Genome-Wide Identification of NLP Gene Families and Haplotype Analysis of SiNLP2 in Foxtail Millet (Setaria italica)
Abstract
1. Introduction
2. Results
2.1. Identification of SiNLP Gene Family Members
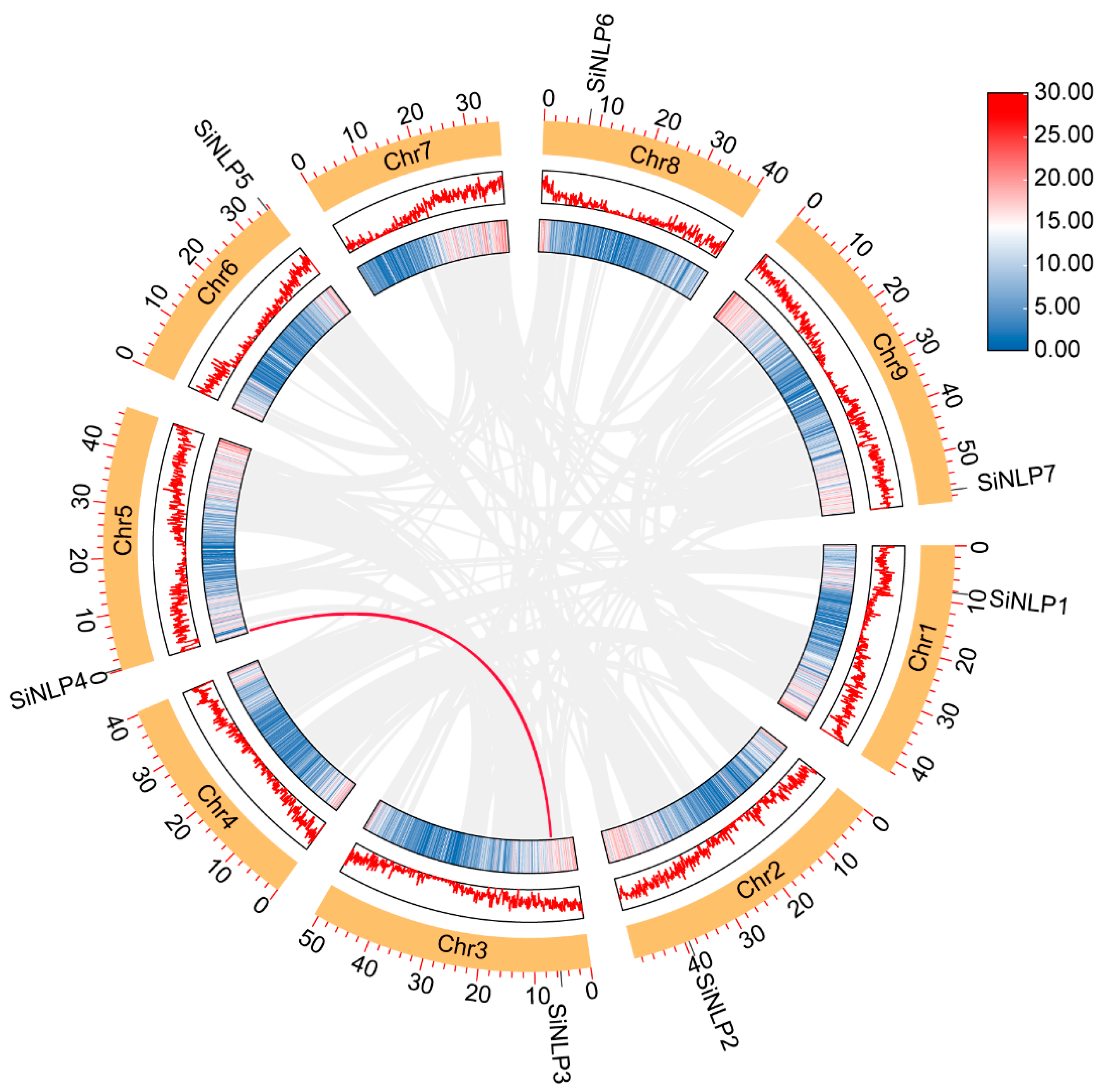
2.2. Characteristics and Chromosome Localization of SiNLP Gene Family Members
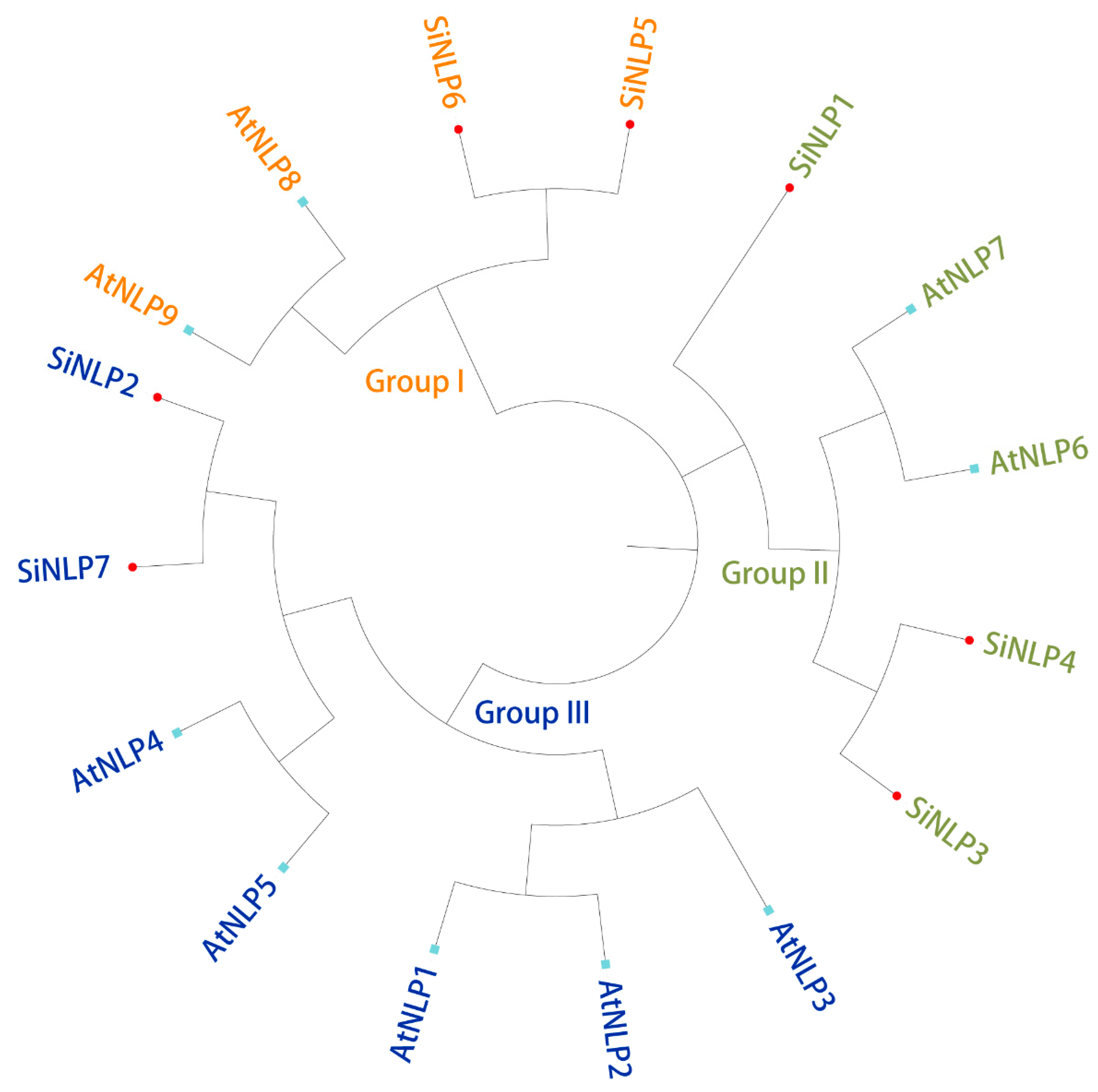
2.3. Phylogenetic and Sequence Analyses of the SiNLP Gene Family
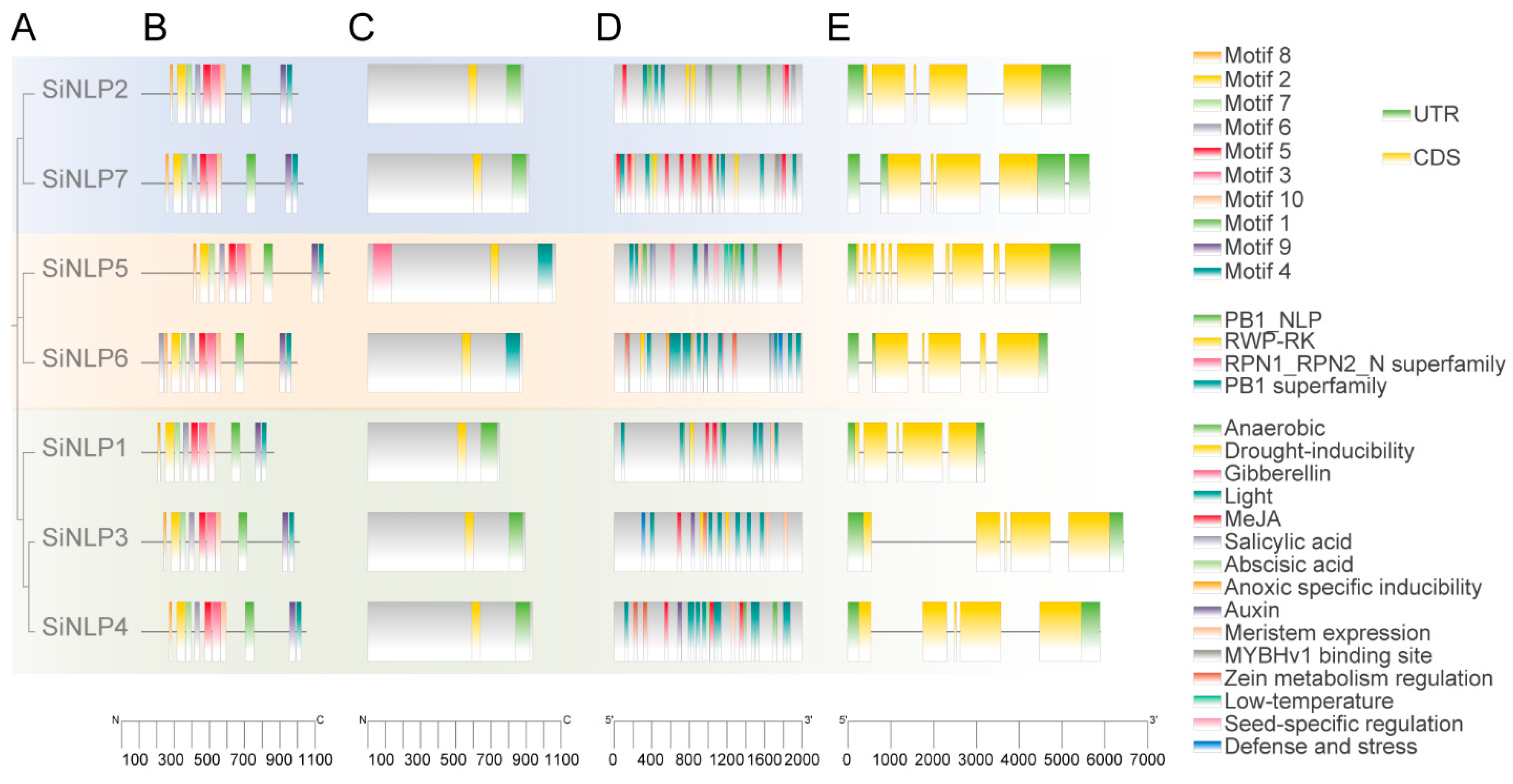
2.4. Analysis of Conserved Motifs, Domains, Gene Structure, and Cis-Acting Elements of the SiNLP Gene Family
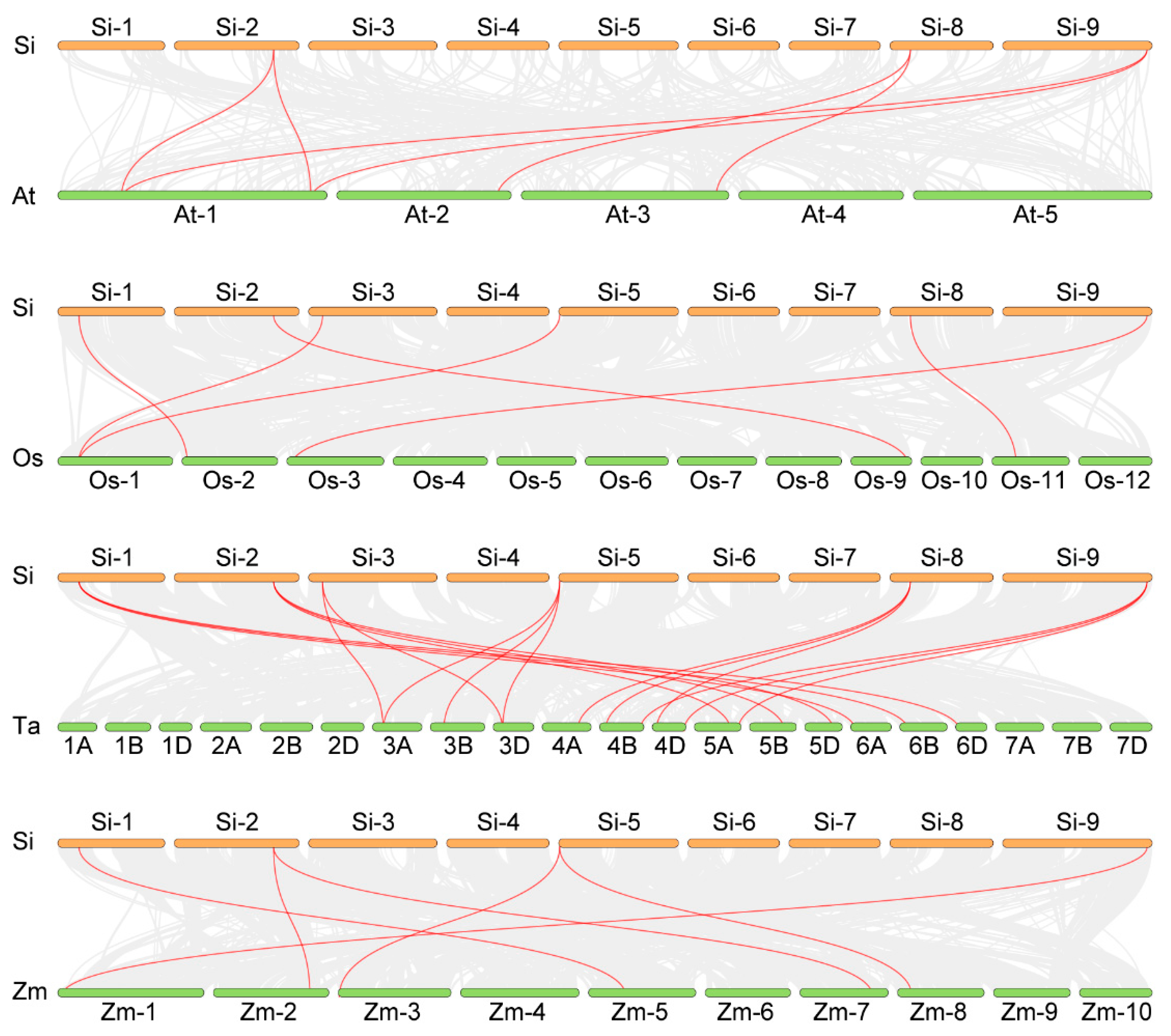
2.5. Collinearity Analysis of the NLP Gene Family
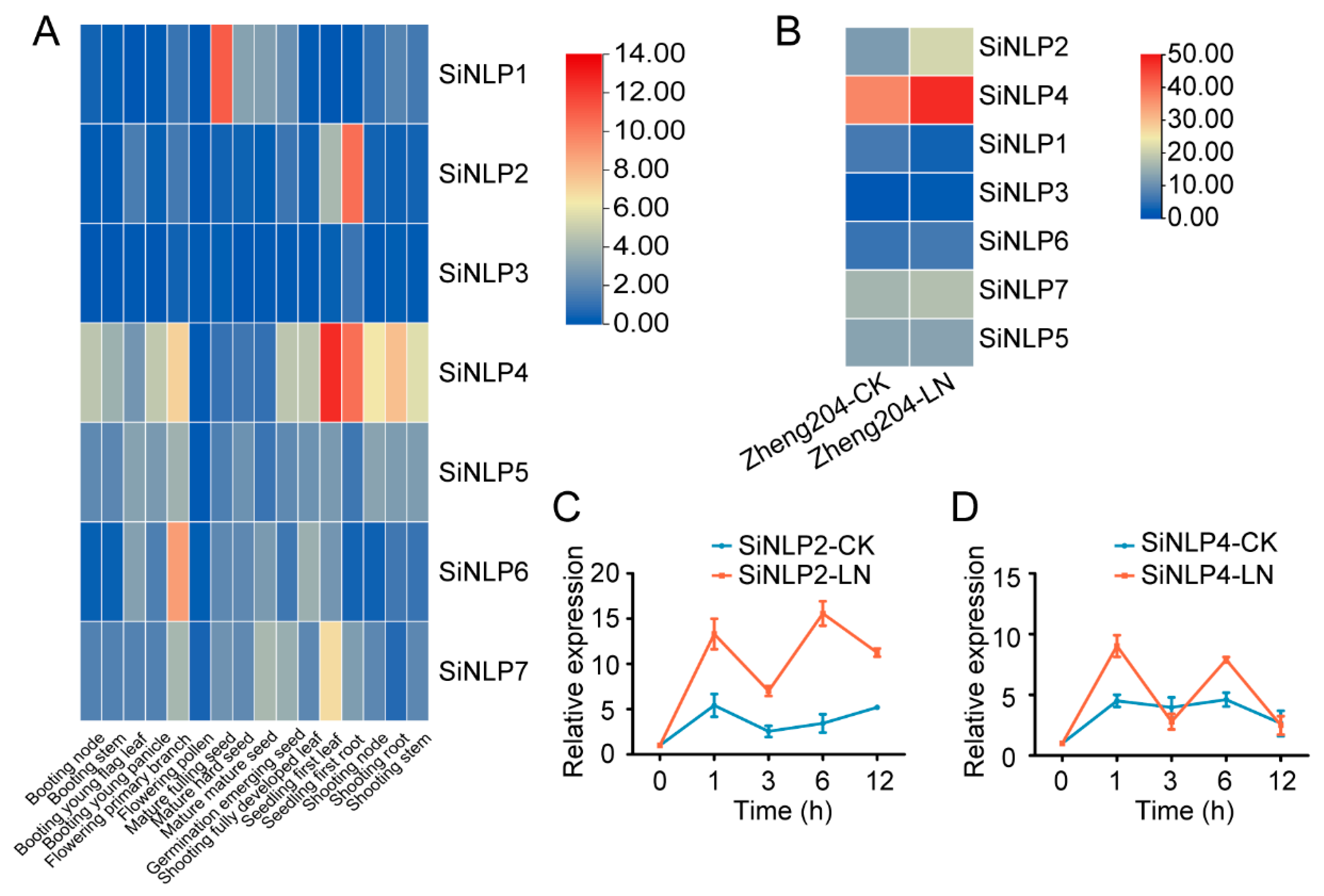
2.6. Expression Profile Analysis of the SiNLP Gene Family and qPCR of Candidate Genes
2.7. The DLR Analysis Between SiNLP2 and Nitrogen Pathway Genes
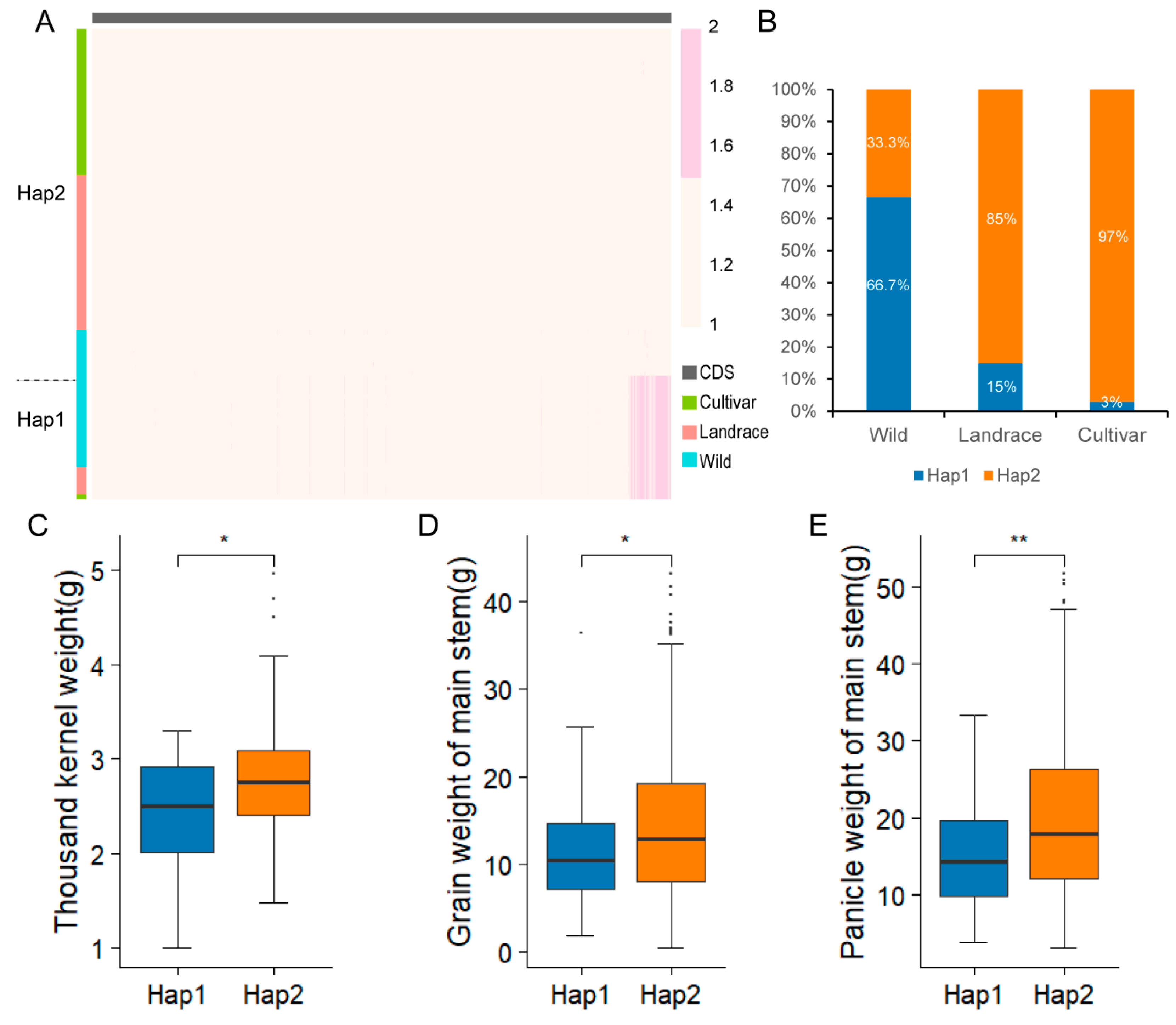
2.8. Haplotype Analysis of the Candidate Gene SiNLP2
3. Discussion
4. Materials and Methods
4.1. Identification and Physicochemical Properties of NLP Transcription Factor Family Members in Millet
4.2. Phylogenetic Analysis of the SiNLP Gene Family
4.3. Analysis of Conserved Motifs, Domains, Cis-Acting Regulatory Elements, and Gene Structure of the SiNLP Gene Family
4.4. Chromosome Localization and Collinearity Analysis of SiNLP Gene Family Members
4.5. Analysis of Expression Profiles of SiNLP Gene Family Members in Different Tissues and Under Low-Nitrogen Stress
4.6. RNA Extraction and RT-qPCR Analysis of Candidate Genes Under Low-Nitrogen Stress
4.7. DLR Assay
4.8. Haplotype Analysis of Candidate Genes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, X.; Cui, Z.; Fan, M.; Vitousek, P.; Zhao, M.; Ma, W.; Wang, Z.; Zhang, W.; Yan, X.; Yang, J.; et al. Producing more grain with lower environmental costs. Nature 2014, 514, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, H.; Tseng, C.S.; Tsay, Y.F. Improving nitrogen use efficiency by manipulating nitrate remobilization in plants. Nat. Plants 2020, 6, 1126–1135. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, K.; Song, W.; Zhong, N.; Wu, Y.; Fu, X. Improving crop nitrogen use efficiency toward sustainable green revolution. Annu. Rev. Plant Biol. 2022, 73, 523–551. [Google Scholar] [CrossRef]
- Crawford, N.M. Nitrate: Nutrient and signal for plant growth. Plant Cell 1995, 7, 859–868. [Google Scholar]
- Liu, K.H.; Tsay, Y.F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003, 22, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Orsel, M.; Krapp, A.; Daniel-Vedele, F. Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiol. 2002, 129, 886–896. [Google Scholar] [CrossRef]
- Krapp, A.; David, L.C.; Chardin, C.; Girin, T.; Marmagne, A.; Leprince, A.S.; Chaillou, S.; Ferrario-Méry, S.; Meyer, C.; Daniel-Vedele, F. Nitrate transport and signalling in Arabidopsis. J. Exp. Bot. 2014, 65, 789–798. [Google Scholar] [CrossRef]
- Chopin, F.; Wirth, J.; Dorbe, M.F.; Lejay, L.; Krapp, A.; Gojon, A.; Daniel-Vedele, F. The Arabidopsis nitrate transporter AtNRT2.1 is targeted to the root plasma membrane. Plant Physiol. Biochem. 2007, 45, 630–635. [Google Scholar] [CrossRef]
- Wirth, J.; Chopin, F.; Santoni, V.; Viennois, G.; Tillard, P.; Krapp, A.; Lejay, L.; Daniel-Vedele, F.; Gojon, A. Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J. Biol. Chem. 2007, 282, 23541–23552. [Google Scholar] [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS Box Gene That Controls Nutrient-Induced Changes in Root Architecture. Science 1998, 279, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty Years from Transport to Signaling Networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Moyano, T.C.; Riveras, E.; Contreras-López, O.; Gutiérrez, R.A. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 2013, 110, 12840–12845. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-López, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; Lejay, L.; et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014, 80, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef]
- Medici, A.; Marshall-Colon, A.; Ronzier, E.; Szponarski, W.; Wang, R.; Gojon, A.; Crawford, N.M.; Ruffel, S.; Coruzzi, G.M.; Krouk, G. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef]
- Xu, N.; Wang, R.; Zhao, L.; Zhang, C.; Li, Z.; Lei, Z.; Liu, F.; Guan, P.; Chu, Z.; Crawford, N.M.; et al. The Arabidopsis NRG2 Protein Mediates Nitrate Signaling and Interacts with and Regulates Key Nitrate Regulators. Plant Cell 2016, 28, 485–504. [Google Scholar] [CrossRef]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.P.; Daniel-Vedele, F.; Fernandez, E.; et al. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef]
- Wang, R.; Xing, X.; Wang, Y.; Tran, A.; Crawford, N.M. A Genetic Screen for Nitrate Regulatory Mutants Captures the Nitrate Transporter Gene NRT1.1. Plant Physiol. 2009, 151, 472–478. [Google Scholar] [CrossRef]
- Liu, K.H.; Liu, M.; Lin, Z.; Wang, Z.F.; Chen, B.; Liu, C.; Guo, A.; Konishi, M.; Yanagisawa, S.; Wagner, G.; et al. NIN-like protein 7 transcription factor is a plant nitrate sensor. Science 2022, 377, 1419–1425. [Google Scholar] [CrossRef]
- Ge, M.; Liu, Y.; Jiang, L.; Wang, Y.; Lv, Y.; Zhou, L.; Liang, S.; Bao, H.; Zhao, H. Genome-wide analysis of maize NLP transcription factor family revealed the roles in nitrogen response. Plant Growth Regul. 2018, 84, 95–105. [Google Scholar] [CrossRef]
- Chardin, C.; Girin, T.; Roudier, F.; Meyer, C.; Krapp, A. The plant RWP-RK transcription factors: Key regulators of nitrogen responses and of gametophyte development. J. Exp. Bot. 2014, 65, 5577–5587. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J. 2010, 63, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.S.; Lagarias, J.C. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 2007, 19, 2124–2139. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H.; Kamakura, S.; Ito, T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci. STKE 2007, 2007, re6. [Google Scholar] [CrossRef]
- Schauser, L.; Wieloch, W.; Stougaard, J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005, 60, 229–237. [Google Scholar] [CrossRef]
- Liu, M.; Zhi, X.; Wang, Y.; Wang, Y. Genome-wide survey and expression analysis of NIN-like Protein (NLP) genes reveals its potential roles in the response to nitrate signaling in tomato. BMC Plant Biol. 2021, 21, 347. [Google Scholar] [CrossRef]
- Kumar, A.; Batra, R.; Gahlaut, V.; Gautam, T.; Kumar, S.; Sharma, M.; Tyagi, S.; Singh, K.P.; Balyan, H.S.; Pandey, R.; et al. Genome-wide identification and characterization of gene family for RWP-RK transcription factors in wheat (Triticum aestivum L.). PLoS ONE 2018, 13, e0208409. [Google Scholar] [CrossRef]
- Liu, M.; Chang, W.; Fan, Y.; Sun, W.; Qu, C.; Zhang, K.; Liu, L.; Xu, X.; Tang, Z.; Li, J.; et al. Genome-wide identification and characterization of NODULE-INCEPTION-LIKE protein (NLP) family genes in Brassica napus. Int. J. Mol. Sci. 2018, 19, 2270. [Google Scholar] [CrossRef]
- Ding, G.; Xiao, G.; Zhu, L. Genome-Wide Identification and Expression Analysis of NLP (NIN-Like Protein) Transcription Factor Gene Family in Cotton. Sci. Agric. Sin. 2023, 56, 3723–3746. [Google Scholar]
- Wu, X.; Xu, Z.; Qu, C.; Li, W.; Sun, Q.; Liu, G. Genome-wide identification and characterization of NLP gene family in Populus trichocarpa. Bull. Bot. Res. 2014, 34, 37–43. (In Chinese) [Google Scholar]
- Yuan, T.; Zhu, C.; Li, Z.; Song, X.; Gao, Z. Identification of NLP transcription factors of Phyllostachys edulis and their expression patterns in response to nitrogen. For. Res. 2021, 34, 39–48. (In Chinese) [Google Scholar]
- Liu, K.H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate-CPK-NLP signalling in central nutrient-growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G. Nitrate signalling: Calcium bridges the nitrate gap. Nat. Plants 2017, 3, 17095. [Google Scholar] [CrossRef]
- Sato, T.; Maekawa, S.; Konishi, M.; Yoshioka, N.; Sasaki, Y.; Maeda, H.; Ishida, T.; Kato, Y.; Yamaguchi, J.; Yanagisawa, S. Direct transcriptional activation of BT genes by NLP transcription factors is a key component of the nitrate response in Arabidopsis. Biochem. Biophys. Res. Commun. 2017, 483, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.H.; Wu, J.; Tang, H.; Yuan, Y.; Wang, S.M.; Wang, Y.P.; Zhu, Q.S.; Li, S.G.; Xiang, C.B. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and -sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 2016, 6, 27795. [Google Scholar] [CrossRef]
- Yan, D.; Easwaran, V.; Chau, V.; Okamoto, M.; Ierullo, M.; Kimura, M.; Endo, A.; Yano, R.; Pasha, A.; Gong, Y.; et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat. Commun. 2016, 7, 13179. [Google Scholar] [CrossRef]
- Duermeyer, L.; Khodapanahi, E.; Yan, D.; Krapp, A.; Nambara, E. Regulation of seed dormancy and germination by nitrate. Seed Sci. Res. 2018, 28, 150–157. [Google Scholar] [CrossRef]
- Ge, M.; Wang, Y.; Liu, Y.; Jiang, L.; He, B.; Ning, L.; Du, H.; Lv, Y.; Zhou, L.; Lin, F.; et al. The NIN-like protein 5 (ZmNLP5) transcription factor is involved in modulating the nitrogen response in maize. Plant J. 2020, 102, 353–368. [Google Scholar] [CrossRef]
- Alfatih, A.; Wu, J.; Zhang, Z.S.; Xia, J.Q.; Jan, S.U.; Yu, L.H.; Xiang, C.B. Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 2020, 71, 6032–6042. [Google Scholar] [CrossRef]
- Jagadhesan, B.; Sathee, L.; Meena, H.S.; Jha, S.K.; Chinnusamy, V.; Kumar, A.; Kumar, S. Genome wide analysis of NLP transcription factors reveals their role in nitrogen stress tolerance of rice. Sci. Rep. 2020, 10, 9368. [Google Scholar] [CrossRef] [PubMed]
- Doust, A.N.; Kellogg, E.A.; Devos, K.M.; Bennetzen, J.L. Foxtail millet: A sequence-driven grass model system. Plant Physiol. 2009, 149, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Brutnell, T.P.; Wang, L.; Swartwood, K.; Goldschmidt, A.; Jackson, D.; Zhu, X.G.; Kellogg, E.; Van Eck, J. Setaria viridis: A model for C4 photosynthesis. Plant Cell 2010, 22, 2537–2544. [Google Scholar] [CrossRef]
- Peng, R.; Zhang, B. Foxtail Millet: A New Model for C4 Plants. Trends Plant Sci. 2021, 26, 199–201. [Google Scholar] [CrossRef]
- Yu, H.; Li, J.; Chang, X.; Dong, N.; Chen, B.; Wang, J.; Zha, L.; Gui, S. Genome-wide identification and expression profiling of the WRKY gene family reveals abiotic stress response mechanisms in Platycodon grandiflorus. Int. J. Biol. Macromol. 2024, 257, 128617. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Li, H.; Zhao, J.J.; Yang, P.; Xiang, X.; Wei, S.Y.; Wang, T.; Shi, Y.J.; Huang, J.; He, F. Genome-wide identification and characterization of the RZFP gene family and analysis of its expression pattern under stress in Populus trichocarpa. Int. J. Biol. Macromol. 2024, 255, 128108. [Google Scholar] [CrossRef]
- Xu, G.; Fan, X.; Miller, A.J. Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef]
- Wang, W.; Hu, B.; Yuan, D.; Liu, Y.; Che, R.; Hu, Y.; Ou, S.; Liu, Y.; Zhang, Z.; Wang, H.; et al. Expression of the Nitrate Transporter Gene OsNRT1.1A/OsNPF6.3 Confers High Yield and Early Maturation in Rice. Plant Cell 2018, 30, 638–651. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Huang, G.; Wu, Z.; Percy, R.G.; Bai, M.; Li, Y.; Frelichowski, J.E.; Hu, J.; Wang, K.; Yu, J.Z.; Zhu, Y. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 2020, 52, 516–524. [Google Scholar] [CrossRef]
- Soyano, T.; Shimoda, Y.; Hayashi, M. NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol. 2015, 56, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. The role of protein-protein interactions mediated by the PB1 domain of NLP transcription factors in nitrate-inducible gene expression. BMC Plant Biol. 2019, 19, 90. [Google Scholar] [CrossRef]
- Liu, J.; Bisseling, T. Evolution of NIN and NIN-like Genes in Relation to Nodule Symbiosis. Genes 2020, 11, 777. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Roudier, F.; Castaings, L.; Bréhaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Z.S.; Xia, J.Q.; Alfatih, A.; Song, Y.; Huang, Y.J.; Wan, G.Y.; Sun, L.Q.; Tang, H.; Liu, Y.; et al. Rice NIN-LIKE PROTEIN 4 plays a pivotal role in nitrogen use efficiency. Plant Biotechnol. J. 2021, 19, 448–461. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Hellens, R.P.; Allan, A.C.; Friel, E.N.; Bolitho, K.; Grafton, K.; Templeton, M.D.; Karunairetnam, S.; Gleave, A.P.; Laing, W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 2005, 1, 13. [Google Scholar] [CrossRef]
- He, Q.; Tang, S.; Zhi, H.; Chen, J.; Zhang, J.; Liang, H.; Alam, O.; Li, H.; Zhang, H.; Xing, L.; et al. A graph-based genome and pan-genome variation of the model plant Setaria. Nat. Genet. 2023, 55, 1232–1242. [Google Scholar] [CrossRef]



| Name | Gene ID | Protein Length (aa) | Molecular Weight (Da) | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular localization |
|---|---|---|---|---|---|---|---|---|
| SiNLP1 | Seita.1G094300.1 | 749 | 79,534.4 | 5.38 | 42.28 | 78.34 | −0.262 | Chloroplast |
| SiNLP2 | Seita.2G298700.1 | 886 | 96,994.84 | 6.29 | 47.06 | 73.97 | −0.413 | Nucleus |
| SiNLP3 | Seita.3G084600.1 | 894 | 98,567.06 | 6 | 44.32 | 84.36 | −0.264 | Nucleus |
| SiNLP4 | Seita.5G004100.1 | 935 | 102,492.49 | 5.75 | 49.19 | 78.3 | −0.36 | Nucleus |
| SiNLP5 | Seita.6G248300.1 | 1069 | 117,460.25 | 5.67 | 51.49 | 74.9 | −0.424 | Nucleus |
| SiNLP6 | Seita.8G074000.1 | 881 | 96,301.53 | 5.83 | 47.53 | 79.68 | −0.329 | Nucleus |
| SiNLP7 | Seita.9G553000.1 | 916 | 102,330.46 | 5.41 | 47.11 | 74.27 | −0.433 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bai, Y.; Wang, J.; Tang, W.; Sun, D.; Wang, S.; Chen, K.; Zhou, Y.; Wang, C.; Chen, J.; Xu, Z.; et al. Genome-Wide Identification of NLP Gene Families and Haplotype Analysis of SiNLP2 in Foxtail Millet (Setaria italica). Int. J. Mol. Sci. 2024, 25, 12938. https://doi.org/10.3390/ijms252312938
Bai Y, Wang J, Tang W, Sun D, Wang S, Chen K, Zhou Y, Wang C, Chen J, Xu Z, et al. Genome-Wide Identification of NLP Gene Families and Haplotype Analysis of SiNLP2 in Foxtail Millet (Setaria italica). International Journal of Molecular Sciences. 2024; 25(23):12938. https://doi.org/10.3390/ijms252312938
Chicago/Turabian StyleBai, Yanming, Juncheng Wang, Wensi Tang, Daizhen Sun, Shuguang Wang, Kai Chen, Yongbin Zhou, Chunxiao Wang, Jun Chen, Zhaoshi Xu, and et al. 2024. "Genome-Wide Identification of NLP Gene Families and Haplotype Analysis of SiNLP2 in Foxtail Millet (Setaria italica)" International Journal of Molecular Sciences 25, no. 23: 12938. https://doi.org/10.3390/ijms252312938
APA StyleBai, Y., Wang, J., Tang, W., Sun, D., Wang, S., Chen, K., Zhou, Y., Wang, C., Chen, J., Xu, Z., Chen, M., Wang, H., & Ma, Y. (2024). Genome-Wide Identification of NLP Gene Families and Haplotype Analysis of SiNLP2 in Foxtail Millet (Setaria italica). International Journal of Molecular Sciences, 25(23), 12938. https://doi.org/10.3390/ijms252312938








