Combined Metabolome and Transcriptome Analyses Reveals Anthocyanin Biosynthesis Profiles Between Purple and White Potatoes
Abstract
1. Introduction
2. Results
2.1. Transcriptome Profiling of the Petal Samples of Dark-Purple and White Potatoes
2.2. DEG Function Analysis
2.3. Major Transcription Factors Were Differentially Regulated Between Purple and White Potatoes
2.4. Metabolome Analysis
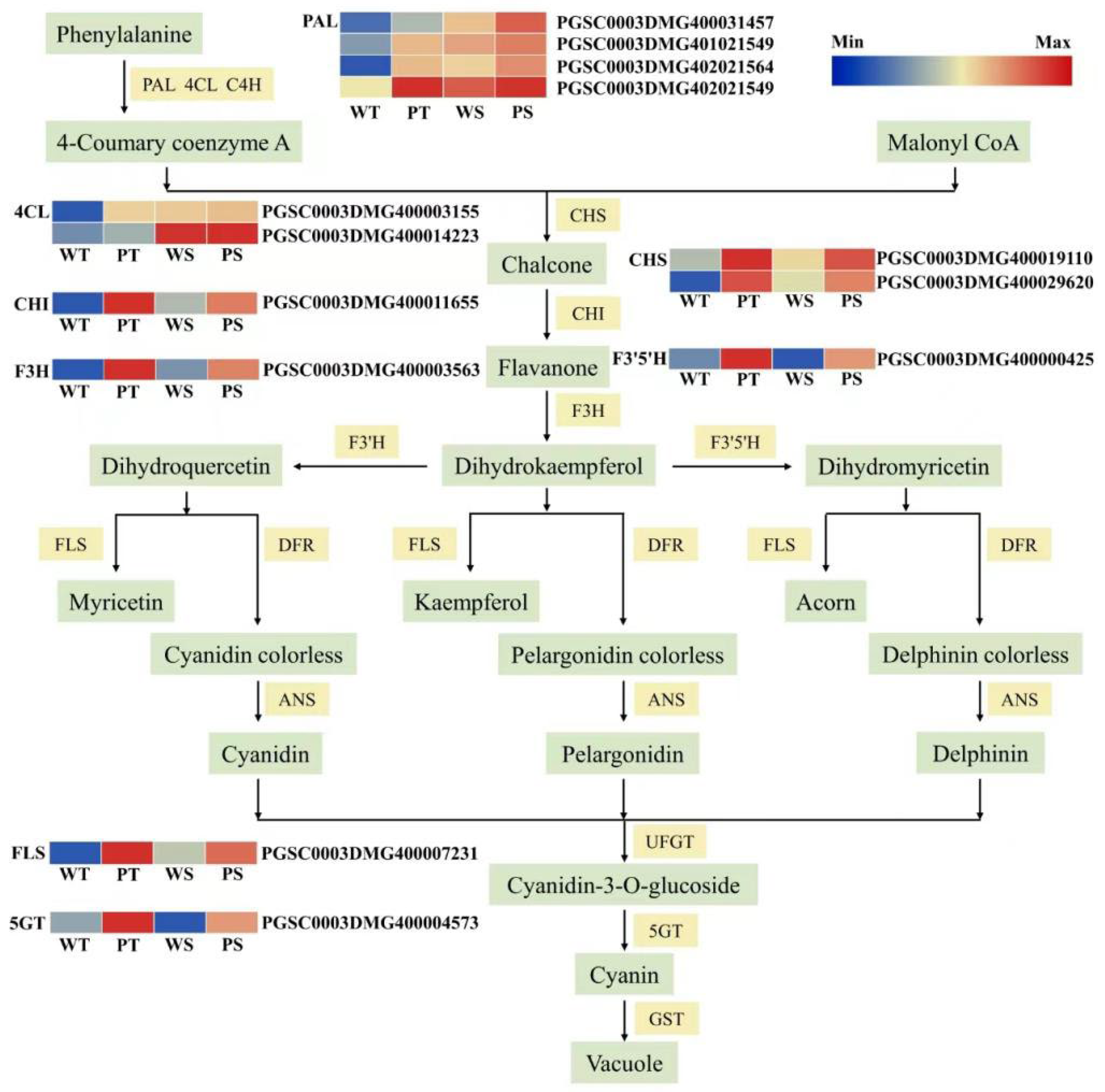
2.5. Differential Expression of Structural Genes Related to Anthocyanidin Biosynthetic Pathways in the Potato 15-12-16 and Xiazhai65
2.6. Integrated Analysis of the Transcriptome and Metabolome
2.7. Validation of the Expression of DEGs by qRT-PCR
2.8. Heterologous Expression of St5GT in Tobacco
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction, Library Construction, and Sequencing
4.3. RNA-Seq Analysis
4.4. The Method of Metabolome Analysis
4.5. Combined Transcriptome and Metabolome Analysis
4.6. Expression Validation by Quantitative Real-Time PCR Analysis
4.7. Tobacco Transformation Assays
4.8. Detection of Anthocyanins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, B. Research on Differential Accumulation and Light-Induced Accumulation of Anthocyanins in Potato Tubers; Huazhong Agricultural University: Wuhan, China, 2019. [Google Scholar]
- Reyes, L.F.; Miller, J.C.; Cisneros-Zevallos, L. Environmental Conditions Influence the Content and Yield of Anthocyanins and Total Phenolics in Purple- and Red-flesh Potatoes during Tuber Development. Am. J. Potato Res. 2004, 81, 187–193. [Google Scholar] [CrossRef]
- Han, K.H.; Shimada, K.; Sekikawa, M.; Fukushima, M. Anthocyanin-rich red potato flakes affect serum lipid peroxidation and hepatic sod mrna level in rats. Biosci. Biotechnol. Biochem. 2007, 71, 1356–1359. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Zheng, Y.L.; Lu, J.; Chen, G.Q.; Sun, Q.J. Purple sweet potato color suppresses lipopolysaccharide-induced acute inflammatory response in mouse brain. Neurochem. Int. 2010, 56, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Kim, S.J.; Shimada, K.I.; Hashimoto, N.; Yamauchi, H.; Fukushima, M. Purple potato flake reduces serum lipid profile in rats fed a cholesterol-rich diet. J. Funct. Foods 2013, 5, 974–980. [Google Scholar] [CrossRef]
- Ferrer, J.L.; Austin, M.B.; Stewart, C.; Noel, J.P. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol. Biochem. 2008, 46, 356–370. [Google Scholar] [CrossRef]
- Nasser, S.; Kemal, K.; Schenk, P.M. Global plant stress signaling: Reactive oxygen species at the cross-road. Front. Plant Sci. 2016, 7, 187. [Google Scholar] [CrossRef]
- Mishra, S.S.B. Recent updates on healthy phytoconstituents in potato: A nutritional depository. Potato Res. 2020, 63, 323–343. [Google Scholar] [CrossRef]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163. [Google Scholar] [CrossRef]
- Martin, C.; Gerats, T. The control of flower coloration. In The Molecular Biology of Flowering; Jordan, B.R., Ed.; CAB Intemational: Wallingford, UK, 1993; pp. 219–255. [Google Scholar]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120432. [Google Scholar] [CrossRef]
- Lu, Q.N. CDNA cloning and expression of anthocyanin biosynthetic genes in wild potato (solanum pinnatisectum). Afr. J. Biotechnol. 2006, 5, 811–818. [Google Scholar]
- Jung, C.S.; Griffiths, H.M.; Jong, D.M.D.; Cheng, S.; Bodis, M.; Kim, T.S.; Jong, W.S.D. The potato developer (d) locus encodes an r2r3 myb transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 2009, 120, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Gong, Y.; Jin, S.; Zhu, Q. Molecular analysis of a UDP-glucose: Flavonoid 3-O-glucosyltransferase (UFGT) gene from purple potato (Solanum tuberosum). Mol. Biol. Rep. 2011, 38, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Wang, Q.Y.; Feng, Z.H.; Wang, B.; Zhang, Y.F.; Yang, Q. Increased accumulation of anthocyanins in transgenic potato tubers by overexpressing the 3GT gene. Plant Biotechnol. Rep. 2012, 6, 69–75. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef]
- D’Amelia, V.; Aversano, R.; Batelli, G.; Caruso, I.; Castellano Moreno, M.; Castro-Sanz, A.B.; Chiaiese, P.; Fasano, C.; Palomba, F.; Carputo, D. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014, 80, 527–540. [Google Scholar] [CrossRef]
- Li, W.; Wang, B.; Wang, M.; Chen, M.; Yang, Q. Cloning and characterization of a potato StAN11 gene involved in anthocyanin biosynthesis regulation. J. Integr. Plant Biol. 2014, 56, 364–372. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Ence 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Chen, B.C.; Wu, X.J.; Dong, Q.J.; Xiao, J.P. Screening and functional analysis of StMYB transcription factors in pigmented potato under low-temperature treatment. BMC Genom. 2024, 25, 283. [Google Scholar] [CrossRef]
- Zhang, Y.; Pu, Y.; Zhang, Y.; Li, K.; Bai, S.; Wang, J.; Xu, M.; Liu, S.; Zhou, Z.; Wu, Y.; et al. Tuber transcriptome analysis reveals a novel WRKY transcription factor StWRKY70 potentially involved in potato pigmentation. Plant Physiol. Biochem. 2024, 213, 108792. [Google Scholar] [CrossRef]
- Stushnoff, C.; Ducreux, L.J.; Hancock, R.D.; Hedley, P.E.; Holm, D.G.; McDougall, G.J.; McNicol, J.W.; Morris, J.; Morris, W.L.; Sungurtas, J.A.; et al. Flavonoid profiling and transcriptome analysis reveals new gene–metabolite correlations in tubers of Solanum tuberosum L. Oxf. Univ. Press. 2010, 61, 1225–1238. [Google Scholar] [CrossRef]
- Liu, Y.; Lin-Wang, K.; Deng, C.; Warran, B.; Wang, L.; Yu, B.; Yang, H.; Wang, J.; Espley, R.V.; Zhang, J. Comparative transcriptome analysis of white and purple potato to identify genes involved in anthocyanin biosynthesis. PLoS ONE 2015, 10, e0129148. [Google Scholar] [CrossRef] [PubMed]
- Kyoungwon, C.; Kwang-Soo, C.; Hwang-Bae, S.; Jin, H.I.; Su-Young, H.; Hyerim, L.; Young-Mi, K.; Hee, N.M. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 2016, 67, 1519–1533. [Google Scholar] [CrossRef]
- Nie, T.K. Explore the Anthocyanin Synthesis Mechanism of Colored Potatoes Based on Full-Length Transcriptome Sequencing and Extensive Targeted Metabolome; Northwest A & F University: Yangling, China, 2020. [Google Scholar]
- Fan, Q.Q. Transcriptomics Study on Anthocyanin Anabolism of Colored Potato Tubers; Hunan Agricultural University: Changsha, China, 2019. [Google Scholar]
- Liu, F.; Yang, Y.; Gao, J.; Ma, C.; Bi, Y. A comparative transcriptome analysis of a wild purple potato and its red mutant provides insight into the mechanism of anthocyanin transformation. PLoS ONE 2018, 13, e0191406. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Zhao, L.; Wu, X.; Song, Z.; Li, Y. Metabolome and transcriptome analyses of the molecular mechanisms of flower color mutation in tobacco. BMC Genom. 2020, 21, 611. [Google Scholar] [CrossRef]
- Pengbo, X.; Christopher, Z.; Yang, L.; Jun, W.; Liancheng, L.; Zhongchi, L.; Run, L.; Hong, L. Transcriptome sequencing reveals role of light in promoting anthocyanin accumulation of strawberry fruit. Plant Growth Regul. 2018, 86, 121–132. [Google Scholar] [CrossRef]
- Tang, R.M.; Dong, H.T.; Wu, W.; Zhao, C.; Jia, X.; Yang, Q.; Zhang, J.; He, L.; Xie, H.; Wu, Z. A Comparative Transcriptome Analysis of Purple and Yellow Fleshed Potato Tubers Reveals Long Non-coding RNAs and their Targets Functioned in Anthocyanin Biosynthesis. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Holton, T.A.; Tanaka, Y. Blue roses: A pigment of our imagination? Trends Biotechnol. (Regul. Ed.) 1994, 12, 40–42. [Google Scholar] [CrossRef]
- Liu, F. Studies on the Transformation Mechanism of Different Anthocyanin glycosides in Potato Tubers; Shandong Normal University: Jinan, China, 2018. [Google Scholar]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef]
- Naing, A.H.; Park, K.I.; Ai, T.N.; Chung, M.Y.; Han, J.S.; Kang, Y.W.; Lim, K.B.; Kim, C.K. Overexpression of snapdragon Delila (Del) gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance. BMC Plant Biol. 2017, 17, 65. [Google Scholar] [CrossRef]
- Ju, Z.; Sun, W.; Meng, X.; Liang, L.; Li, Y.; Zhou, T.; Shen, H.; Xiang, G.; Wang, L. Isolation and functional characterization of two 5- o -glucosyltransferases related to anthocyanin biosynthesis from freesia hybrida. Plant Cell Tissue Organ. Cult. 2018, 135, 99–110. [Google Scholar] [CrossRef]
- Yamazaki, M.; Yamagishi, E.; Gong, Z.; Fukuchi-Mizutani, M.; Fukui, Y.; Tanaka, Y.; Kusumi, T.; Yamaguchi, M.; Saito, K. Two flavonoid glucosyltransferases from Petunia hybrida: Molecular cloning, biochemical properties and developmentally regulated expression. Plant Mol. Biol. 2002, 48, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Teusch, M.; Forkmann, G.; Seyffert, W. Genetic control of udp-glucose: Anthocyanin 5-o-glucosyltransferase from flowers of matthiola incana r.br. Planta 1986, 168, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Lorenc-Kukuła, K.; Jafra, S.; Oszmiański, J.; Szopa, J. Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J. Agric. Food Chem. 2005, 53, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.Y.; Zheng, G.; Zhu, S.; Qian, J.; Liang, L. Integrative Analysis of Metabolome and Transcriptome Reveals the Mechanism of Color Formation in Liriope spicata Fruit. Metabolites 2022, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Gisbert, C.; Dumm, J.M.; Prohens, J.; Vilanova, S.; Stommel, J.R. A Spontaneous eggplant (Solanum melongena L.) color mutant conditions anthocyanin-free fruit pigmentation. HortScience 2016, 51, 793–798. [Google Scholar] [CrossRef]
- Kiferle, C.; Fantini, E.; Bassolino, L.; Povero, G.; Spelt, C.; Buti, S.; Giuliano, G.; Quattrocchio, F.; Koes, R.; Perata, P.J.P.O. Tomato R2R3-MYB proteins SlANT1 and SlAN2: Same protein activity, different roles. PLoS ONE 2015, 10, e0136365. [Google Scholar] [CrossRef]
- Dasgupta, K.; Thilmony, R.; Stover, E.; Oliveira, M.L.; Thomson, J. Novel R2R3-MYB transcription factors from Prunus americana regulate differential patterns of anthocyanin accumulation in tobacco and citrus. GM Crops Food 2017, 8, 85–105. [Google Scholar] [CrossRef]
- Xie, Y.; Tan, H.; Ma, Z.; Huang, J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant. 2016, 9, 711–721. [Google Scholar] [CrossRef]
- Boase, M.R.; Brendolise, C.; Wang, L.; Ngo, H.; Espley, R.V.; Hellens, R.P.; Schwinn, K.E.; Davies, K.M.; Albert, N.W. Failure to launch: The self-regulating Md-MYB10 R6 gene from apple is active in flowers but not leaves of Petunia. Plant Cell Rep. 2015, 34, 1817–1823. [Google Scholar] [CrossRef]
- Lai, B.; Li, X.J.; Hu, B.; Qin, Y.H.; Huang, X.M.; Wang, H.C.; Hu, G.B. LcMYB1 is a key determinant of differential anthocyanin accumulation among genotypes, tissues, developmental phases and ABA and light stimuli in Litchi chinensis. PLoS ONE 2014, 9, e86293. [Google Scholar] [CrossRef]
- Allan, A.C.; Hellens, R.P.; Laing, W.A. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Kui, L.W.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Botany. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Lamb, D.C. Activation tagging identifies a conserved myb regulator of phenylpropanoid biosynthesis. Plant Cell 2001, 12, 2383–2394. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Qiu, J.; Sun, S.; Luo, S.; Zhang, J.; Xiao, X.; Zhang, L.; Wang, F.; Liu, S. Arabidopsis AtPAP1 transcription factor induces anthocyanin production in transgenic Taraxacum brevicorniculatum. Plant Cell Rep. 2014, 33, 669–680. [Google Scholar] [CrossRef]
- Payyavula, R.S.; Singh, R.K.; Navarre, D.A. Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 2013, 64, 5115–5131. [Google Scholar] [CrossRef]
- Cheng, Y.; Liu, L.; Yuan, C.; Guan, J. Molecular characterization of ethylene-regulated anthocyanin biosynthesis in plums during fruit ripening. Plant Mol. Biol. Rep. 2016, 34, 777–785. [Google Scholar] [CrossRef]
- Ishida, T.; Hattori, S.; Sano, R.; Inoue, K.; Shirano, Y.; Hayashi, H.; Shibata, D.; Sato, S.; Kato, T.; Tabata, S.; et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. Plant Cell 2007, 19, 2531–2543. [Google Scholar] [CrossRef]
- Mao, Z.; Jiang, H.; Wang, S.; Wang, Y.; Yu, L.; Zou, Q.; Liu, W.; Jiang, S.; Wang, N.; Zhang, Z.; et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021, 306, 110848. [Google Scholar] [CrossRef]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17056. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, U.; Sagasser, M.; Mehrtens, F.; Stracke, R.; Weisshaar, B. Differential combinatorial interactions of cisacting elements recognized by R2R3-MYB, BZIP, and BHLH factors control lightresponsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005, 57, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Liu, Z.; Wang, L.; Lin-Wang, K.; Zhu, J.; Bi, Z.; Sun, C.; Zhang, J.; Bai, J. Integrative analysis of metabolome and transcriptome reveals a dynamic regulatory network of potato tuber pigmentation. iScience 2022, 26, 105903. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.H.; Kim, H.S.; Choi, K.H.; Joung, Y.H.; Byun, S.M. Cloning and characterization of one member of the chalcone synthase gene family from Solanum tuberosum L. Biosci. Biotechnol. Biochem. 1996, 60, 1907–1910. [Google Scholar] [CrossRef]
- Jung, C.S.; Griffiths, H.M.; Jong, D.M.D.; Cheng, S.; Bodis, M.; Jong, W.S.D. The potato p locus codes for flavonoid 3′,5′-hydroxylase. Theor. Appl. Genet. 2005, 111, 269–275. [Google Scholar] [CrossRef]
- Lu, D.; Zhiqiang, H.; Di, L.; Pengfang, Z.; Shengjin, L.; Na, L.; Hongyu, M. Transcriptome analysis of chrysanthemum in responses to white rust. Sci. Hortic. 2018, 233, 421–430. [Google Scholar] [CrossRef]
- Bolser, D.M.; Staines, D.; Perry, E.; Kersey, P.J. Ensembl Plants: Integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2017, 1533, 1–31. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Perez-Rodriguez, P.; Riano-Pachon, D.M.; Correa, L.G.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gong, L.; Guo, Z.L.; Wang, W.S.; Zhang, H.Y.; Liu, X.Q.; Yu, S.B.; Xiong, L.Z.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-Time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tan, L.; Li, Y.L.; Gao, X.Z.; Zhao, J.; Wang, T.; Jiang, G.J.; Wang, H.L.; Wang, H. Determination of anthocyanidins in different plant origin foods by improved PH differential method. Food Ferment. Ind. 2022, 48, 276–285. [Google Scholar]
- Mehrtens, F.; Kranz, H.; Bednarek, P.; Weisshaar, B. The arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005, 138, 1083–1096. [Google Scholar] [CrossRef]








| Sample | Raw Reads | Clean Reads | Clean Base (G) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| PS-1 | 48860478 | 47610640 | 7.14 | 0.02 | 98.27 | 94.65 | 42.18 |
| PS-2 | 45029580 | 44036178 | 6.61 | 0.02 | 98.27 | 94.7 | 42.47 |
| PS-3 | 55048496 | 53272348 | 7.99 | 0.02 | 98.23 | 94.61 | 43.43 |
| PT-1 | 43451020 | 41554096 | 6.23 | 0.02 | 98.32 | 94.78 | 42.8 |
| PT-2 | 46305534 | 44798410 | 6.72 | 0.02 | 98.51 | 95.13 | 42.18 |
| PT-3 | 44340698 | 42478042 | 6.37 | 0.02 | 98.36 | 94.84 | 42.02 |
| WS-1 | 45139190 | 43512134 | 6.53 | 0.02 | 98.29 | 94.64 | 41.23 |
| WS-2 | 50208396 | 48873826 | 7.33 | 0.02 | 98.38 | 94.91 | 41.45 |
| WS-3 | 43767374 | 41440758 | 6.22 | 0.02 | 98.35 | 94.91 | 41.59 |
| WT-1 | 43302236 | 41430580 | 6.21 | 0.02 | 98.4 | 95.01 | 42.09 |
| WT-3 | 46903456 | 45435876 | 6.82 | 0.02 | 98.38 | 94.93 | 42.15 |
| Compounds | RT/min | M/Z | PS | PT | WS | WT |
|---|---|---|---|---|---|---|
| Cyanidin 3,5-O-diglucoside | 4.28 | 611.2 | 0.0168 ± 0.0013 a | 0.0162 ± 0.0015 a | N/A | N/A |
| Cyanidin 3-O-arabinoside | 6.17 | 419.1 | N/A | 0.0024 ± 0.0012 a | N/A | N/A |
| Cyanidin 3-O-galactoside | 5.18 | 449.1 | 0.0259 ± 0.0033 a | 0.0276 ± 0.0059 a | 0.0130 ± 0.0089 a | 0.0151 ± 0.0105 a |
| Cyanidin 3-O-glucoside | 5.75 | 449.1 | 0.0061 ± 0.0017 b | 0.0800 ± 0.0087 a | 0.0054 ± 0.0044 b | N/A |
| Cyanidin 3-O-rutinoside | 6.36 | 595.17 | 0.5193 ± 0.2638 b | 5.4433 ± 0.9150 a | 0.0068 ± 0.0097 b | N/A |
| Cyanidin 3-O-sophoroside | 5.15 | 611.2 | N/A | 0.0101 ± 0.0020 a | N/A | N/A |
| Cyanidin-3-O-(coumaryl)-glucoside | 11.24 | 595.15 | N/A | 0.0056 ± 0.0013 a | N/A | N/A |
| Cyanidin-3-O-xyloside | 7.9 | 419.1 | N/A | 0.0002 ± 0.0001 a | N/A | N/A |
| Peonidin 3-O-glucoside | 7.44 | 463.3 | 0.0669 ± 0.0065 a | 0.0073 ± 0.0032 c | 0.0337 ± 0.0119 b | N/A |
| Peonidin 3,5-O-diglucoside | 5.66 | 625.2 | 0.1307 ± 0.0240 a | 0.0325 ± 0.0024 b | N/A | N/A |
| Peonidin 3-O-rutinoside | 7.98 | 609.5 | 0.6447 ± 0.2984 a | 0.7067 ± 0.0379 a | N/A | N/A |
| Peonidin-3-(caffeoyl-glucosylglucoside)-5-glucoside | 10.25 | 949.26 | 0.0119 ± 0.0012 a | N/A | N/A | N/A |
| Delphinidin 3-O-arabinoside | 5.26 | 435.5 | N/A | 0.0007 ± 0.0005 a | N/A | N/A |
| Delphinidin 3-O-glucoside | 4.73 | 465.1 | 0.0447 ± 0.0204 b | 1.2633 ± 0.0379 a | 0.0360 ± 0.0330 b | N/A |
| Delphinidin 3-O-rutinoside | 5.3 | 611.1 | 0.0865 ± 0.0126 b | 10.9133 ± 0.9801 a | N/A | N/A |
| Delphinidin 3-O-sophoroside | 4.34 | 627.15 | 0.0089 ± 0.0012 b | N/A | 0.0177 ± 0.0026 a | N/A |
| Delphinidin-3-O-(6-O-acetyl)-glucoside | 9.02 | 507.11 | 0.0386 ± 0.0228 a | N/A | N/A | N/A |
| Delphinidin-3-O-(coumaryl)-glucoside | 10.43 | 611.14 | N/A | 0.0109 ± 0.0032 a | N/A | N/A |
| Pelargonidin 3-O-glucoside | 6.72 | 433.2 | N/A | 0.0065 ± 0.0012 a | N/A | N/A |
| Pelargonidin 3-O-rutinoside | 7.39 | 579.06 | 0.1957 ± 0.0885 b | 0.9173 ± 0.0386 a | 0.0210 ± 0.0254 c | N/A |
| Pelargonidin 3-O-sambubioside | 6.98 | 565.2 | N/A | 0.0033 ± 0.0008 a | N/A | N/A |
| Pelargonidin-3-O-5-O-(6-O-coumaryl)-diglucoside | 11.05 | 741.2 | 0.0040 ± 0.0032 b | 0.0160 ± 0.0025 a | N/A | N/A |
| Quercetin 3-O-glucoside | 8.91 | 465.1 | 0.2983 ± 0.0480 a | N/A | N/A | 0.0073 ± 0.0018 b |
| Rutin | 8.97 | 611.2 | 0.4180 ± 0.1113 a | 0.3663 ± 0.1190 a | N/A | N/A |
| Dihydromyricetin | 3.6 | 321.1 | N/A | 0.1697 ± 0.0091 a | N/A | N/A |
| Kaempferol-3-O-rutinoside | 10.67 | 595.2 | 0.6143 ± 0.0593 a | 0.1647 ± 0.0471 b | 0.0327 ± 0.0390 c | N/A |
| Malvidin 3,5-diglucoside | 6.02 | 655.4 | 0.4383 ± 0.1222 a | 0.0245 ± 0.0081 b | N/A | N/A |
| Malvidin 3-O-glucoside | 7.96 | 493.1 | 0.9527 ± 0.7722 a | 0.0099 ± 0.0054 a | 0.5053 ± 0.4425 a | N/A |
| Malvidin 3-O-rutinoside | 8.39 | 639.06 | 0.7993 ± 0.3656 a | 0.2690 ± 0.0193 b | N/A | N/A |
| Petunidin 3,5-diglucoside | 4.82 | 641.2 | 0.4970 ± 0.1206 a | 0.5580 ± 0.0520 a | N/A | N/A |
| Petunidin 3-O-arabinoside | 6.99 | 449.1 | N/A | 0.0060 ± 0.0024 a | N/A | N/A |
| Petunidin 3-O-glucoside | 6.5 | 479.1 | 0.1437 ± 0.0244 a | 0.0793 ± 0.0155 b | 0.0323 ± 0.0339 bc | N/A |
| Petunidin 3-O-rutinoside | 7 | 625.06 | 1.9733 ± 0.3050 b | 15.1333 ± 0.5132 a | N/A | N/A |
| Petunidin 3-O-sophoroside | 5.88 | 641.11 | N/A | 0.4613 ± 0.0268 a | N/A | N/A |
| Gene ID | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PGSC0003DMG400004573 qSt5GT | TGGATGGAGGGGTAAAAGGGGAAG | ACCTTCTTTCACAGCTTCTCTAGCC |
| PGSC0003DMG400004573 attB-St5GT | AAAAAGCAGGCTTCATGGTGAAGCCTCATGTTAT | AGAAAGCTGGGTCTCAATAACCTTTGGCAATTTCTT |
| GenBank No. X83206 | AGATGCTTACGCTGGATGGAATGC | TTCCGGTGTGGTTGGATTCTGTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, M.; Ma, X.; Zhou, Y.; Wang, F.; Fang, G.; Wang, J. Combined Metabolome and Transcriptome Analyses Reveals Anthocyanin Biosynthesis Profiles Between Purple and White Potatoes. Int. J. Mol. Sci. 2024, 25, 12884. https://doi.org/10.3390/ijms252312884
He M, Ma X, Zhou Y, Wang F, Fang G, Wang J. Combined Metabolome and Transcriptome Analyses Reveals Anthocyanin Biosynthesis Profiles Between Purple and White Potatoes. International Journal of Molecular Sciences. 2024; 25(23):12884. https://doi.org/10.3390/ijms252312884
Chicago/Turabian StyleHe, Miaomiao, Xinping Ma, Yun Zhou, Fang Wang, Guonan Fang, and Jian Wang. 2024. "Combined Metabolome and Transcriptome Analyses Reveals Anthocyanin Biosynthesis Profiles Between Purple and White Potatoes" International Journal of Molecular Sciences 25, no. 23: 12884. https://doi.org/10.3390/ijms252312884
APA StyleHe, M., Ma, X., Zhou, Y., Wang, F., Fang, G., & Wang, J. (2024). Combined Metabolome and Transcriptome Analyses Reveals Anthocyanin Biosynthesis Profiles Between Purple and White Potatoes. International Journal of Molecular Sciences, 25(23), 12884. https://doi.org/10.3390/ijms252312884





