Characteristics and Genomic Localization of Nurse Shark (Ginglymostoma cirratum) IgNAR
Abstract
1. Introduction
2. Results
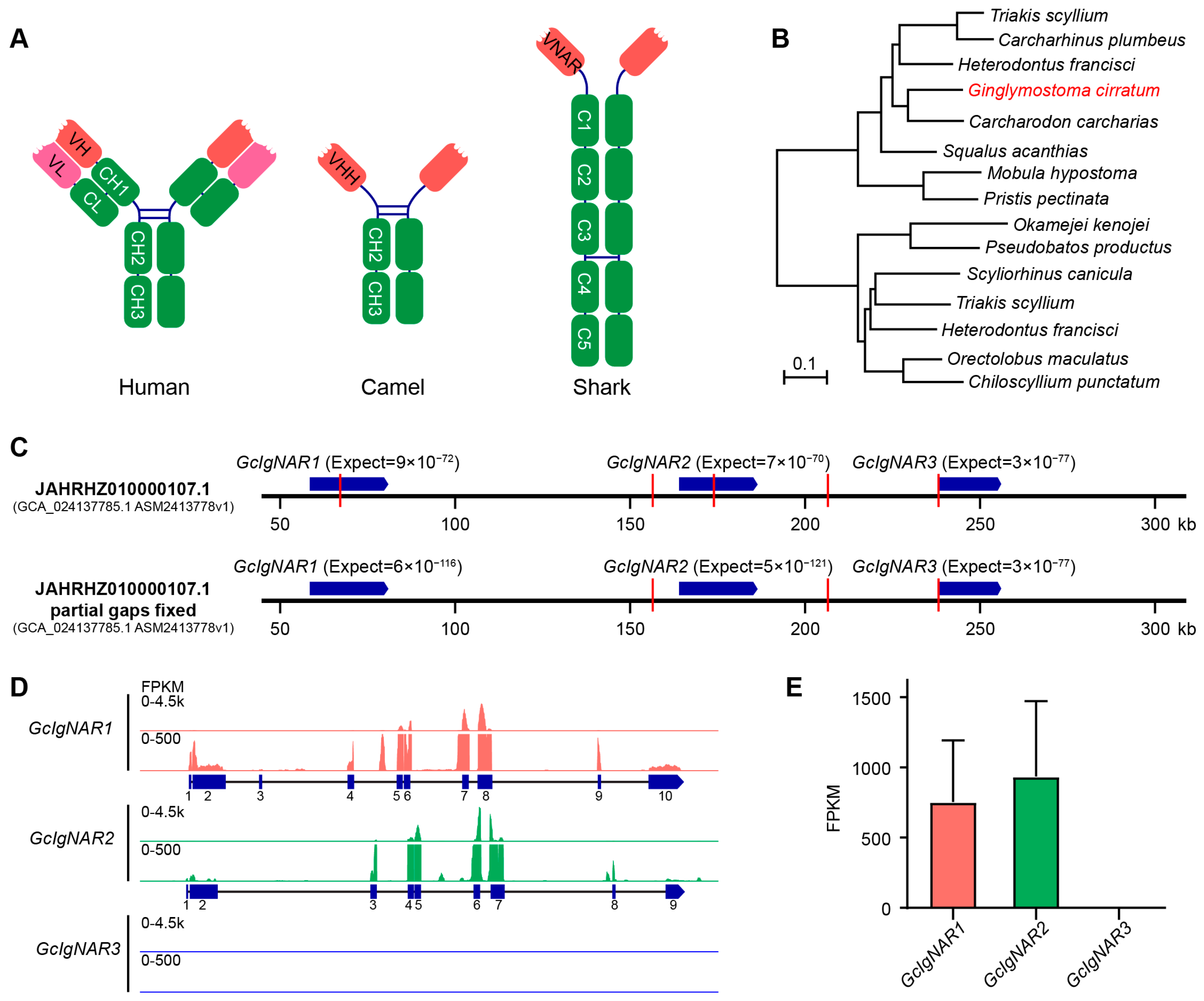
2.1. Three IgNAR Loci Discovered in Nurse Shark Genome Scaffold JAHRHZ010000107.1, with Only Two Expressed
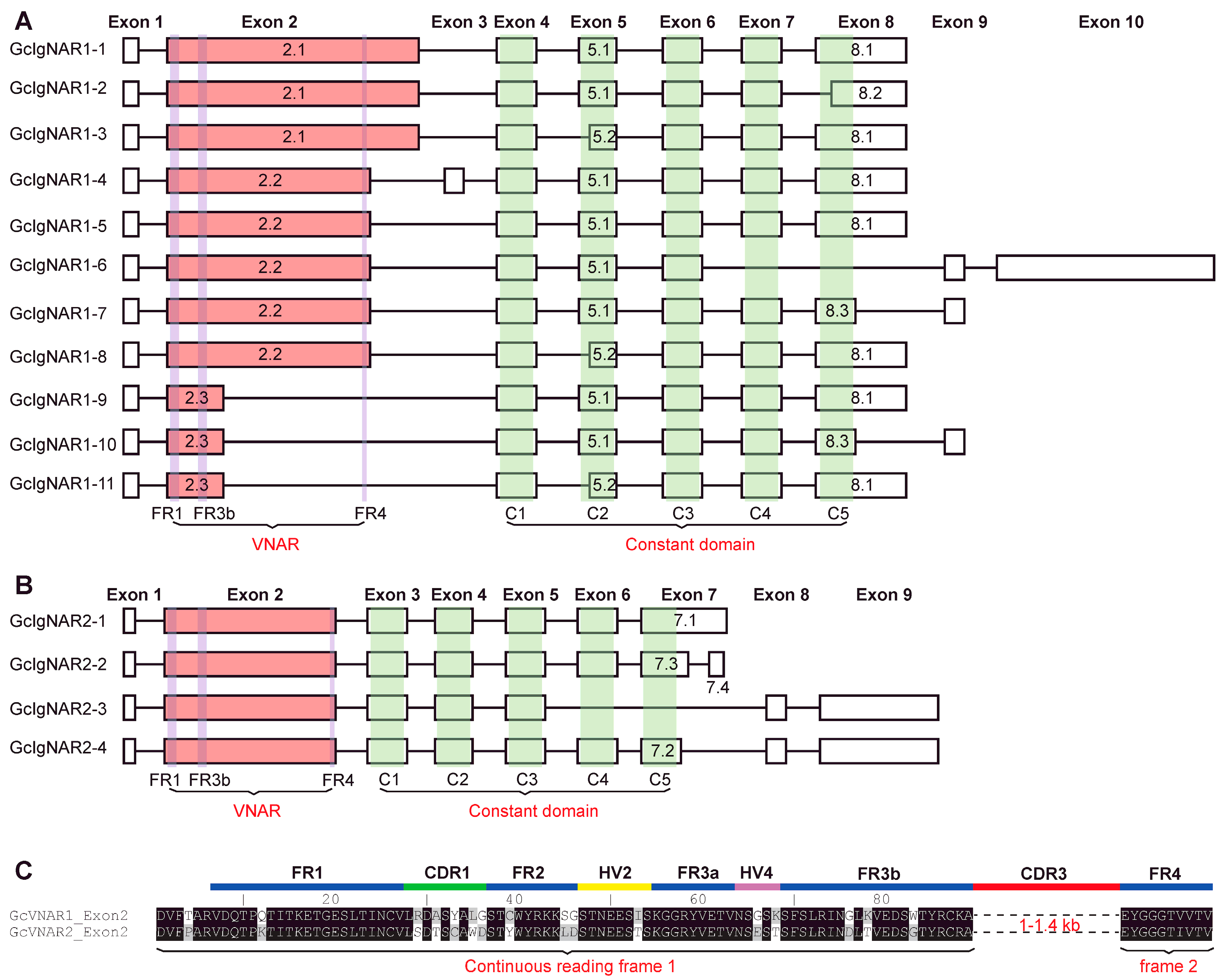
2.2. GcIgNAR1 and GcIgNAR2 Transcripts Respectively Exhibit at Least Eleven and Four Different Isoforms
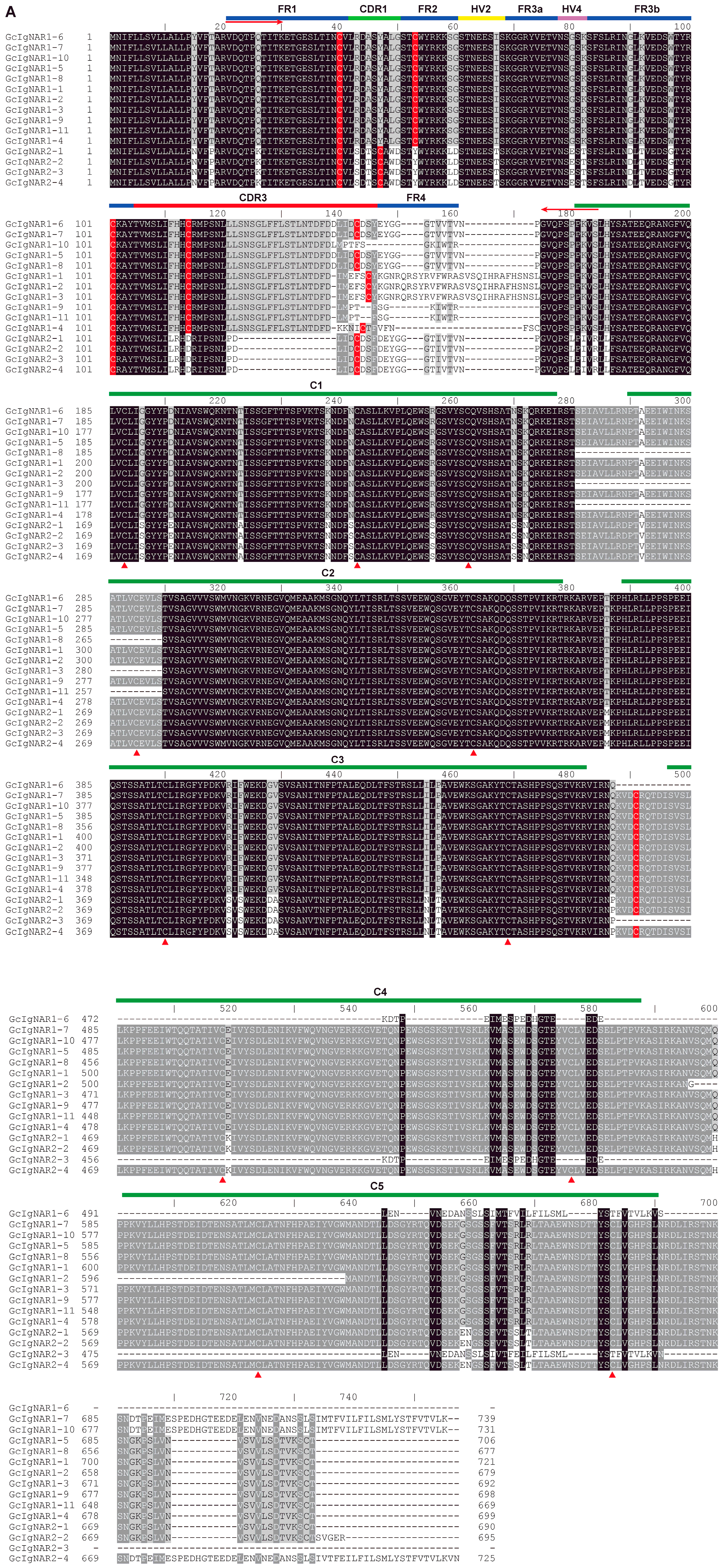
2.3. The Characteristics of GcIgNAR1 and GcIgNAR2 Isoforms
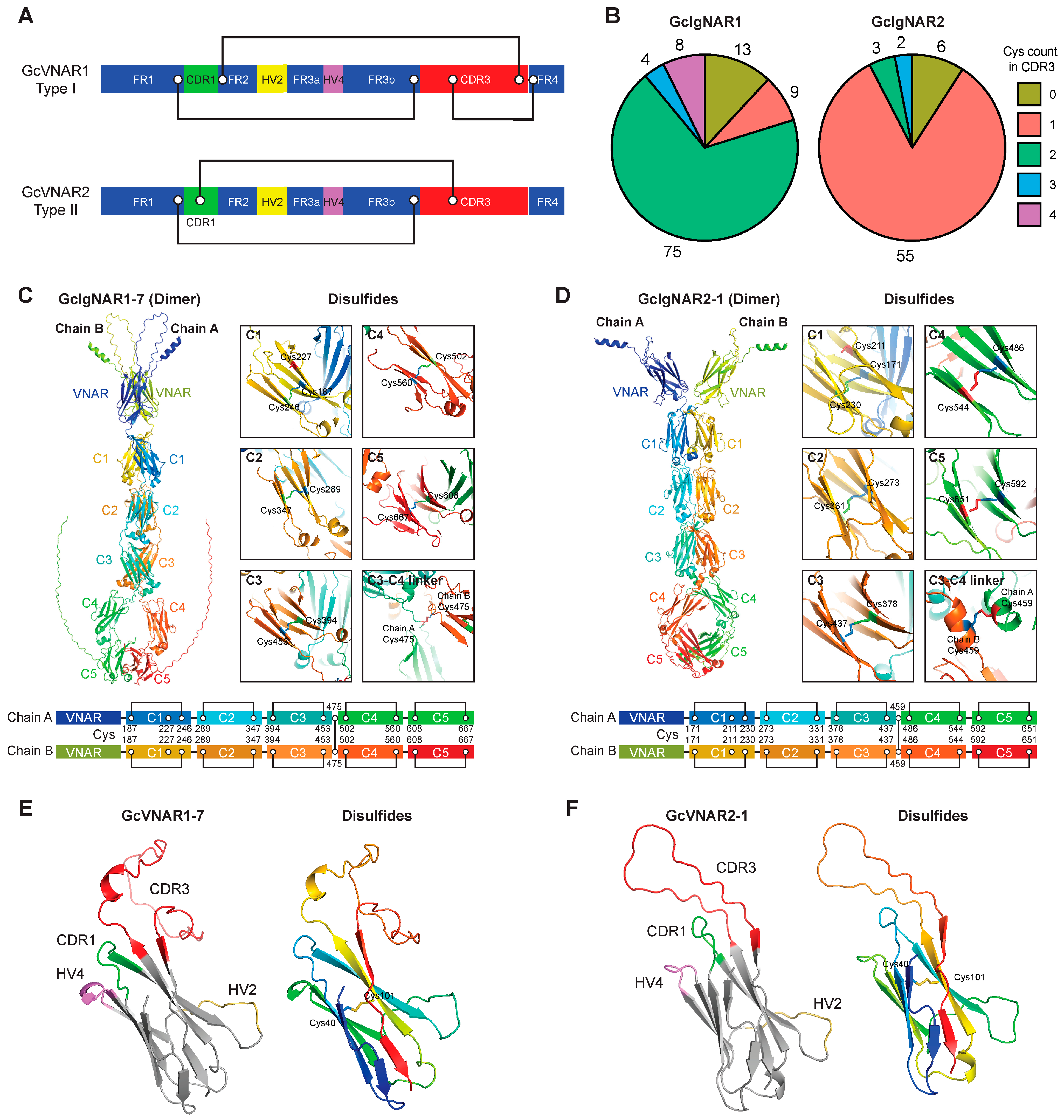
2.4. GcIgNAR1 and GcIgNAR2 Belong to Type I and Type II IgNAR, Respectively
3. Discussion
4. Materials and Methods
4.1. Whole Genome Sequencing Data Analysis and Missing Regions Fixation
4.2. RNA-Seq Data Analysis and Exon Extraction
4.3. Protein Structure Prediction Using AlphaFold3
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schroeder, H.W., Jr.; Cavacini, L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010, 125 (Suppl. S2), S41–S52. [Google Scholar] [CrossRef] [PubMed]
- Dudley, D.J. The immune system in health and disease. Bailliere’s Clin. Obstet. Gynaecol. 1992, 6, 393–416. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. B cells and antibodies. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; Volume 24, pp. 3486–3494. [Google Scholar]
- Kavathas, P.B.; Krause, P.J.; Ruddle, N.H. Adaptive immunity: Antigen recognition by T and B lymphocytes. In Immunoepidemiology; Springer: Cham, Switzerland, 2019; pp. 55–74. [Google Scholar]
- Rees, A.R. Understanding the human antibody repertoire. mAbs 2020, 12, e1729683-4. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, G.; Ippolito, G.C.; Beausang, J.; Busse, C.E.; Wardemann, H.; Quake, S.R. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat. Biotechnol. 2014, 32, 158–168. [Google Scholar] [CrossRef] [PubMed]
- de Los Rios, M.; Criscitiello, M.F.; Smider, V.V. Structural and genetic diversity in antibody repertoires from diverse species. Curr. Opin. Struct. Biol. 2015, 33, 27–41. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Lefranc, G. Immunoglobulins or antibodies: IMGT® bridging genes, structures and functions. Biomedicines 2020, 8, 319. [Google Scholar] [CrossRef]
- Pan, S.; Manabe, N.; Yamaguchi, Y. 3D structures of IgA, IgM, and components. Int. J. Mol. Sci. 2021, 22, 12776. [Google Scholar] [CrossRef]
- Oreste, U.; Ametrano, A.; Coscia, M.R. On origin and evolution of the antibody molecule. Biology 2021, 10, 140. [Google Scholar] [CrossRef]
- Zavrtanik, U.; Lukan, J.; Loris, R.; Lah, J.; Hadži, S. Structural basis of epitope recognition by heavy-chain camelid antibodies. J. Mol. Biol. 2018, 430, 4369–4386. [Google Scholar] [CrossRef]
- Arbabi-Ghahroudi, M. Camelid single-domain antibodies: Promises and challenges as lifesaving treatments. Int. J. Mol. Sci. 2022, 23, 5009. [Google Scholar] [CrossRef]
- Wilkes, J. Synthetic Biology Applications of Single Domain Antibodies. Ph.D. Thesis, The University of Manchester, Manchester, UK, 2019. [Google Scholar]
- Könning, D.; Zielonka, S.; Grzeschik, J.; Empting, M.; Valldorf, B.; Krah, S.; Schröter, C.; Sellmann, C.; Hock, B.; Kolmar, H. Camelid and shark single domain antibodies: Structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2017, 45, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.K.; Desmyter, A.; Muyldermans, S. Functional heavy-chain antibodies in Camelidae. In Advances in Immunology; Elsevier: Amsterdam, The Netherlands, 2001; Volume 79, pp. 261–296. [Google Scholar]
- Vincke, C.; Muyldermans, S. Introduction to heavy chain antibodies and derived Nanobodies. In Single Domain Antibodies: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2012; pp. 15–26. [Google Scholar]
- Bannas, P.; Hambach, J.; Koch-Nolte, F. Nanobodies and nanobody-based human heavy chain antibodies as antitumor therapeutics. Front. Immunol. 2017, 8, 1603. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Song, H.; Chen, Q.; Yu, J.; Xian, M.; Nian, R.; Feng, D. Recent advances in the selection and identification of antigen-specific nanobodies. Mol. Immunol. 2018, 96, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Aristiguieta, A.C.A. Nanobodies targeting SARS-CoV-2. In Biomedical Innovations to Combat COVID-19; Elsevier: Amsterdam, The Netherlands, 2022; pp. 231–240. [Google Scholar]
- Wang, Y.; Fan, Z.; Shao, L.; Kong, X.; Hou, X.; Tian, D.; Sun, Y.; Xiao, Y.; Yu, L. Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. Int. J. Nanomed. 2016, 11, 3287–3303. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Sapienza, G.; Rossotti, M.A.; Rosa, S.T.-D. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front. Immunol. 2017, 8, 977. [Google Scholar] [CrossRef]
- Matz, H.; Dooley, H. Shark IgNAR-derived binding domains as potential diagnostic and therapeutic agents. Dev. Comp. Immunol. 2019, 90, 100–107. [Google Scholar] [CrossRef]
- Zielonka, S.; Empting, M.; Grzeschik, J.; Könning, D.; Barelle, C.J.; Kolmar, H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. mAbs 2015, 7, 15–25. [Google Scholar] [CrossRef]
- Juma, S.N.; Gong, X.; Hu, S.; Lv, Z.; Shao, J.; Liu, L.; Chen, G. Shark new antigen receptor (IgNAR): Structure, characteristics and potential biomedical applications. Cells 2021, 10, 1140. [Google Scholar] [CrossRef]
- Khalid, Z.; Chen, Y.; Yu, D.; Abbas, M.; Huan, M.; Naz, Z.; Mengist, H.M.; Cao, M.-J.; Jin, T. IgNAR antibody: Structural features, diversity and applications. Fish Shellfish. Immunol. 2022, 121, 467–477. [Google Scholar] [CrossRef]
- Zielonka, S.; Weber, N.; Becker, S.; Doerner, A.; Christmann, A.; Christmann, C.; Uth, C.; Fritz, J.; Schäfer, E.; Steinmann, B. Shark attack: High affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J. Biotechnol. 2014, 191, 236–245. [Google Scholar] [CrossRef]
- Li, D.; English, H.; Hong, J.; Liang, T.; Merlino, G.; Day, C.-P.; Ho, M. A novel PD-L1-targeted shark VNAR single-domain-based CAR-T cell strategy for treating breast cancer and liver cancer. Mol. Ther.-Oncolytics 2022, 24, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Xi, X.; Wang, Y.; An, G.; Feng, S.; Zhu, Q.; Wu, Z.; Chen, J.; Zuo, Z.; Wang, Q.; Wang, M.-W. A novel shark VNAR antibody-based immunotoxin targeting TROP-2 for cancer therapy. Acta Pharm. Sin. B, 2024; in press. [Google Scholar] [CrossRef]
- Chen, W.-H.; Hajduczki, A.; Martinez, E.J.; Bai, H.; Matz, H.; Hill, T.M.; Lewitus, E.; Chang, W.C.; Dawit, L.; Peterson, C.E. Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity. Nat. Commun. 2023, 14, 580. [Google Scholar] [CrossRef] [PubMed]
- Cheong, W.S.; Leow, C.Y.; Majeed, A.B.A.; Leow, C.H. Diagnostic and therapeutic potential of shark variable new antigen receptor (VNAR) single domain antibody. Int. J. Biol. Macromol. 2020, 147, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Stanfield, R.L.; Greenberg, A.S.; Flajnik, M.F. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): Identification of a new locus preferentially expressed in early development. Immunogenetics 2002, 54, 501–512. [Google Scholar] [CrossRef]
- Rumfelt, L.L.; Lohr, R.L.; Dooley, H.; Flajnik, M.F. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004, 5, 8. [Google Scholar] [CrossRef]
- Jia, L.; Wang, Y.; Shen, Y.; Zhong, B.; Luo, Z.; Yang, J.; Chen, G.; Jiang, X.; Chen, J.; Lyu, Z. IgNAR characterization and gene loci identification in whitespotted bamboo shark (Chiloscyllium plagiosum) genome. Fish Shellfish. Immunol. 2023, 133, 108535. [Google Scholar] [CrossRef]
- Suzuki, R.; Saito, K.; Matsuda, M.; Sato, M.; Kanegae, Y.; Shi, G.; Watashi, K.; Aizaki, H.; Chiba, J.; Saito, I. Single-domain intrabodies against hepatitis C virus core inhibit viral propagation and core-induced NFκB activation. J. Gen. Virol. 2016, 97, 887–892. [Google Scholar] [CrossRef]
- Roux, K.H.; Greenberg, A.S.; Greene, L.; Strelets, L.; Avila, D.; McKinney, E.C.; Flajnik, M.F. Structural analysis of the nurse shark (new) antigen receptor (NAR): Molecular convergence of NAR and unusual mammalian immunoglobulins. Proc. Natl. Acad. Sci. USA 1998, 95, 11804–11809. [Google Scholar] [CrossRef]
- Cabanillas-Bernal, O.; Dueñas, S.; Ayala-Avila, M.; Rucavado, A.; Escalante, T.; Licea-Navarro, A.F. Synthetic libraries of shark vNAR domains with different cysteine numbers within the CDR3. PLoS ONE 2019, 14, e0213394. [Google Scholar] [CrossRef]
- Feige, M.J.; Gräwert, M.A.; Marcinowski, M.; Hennig, J.; Behnke, J.; Ausländer, D.; Herold, E.M.; Peschek, J.; Castro, C.D.; Flajnik, M. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl. Acad. Sci. USA 2014, 111, 8155–8160. [Google Scholar] [CrossRef] [PubMed]
- Barelle, C.; Gill, D.S.; Charlton, K. Shark novel antigen receptors—The next generation of biologic therapeutics? In Pharmaceutical Biotechnology; Springer: New York, NY, USA, 2009; pp. 49–62. [Google Scholar]
- Zhang, W.; Qin, L.; Cai, X.; Juma, S.N.; Xu, R.; Wei, L.; Wu, Y.; Cui, X.; Chen, G.; Liu, L. Sequence structure character of IgNAR Sec in whitespotted bamboo shark (Chiloscyllium plagiosum). Fish Shellfish. Immunol. 2020, 102, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zan, H.; Xu, Z.; Casali, P. Epigenetics of the antibody response. Trends Immunol. 2013, 34, 460–470. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, W.; Zheng, K.; Sun, S.; Zhong, B.; Luo, Z.; Yang, J.; Jia, L.; Yang, L.; Shang, W.; Jiang, X.; et al. Characteristics and Genomic Localization of Nurse Shark (Ginglymostoma cirratum) IgNAR. Int. J. Mol. Sci. 2024, 25, 12879. https://doi.org/10.3390/ijms252312879
Tang W, Zheng K, Sun S, Zhong B, Luo Z, Yang J, Jia L, Yang L, Shang W, Jiang X, et al. Characteristics and Genomic Localization of Nurse Shark (Ginglymostoma cirratum) IgNAR. International Journal of Molecular Sciences. 2024; 25(23):12879. https://doi.org/10.3390/ijms252312879
Chicago/Turabian StyleTang, Wenjie, Kaixi Zheng, Shengjie Sun, Bo Zhong, Zhan Luo, Junjie Yang, Lei Jia, Lan Yang, Wenna Shang, Xiaofeng Jiang, and et al. 2024. "Characteristics and Genomic Localization of Nurse Shark (Ginglymostoma cirratum) IgNAR" International Journal of Molecular Sciences 25, no. 23: 12879. https://doi.org/10.3390/ijms252312879
APA StyleTang, W., Zheng, K., Sun, S., Zhong, B., Luo, Z., Yang, J., Jia, L., Yang, L., Shang, W., Jiang, X., Lyu, Z., Chen, J., & Chen, G. (2024). Characteristics and Genomic Localization of Nurse Shark (Ginglymostoma cirratum) IgNAR. International Journal of Molecular Sciences, 25(23), 12879. https://doi.org/10.3390/ijms252312879




