooijmans, R.P.M.A.; Ryder, O.A.; Austerlitz, F.; Groenen, M.A.M.; Cara, M.A.R. Using genome-wide measures of coancestry to maintain diversity and fitness in endangered and domestic pig populations. Genome Res. 2015, 25, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, S.; Ciani, E.; Sardina, M.T.; Sottile, G.; Pilla, F.; Portolano, B.; Bi. Ov. Ita Consortium. Runs of homozygosity reveal genome-wide autozygosity in Italian sheep breeds. Anim. Genet. 2018, 49, 71–81. [Google Scholar] [CrossRef]
- Liao, R.; Zhang, X.; Chen, Q.; Wang, Z.; Wang, Q.; Yang, C.; Pan, Y. Genome-wide association study reveals novel variants for growth and egg traits in Dongxiang blue-shelled and White Leghorn chickens. Anim. Genet. 2016, 47, 588–596. [Google Scholar] [CrossRef]
- Tarsani, E.; Kranis, A.; Maniatis, G.; Avendano, S.; Hager-Theodorides, A.L.; Kominakis, A. Discovery and characterization of functional modules associated with body weight in broilers. Sci. Rep. 2019, 9, 9125. [Google Scholar] [CrossRef]
- Cao, X.; Wang, Y.; Shu, D.; Qu, H.; Luo, C.; Hu, X. Food intake-related genes in chicken determined through combinatorial genome-wide association study and transcriptome analysis. Anim. Genet. 2020, 51, 741–751. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, X.; Wen, J.; Wang, C.; Zhang, X.; Ren, X.; Zhang, J.; Li, H.; Muhatai, G.; Qu, L. Comparative population genomics analysis uncovers genomic footprints and genes influencing body weight trait in Chinese indigenous chicken. Poult. Sci. 2023, 102, 103031. [Google Scholar] [CrossRef]
- Deng, X.M.; Li, J.Y.; Li, N.; Wu, C.X. Genetic analysis of important growth trait based on F-2 resource population in chicken. Yi Chuan Xue Bao 2001, 28, 801–807. [Google Scholar] [PubMed]
- Yang, L.; Wang, X.; He, T.; Xiong, F.; Chen, X.; Chen, X.; Jin, S.; Geng, Z. Association of residual feed intake with growth performance, carcass traits, meat quality, and blood variables in native chickens. J. Anim. Sci. 2020, 98, skaa121. [Google Scholar] [CrossRef] [PubMed]
- Hillel, J.; Groenen, M.A.M.; Tixier-Boichard, M.; Korol, A.B.; David, L.; Kirzhner, V.M.; Burke, T.; Barre-Dirie, A.; Crooijmans, R.P.M.A.; Elo, K.; et al. Biodiversity of 52 chicken populations assessed by microsatellite typing of DNA pools. Genet. Sel. Evol. 2003, 35, 533–557. [Google Scholar] [CrossRef] [PubMed]
- Azimu, W.; Manatbay, B.; Li, Y.; Kaimaerdan, D.; Wang, H.E.; Reheman, A.; Muhatai, G. Genetic diversity and population structure analysis of eight local chicken breeds of Southern Xinjiang. Br. Poult. Sci. 2018, 59, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Kirikci, K.; Cam, M.A.; Mercan, L. Genetic diversity and relationship among indigenous Turkish Karayaka sheep subpopulations. Arch. Anim. Breed. 2020, 63, 269–275. [Google Scholar] [CrossRef]
- Talebi, R.; Szmatoła, T.; Mészáros, G.; Qanbari, S. Runs of Homozygosity in Modern Chicken Revealed by Sequence Data. G3 Genes Genomes Genet. 2020, 10, 4615–4623. [Google Scholar] [CrossRef]
- Purfield, D.C.; Berry, D.P.; McParland, S.; Bradley, D.G. Runs of homozygosity and population history in cattle. BMC Genet. 2012, 13, 70. [Google Scholar] [CrossRef]
- Bortoluzzi, C.; Crooijmans, R.P.M.A.; Bosse, M.; Hiemstra, S.J.; Groenen, M.A.M.; Megens, H.J. The effects of recent changes in breeding preferences on maintaining traditional Dutch chicken genomic diversity. Heredity 2018, 121, 564–578. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Kudinov, A.A.; Larkina, T.A.; Mitrofanova, O.V.; Dysin, A.P.; Terletsky, V.P.; Tyshchenko, V.I.; Griffin, D.K.; Romanov, M.N. Genetic Variability in Local and Imported Germplasm Chicken Populations as Revealed by Analyzing Runs of Homozygosity. Animals 2020, 10, 1887. [Google Scholar] [CrossRef]
- He, H.; Yin, H.; Yu, X.; Zhang, Y.; Ma, M.; Li, D.; Zhu, Q. PDLIM5 Affects Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via the p38-MAPK Pathway. Animals 2021, 11, 1016. [Google Scholar] [CrossRef]
- Tian, S.; Tang, W.; Zhong, Z.; Wang, Z.; Xie, X.; Liu, H.; Chen, F.; Liu, J.; Han, Y.; Qin, Y.; et al. Identification of Runs of Homozygosity Islands and Functional Variants in Wenchang Chicken. Animals 2023, 13, 1645. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, P.; Ostkotte, D.; Nolte, H.; Gerl, M.J.; Jais, A.; Brunner, H.L.; Sprenger, H.G.; Awazawa, M.; Nicholls, H.T.; Turpin-Nolan, S.M.; et al. CerS6-Derived Sphingolipids Interact with Mff and Promote Mitochondrial Fragmentation in Obesity. Cell 2019, 177, 1536–1552. [Google Scholar] [CrossRef]
- Peng, H.; Chiu, T.Y.; Liang, Y.J.; Lee, C.J.; Liu, C.S.; Suen, C.S.; Yen, J.J.Y.; Chen, H.T.; Hwang, M.J.; Hussain, M.M.; et al. PRAP1 is a novel lipid-binding protein that promotes lipid absorption by facilitating MTTP-mediated lipid transport. J. Biol. Chem. 2021, 296, 100052. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ahn, D.; Chung, S.J. Chebulinic Acid Suppresses Adipogenesis in 3T3-L1 Preadipocytes by Inhibiting PPP1CB Activity. Int. J. Mol. Sci. 2022, 23, 865. [Google Scholar] [CrossRef]
- Weng, Z.; Xu, Y.; Li, W.; Chen, J.; Zhong, M.; Zhong, F.; Du, B.; Zhang, B.; Huang, X. Genomic variations and signatures of selection in Wuhua yellow chicken. PLoS ONE 2020, 15, e0241137. [Google Scholar] [CrossRef]
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. [Google Scholar] [CrossRef]
- Raza, I.G.A.; Clarke, A.J. B Cell Metabolism and Autophagy in Autoimmunity. Front. Immunol. 2021, 12, 681105. [Google Scholar] [CrossRef] [PubMed]
- Abasht, B.; Lamont, S.J. Genome-wide association analysis reveals cryptic alleles as an important factor in heterosis for fatness in chicken F2 population. Anim. Genet. 2007, 38, 491–498. [Google Scholar] [CrossRef]
- Yuan, Y.; Peng, D.; Gu, X.; Gong, Y.; Sheng, Z.; Hu, X. Polygenic Basis and Variable Genetic Architectures Contribute to the Complex Nature of Body Weight —A Genome-Wide Study in Four Chinese Indigenous Chicken Breeds. Front. Genet. 2018, 9, 229. [Google Scholar] [CrossRef]
- Held, R.G.; Kaeser, P.S. ELKS active zone proteins as multitasking scaffolds for secretion. Open Biol. 2018, 8, 170258. [Google Scholar] [CrossRef]
- Mitchell, J.A.; Chesi, A.; Elci, O.; McCormack, S.E.; Roy, S.M.; Kalkwarf, H.J.; Lappe, J.M.; Gilsanz, V.; Oberfield, S.E.; Shepherd, J.A.; et al. Physical Activity Benefits the Skeleton of Children Genetically Predisposed to Lower Bone Density in Adulthood. J. Bone Miner. Res. 2016, 31, 1504–1512. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, H.; Wang, S.; Hu, X.; Gao, Y.; Wang, Q.; Li, N.; Wang, Y.; Zhang, H. Mapping quantitative trait loci affecting body weight and abdominal fat weight on chicken chromosome one. Poult. Sci. 2007, 86, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Besnier, F.; Wahlberg, P.; Rönnegård, L.; Ek, W.; Andersson, L.; Siegel, P.B.; Carlborg, O. Fine mapping and replication of QTL in outbred chicken advanced intercross lines. Genet. Sel. Evol. 2011, 43, 3. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Luo, C.; Zhang, C.; Zhang, R.; Tang, J.; Nie, Q.; Ma, L.; Hu, X.; Li, N.; Da, Y.; et al. Genome-Wide Association Study Identified a Narrow Chromosome 1 Region Associated with Chicken Growth Traits. PLoS ONE 2012, 7, e30910. [Google Scholar] [CrossRef] [PubMed]
- Romero, L.F.; Zuidhof, M.J.; Renema, R.A.; Robinson, F.E.; Naeima, A. Nonlinear mixed models to study metabolizable energy utilization in broiler breeder hens. Poult. Sci. 2009, 88, 1310–1320. [Google Scholar] [CrossRef]
- Xu, X.; Bi, X.; Wang, J.; Gui, R.; Li, T.; Li, L.; Wang, B. Identification of KANSL1 as a novel pathogenic gene for developmental dysplasia of the hip. J. Mol. Med. 2022, 100, 1159–1168. [Google Scholar] [CrossRef]
- Meunier, S.; Shvedunova, M.; Van Nguyen, N.; Avila, L.; Vernos, I.; Akhtar, A. An epigenetic regulator emerges as microtubule minus-end binding and stabilizing factor in mitosis. Nat. Commun. 2015, 6, 7889. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Bai, X.; Wang, Y.; Leng, L.; Zhang, H.; Li, Y.; Cao, Z.; Luan, P.; Xiao, F.; et al. Genetic parameters estimation and genome-wide association studies for internal organ traits in an F2 chicken population. J. Anim. Breed. Genet. 2022, 139, 434–446. [Google Scholar] [CrossRef]
- Dennis, G.; Kubagawa, H.; Cooper, M.D. Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc. Natl. Acad. Sci. USA 2000, 97, 13245–13250. [Google Scholar] [CrossRef]
- Sparling, B.A.; Ng, T.T.; Carlo-Allende, A.; McCarthy, F.M.; Taylor, R.L.; Drechsler, Y. Immunoglobulin-like receptors in chickens: Identification, functional characterization, and renaming to cluster homolog of immunoglobulin-like receptors. Poult. Sci. 2024, 103, 103292. [Google Scholar] [CrossRef]
- Sa, M.; Yoo, E.S.; Koh, W.; Park, M.G.; Jang, H.J.; Yang, Y.R.; Bhalla, M.; Lee, J.H.; Lim, J.; Won, W.; et al. Hypothalamic GABRA5-positive neurons control obesity via astrocytic GABA. Nat. Metab. 2023, 5, 1506–1525. [Google Scholar] [CrossRef] [PubMed]
- Carmelo, V.A.O.; Kadarmideen, H.N. Genome Regulation and Gene Interaction Networks Inferred from Muscle Transcriptome Underlying Feed Efficiency in Pigs. Front. Genet. 2020, 11, 650. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Jang, S.S.; Kang, D.H.; Kim, Y.K.; Kim, H.J.; Chung, K.Y.; Choi, I.; Lee, S.H. Identification of potential biomarkers associated with meat tenderness in Hanwoo (Korean cattle): An expression quantitative trait loci analysis. Anim. Genet. 2023, 54, 786–791. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.U.; Bai, G.; Pang, Y. Roles of NPAS2 in circadian rhythm and disease. Acta Biochim. Biophys. Sin. 2021, 53, 1257–1265. [Google Scholar] [CrossRef]
- Hieke, A.-S.C.; Hubert, S.M.; Athrey, G. Circadian disruption and divergent microbiota acquisition under extended photoperiod regimens in chicken. PeerJ 2019, 7, e6592. [Google Scholar] [CrossRef]
- Jiang, S.; Fu, Y.; Cheng, H. Daylight exposure and circadian clocks in broilers: Part I—Photoperiod effect on broiler behavior, skeletal health, and fear response. Poult. Sci. 2023, 102, 103162. [Google Scholar] [CrossRef]
- Nagayama, T.; Okuhara, S.; Ota, M.S.; Tachikawa, N.; Kasugai, S.; Iseki, S. FGF18 accelerates osteoblast differentiation by upregulating Bmp2 expression. Congenit. Anom. 2013, 53, 83–88. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, H.; Li, Z.; Zheng, X.; Jia, X.; Nie, Q.; Zhang, X. Comparison of the genome-wide DNA methylation profiles between fast-growing and slow-growing broilers. PLoS ONE 2013, 8, e56411. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Guo, Q.; Ma, X.; Zhang, J.; Huang, X.; Zhang, X.; Zhang, S. Association of ARHGEF11 R1467H polymorphism with risk for type 2 diabetes mellitus and insulin resistance in Chinese population. Mol. Biol. Rep. 2011, 38, 2499–2505. [Google Scholar] [CrossRef]
- Tarlow, D.M.; Watkins, P.A.; Reed, R.E.; Miller, R.S.; Zwergel, E.E.; Lane, M.D. Lipogenesis and the synthesis and secretion of very low density lipoprotein by avian liver cells in nonproliferating monolayer culture. Hormonal effects. J. Cell Biol. 1977, 73, 332–353. [Google Scholar] [CrossRef]
- Hermier, D. Lipoprotein metabolism and fattening in poultry. J. Nutr. 1997, 127 (Suppl. S5), 805S–808S. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Ruano-Rubio, V.; Depristo, M.A.; Fennell, T.J.; Banks, E. Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 2017. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh, B.I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Gao, X.; Starmer, J.; Martin, E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008, 32, 361–369. [Google Scholar] [CrossRef]

| MAF | He | Ho | PIC | Ho |
|---|---|---|---|---|
| 0.238584 | 0.32583 | 0.320666 | 0.325829 | 0.320666 |
| Max | Min | Aver | Std | |
|---|---|---|---|---|
| ROH length (Kb) | 3297.98 | 500.00 | 708.71 | 223.51 |
| Total length of individual ROHs (Kb) | 89,803.00 | 570.58 | 17,403.19 | 11,183.42 |
| Number of ROHs | 118.00 | 1.00 | 24.56 | 15.24 |
| FROH | 0.086274 | 0.000548 | 0.017 | 0.011 |
| Length | Number | Max | Min | Aver | Std | |
|---|---|---|---|---|---|---|
| <1 Mb | 6,739,650 | 10,319 | 999.95 | 500.004 | 14,556.48 | 9059.392 |
| 1–3 Mb | 1,322,810 | 1071 | 2693.12 | 1000.601 | 3584.85 | 2847.158 |
| >3 Mb | 12,620 | 4 | 3297.98 | 3017.774 | 3155.02 | 153.687 |
| Function of Some Candidate Genes | Gene Names |
|---|---|
| Stress resistance | CD38, BTBD9, ADH6, CISD2, BDH2, SLC39A8, NFKB1, TLR3, FAM149A, BANK1, GRPEL1, SORCS2, PFKP, ACOX3, HSPA2, LRFN2, TMEM179, SIVA1, INF2, FASTKD1 |
| Muscle development | CPEB2, NCAPG, LAP3, LDB2, ZNF318, QKI, PDLIM5, NEUROG2, PITX2, WDR37, MYF6, MYF5, SYNE2, PPP2R5E, BRE, AKT1, JAG2, PDK1, METTL8, SLC25A12 |
| Bone growth | NKX3-2, CYTL1, SLC35B2, FAM98A, CAPN11, DLK2, SNORD71, ALPK1, EMCN, PPP3CA, DIP2C, RPS6KA2, CPZ, GPHB5, DLX2, GALNT3 |
| Energy metabolism | HS3ST1, NSG1, GLO1, SLC29A1, PHLPP2, ELOVL6, EGF, PITRM1, FOSL2, G6PC2, CSRNP3 |
| Fat deposition | FBXL5, RASGRP3, JAKMIP1, COG8, PLA2G12A, MTTP, KLF6, ACSS3, RHOJ, PPP1CB, GPR132, PPIG, LRP2, CERS6 |
| Chr | SNP ID | Ref/Alt | Func.refGene | Ensembl ID | Gene_Name | Phenotypes |
|---|---|---|---|---|---|---|
| 1 | 1:60821062 | A/G | intronic | ENSGALG00010012399 | ERC1 | BW1, BW2 |
| 1 | 1:169293230 | T/C | ncRNA_intronic | ENSGALG00010004476 | NA | BW3 |
| 1 | 1:84523514 | G/A | intronic | ENSGALG00010005423; ENSGALG00010005474 | ST3GAL6; COL8A1 | ADG2 |
| 1 | 1:172549299 | T/G | intergenic | ENSGALG00010006408; ENSGALG00010007373 | NA;NA | ADG5, FCR5 |
| 28 | 28:992707 | A/G | intronic | ENSGALG00010028357 | SPPL2C | FCR1 |
| 13 | 13:2852686 | T/C | intronic | ENSGALG00010010399 | FGF18 | FCR2 |
| 13 | 13:2856342 | G/A | intronic | ENSGALG00010010399 | FGF18 | FCR2 |
| 14 | 14:6259710 | G/A | intronic | ENSGALG00010017898 | NHERF2 | FCR2 |
| 18 | 18:250974 | T/C | intergenic | NONE; ENSGALG00010030050 | NA; NA | FCR2 |
| 41 | 41:76970673 | T/C | intergenic | NONE;NONE | NA; NA | FCR2 |
| 1 | 1:2782842 | C/T | exonic | ENSGALG00010007596 | NA | FCR3 |
| 1 | 1:119168544 | A/G | intergenic | ENSGALG00010003392; ENSGALG00010003275 | NA; MBTPS2 | FCR3 |
| 1 | 1:126186942 | C/T | intergenic | ENSGALG00010001468; ENSGALG00010003235 | ANOS1; gga-mir-7448 | FCR3 |
| 1 | 1:130412664 | T/C | UTR3 | ENSGALG00010002753 | SLC9A7 | FCR3 |
| 1 | 1:130442457 | A/G | intergenic | ENSGALG00010003157; ENSGALG00010002800 | NA; TUBGCP5 | FCR3 |
| 1 | 1:130442500 | A/G | intergenic | ENSGALG00010003157; ENSGALG00010002800 | NA; TUBGCP5 | FCR3 |
| 1 | 1:130619948 | G/T | intronic | ENSGALG00010002886 | HERC2 | FCR3 |
| 1 | 1:131241497 | A/G | intronic | ENSGALG00010002907 | GABRG3 | FCR3 |
| 1 | 1:131389633 | A/G | intronic | ENSGALG00010002927; ENSGALG00010002942 | GABRA5; GABRB3 | FCR3 |
| 1 | 1:132406216 | C/A | upstream | ENSGALG00010015669 | INPP4A | FCR3 |
| 1 | 1:132427487 | C/T | intronic | ENSGALG00010015716 | MGAT4A | FCR3 |
| 1 | 1:133078359 | T/C | intronic | ENSGALG00010015886 | AFF3 | FCR3 |
| 1 | 1:133453381 | T/A | intronic | ENSGALG00010013436 | NPAS2 | FCR3 |
| 1 | 1:139215439 | G/T | intergenic | ENSGALG00010014379; ENSGALG00010014396 | CARS2; ING1 | FCR3 |
| 1 | 1:162102310 | A/G | intronic | ENSGALG00010002381 | TDRD3 | FCR3 |
| 5 | 5:49414192 | C/T | exonic | ENSGALG00010015137 | AMN | FCR3 |
| 6 | 6:34082468 | G/A | intronic | ENSGALG00010002712 | PTPRE | FCR3 |
| 9 | 9:20447352 | G/A | ncRNA_intronic | ENSGALG00010001602 | NA | FCR3 |
| 16 | 16:291729 | C/T | intronic | ENSGALG00010003661 | NA | FCR3 |
| 16 | 16:1610013 | C/A | intergenic | ENSGALG00010002238; ENSGALG00010002354 | NA; NA | FCR3 |
| 18 | 18:6176886 | T/G | ncRNA_intronic | ENSGALG00010030069 | NA | FCR3 |
| 18 | 18:6204681 | G/A | ncRNA_intronic | ENSGALG00010030069 | NA | FCR3 |
| 25 | 25:1112852 | C/G | intronic | ENSGALG00010027826 | ARHGEF11 | FCR3 |
| 25 | 25:1187425 | A/G | exonic | ENSGALG00010027624 | NA | FCR3 |
| 25 | 25:1296967 | C/T | intronic | ENSGALG00010027728 | CRNN | FCR3 |
| 25 | 25:1526371 | G/A | intronic | ENSGALG00010028545; ENSGALG00010028558 | NA; NA | FCR3 |
| 25 | 25:1574011 | A/G | intronic | ENSGALG00010028882; ENSGALG00010028886 | NA;NA | FCR3 |
| 25 | 25:2434093 | C/T | upstream;downstream | ENSGALG00010028878; ENSGALG00010028875 | IGSF9; DUSP23 | FCR3 |
| 33 | 33:1553885 | G/A | exonic | ENSGALG00010001909 | NA | FCR3 |
| 33 | 33:2515964 | T/G | exonic | ENSGALG00010002057 | NA | FCR3 |
| 41 | 41:22283106 | T/C | intergenic | NONE; NONE | NA; NA | FCR3 |
| 41 | 41:74728417 | G/T | intergenic | NONE; NONE | NA; NA | FCR3 |
| 41 | 41:76016616 | C/T | intergenic | NONE; NONE | NA; NA | FCR3 |
| 41 | 41:76442122 | T/C | intergenic | NONE; NONE | NA; NA | FCR3 |
| 41 | 41:77227385 | G/A | intergenic | NONE; NONE | NA; NA | FCR3 |
| 41 | 41:81787412 | A/G | intergenic | NONE; NONE | NA; NA | FCR3 |
| 41 | 41:82785087 | T/C | intergenic | NONE; NONE | NA; NA | FCR3 |
| 27 | 27:2560032 | G/A | intronic | ENSGALG00010024809 | KANSL1 | RFI1 |
| 31 | 31:2019899 | G/A | intronic | ENSGALG00010005410; ENSGALG00010005529 | NA; CHIR3B8 | RFI1 |
| 2 | 2:72477770 | G/C | intergenic | ENSGALG00010000898; ENSGALG00010000899 | NA; NA | RFI2 |
| 1 | 1:23290767 | G/A | intergenic | ENSGALG00010001979; ENSGALG00010001264 | NA; FAM3C | RFI3 |
| 1 | 1:23332705 | T/G | intergenic | ENSGALG00010001979; ENSGALG00010001264 | NA; FAM3C | RFI4, RFI5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Wang, Y.; Huang, Y.; Wang, Z.; Wang, Q.; Wang, H. Genetic Diversity Analysis and Identification of Candidate Genes for Growth Traits in Chengkou Mountain Chicken. Int. J. Mol. Sci. 2024, 25, 12939. https://doi.org/10.3390/ijms252312939
Liu L, Wang Y, Huang Y, Wang Z, Wang Q, Wang H. Genetic Diversity Analysis and Identification of Candidate Genes for Growth Traits in Chengkou Mountain Chicken. International Journal of Molecular Sciences. 2024; 25(23):12939. https://doi.org/10.3390/ijms252312939
Chicago/Turabian StyleLiu, Lingbin, Yi Wang, Yu Huang, Zhen Wang, Qigui Wang, and Haiwei Wang. 2024. "Genetic Diversity Analysis and Identification of Candidate Genes for Growth Traits in Chengkou Mountain Chicken" International Journal of Molecular Sciences 25, no. 23: 12939. https://doi.org/10.3390/ijms252312939
APA StyleLiu, L., Wang, Y., Huang, Y., Wang, Z., Wang, Q., & Wang, H. (2024). Genetic Diversity Analysis and Identification of Candidate Genes for Growth Traits in Chengkou Mountain Chicken. International Journal of Molecular Sciences, 25(23), 12939. https://doi.org/10.3390/ijms252312939





