Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques
Abstract
1. Introduction
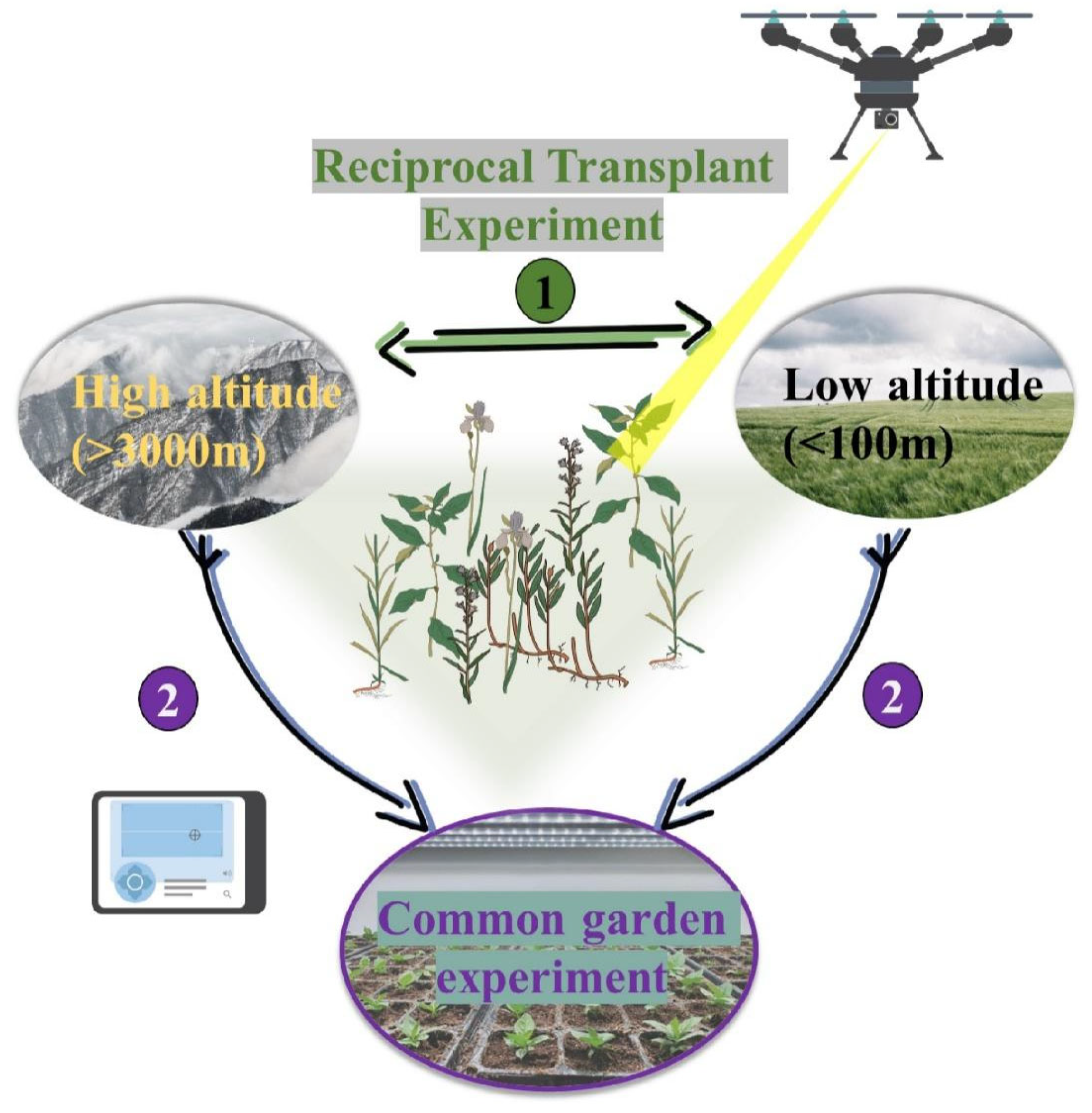
2. Transplant Experiment
3. Genomics Facilitates the Molecular Basis of Adaptive Evolution
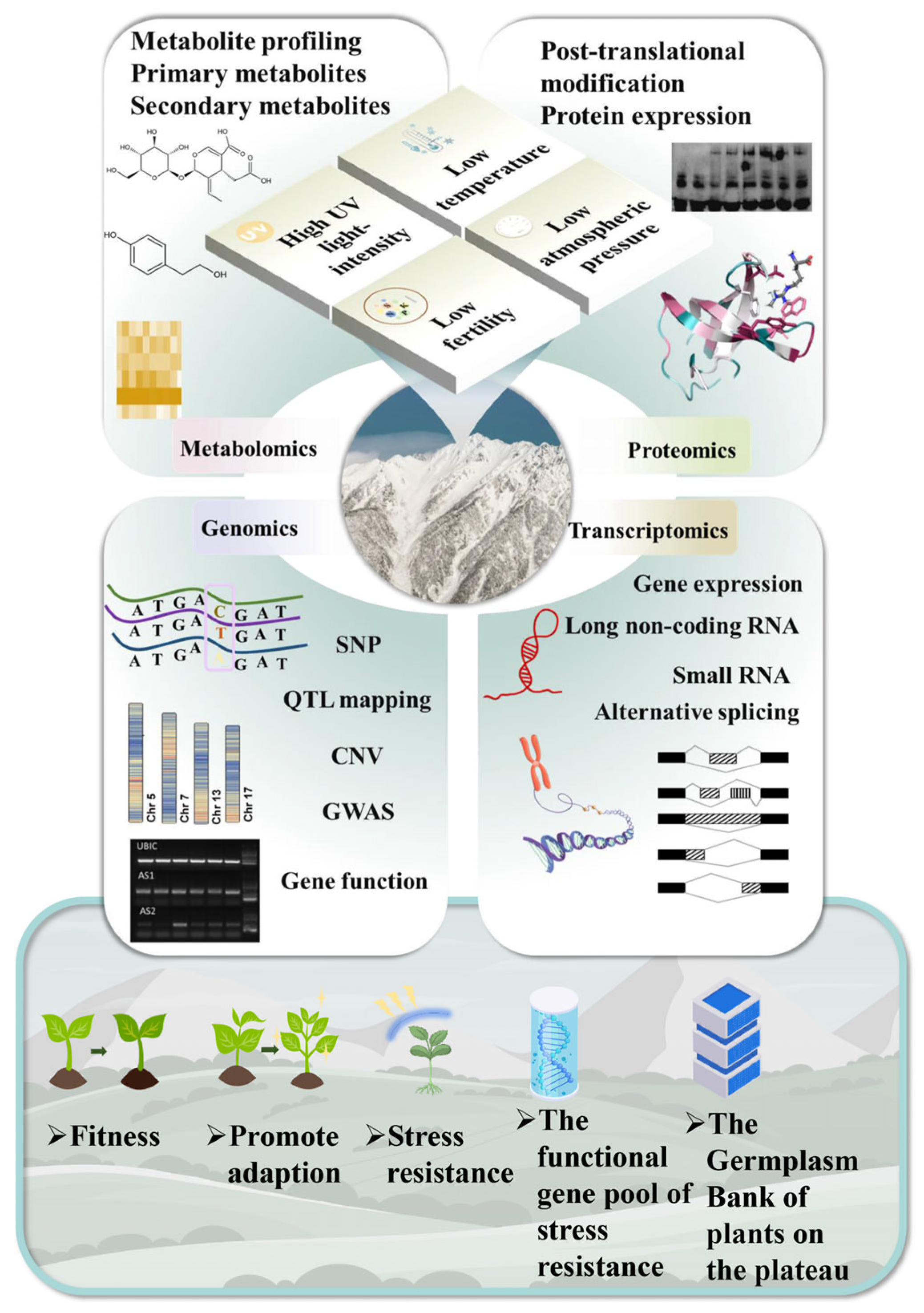
4. The Role of Multi-Omics in the Adaptive Evolution of Alpine Plants
5. Conclusions and Perspectives
| Plant Species | Altitude | Omics Approach | Effect | Reference |
|---|---|---|---|---|
| Crucihimalaya himalaica | Seedlings of C. himalaica were sampled from an altitude of 4010 m. | Genomic | Gene families showing dramatic changes in size and genes showing signs of positive selection are likely candidates for C. himalaica’s adaptation to intense radiation, low temperature, and pathogen–depauperate environments in the QTP. Loss of function at the S-locus, the reason for the transition to self-fertilization of C. himalaica, might have enabled its QTP occupation. | [54] |
| Prunus sp. | A total of 377 accessions of Prunus germplasm along altitude gradients ranging from 2067 to 4492 m in the Himalayas were collected and sequenced. | Genomic and metabolomic | A total of 379 metabolites had significant genetic correlations with altitudes; in particular, phenylpropanoids were positively correlated with altitudes. Specific SINE insertions change the expression of altitude-related genes. | [57] |
| Saussurea sp. | Three (S. pachyneura, S. salwinensis, and S. velutina) were collected between 4550 and 4620 m, and the other two (S. amurensis and S. amara) were collected between 150 and 350 m | Transcriptomic | Gene families specific to alpine species were identified, which involve oxidoreductase activity, pectin metabolism, lipid transport, and polysaccharide metabolism, potentially aiding in the defense against hypoxia and the freezing temperatures of the QTP. Also, hundreds of genes under positive selection were discovered, related to DNA repair, membrane transport, UV-B and hypoxia responses, reproduction, and nutrient metabolism, likely contributing to Saussurea’s adaptation to high-altitude environments. | [73] |
| Potentilla bifurca | Sample selected from two altitude ranges −3215 and 1725 masl | Transcriptomic | Fifty differentially expressed genes (DEG), including peroxidase, superoxide dismutase protein, and the ubiquitin-conjugating enzyme responded to abiotic stresses; a large number of DEGs encode key enzymes involved in secondary metabolites, including phenylpropane and flavonoids; 298 potential genomic SSRs were identified for genetic diversity assessment. | [76] |
| Rhododendron sp. | Four colored species, Rhododendron fastigiatum Franch (4194 m), Rhododendron lacteum Franch (3927 m), Rhododendron facetum I. B. Balfour & Kingdon Ward (2817 m) and Rhododendron pachypodum Balf. f. et W. W. Smith (2413 m), were collected. | Transcriptomic and metabolomic | Genes related to carbohydrates, fatty acids, amino acids and flavonoids biosynthesis play important roles in the altitude adaptability. | [77] |
| Potentilla saundersiana | Samples from 4350 to 5200 m in altitude. | Proteomic | Proteins involved in antioxidative activity, primary metabolites, epigenetic regulation, and protein post-translational modification play important roles in conferring tolerance to alpine environments. | [88] |
| Sinopodophyllum hexandrum | Leaves from 3-year-old plants were collected from 3300 and 2300 m. | Proteomic and transcriptomic | Nine DEPs and 41 DEGs were identified as being involved in flavonoid biosynthesis and the light response at 3300 m. | [94] |
| Herpetospermum pedunculosum | Samples from 2800 m, 3000 m, 3100 m and 3300 m. | Proteomic | High level of expression of some proteins, such as oxygen-evolving enhancer proteins, calreticulins, and S-adenosyl-l-homocysteine hydrolase, might confer greater tolerance in H. pedunculosum to the complex environment associated with high altitudes. | [104] |
| Cannabis sativa | Raw inflorescences material obtained from plants cultivated in 1200 m and 130 m altitude. | Metabolomic | All plants grown at altitude exhibited a higher total amount of terpenes when compared with plains counterparts, with β-Myrcene, trans-Caryophyllene and α-Humulene as the main contributors. | [92] |
| Draba oreades | Samples were collected at altitudes of 3800 m, 4000 m and 4200 m. | Metabolomic | Phenylalanine, tyrosine, and tryptophan biosynthesis and phenylalanine metabolism related to the biosynthesis of flavonoids were up-regulated in the high-altitude group, and 10 important metabolites were identified as potential biomarkers. | [93] |
| Cyclocarya paliurus | Mature leaves with the largest leaf area at F4 stage from 280 m and 920 m. | Metabolomic and transcriptomic | High altitude induces more flavonoid accumulation than low altitude, which may be contributed by the up-regulation of genes involved in energy and protein synthesis. | [105] |
Author Contributions
Funding
Conflicts of Interest
References
- Ruddiman, F.W.; Kutzbach, E.J. Plateau uplift and climatic change. Sci. Am. 1991, 264, 66–75. [Google Scholar] [CrossRef]
- Qiao, Q.; Wang, Q.; Han, X.; Guan, Y.; Sun, H.; Zhong, Y.; Huang, J.; Zhang, T. Transcriptome sequencing of Crucihimalaya himalaica (Brassicaceae) reveals how Arabidopsis close relative adapt to the Qinghai-Tibet Plateau. Sci. Rep. 2016, 6, 21729. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Niu, Y.; Chen, Y.-S.; Song, B.; Liu, C.-Q.; Peng, D.-L.; Chen, J.-G.; Yang, Y. Survival and reproduction of plant species in the Qinghai-Tibet Plateau. J. Syst. Evol. 2014, 52, 378–396. [Google Scholar] [CrossRef]
- Dvorsky, M.; Dolezal, J.; Kopecky, M.; Chlumska, Z.; Janatkova, K.; Altman, J.; de Bello, F.; Rehakova, K. Testing the Stress-Gradient Hypothesis at the Roof of the World: Effects of the Cushion Plant Thylacospermum caespitosum on Species Assemblages. PLoS ONE 2013, 8, e53514. [Google Scholar] [CrossRef]
- Ma, W.; Shi, P.; Li, W.; He, Y.; Zhang, X.; Shen, Z.; Chai, S. Changes in individual plant traits and biomass allocation in alpine meadow with elevation variation on the Qinghai-Tibetan Plateau. Sci. China-Life Sci. 2010, 53, 1142–1151. [Google Scholar] [CrossRef]
- Yin, Z.-Y.; Li, M.; Zhang, Y.; Shao, X. Growth-climate relationships along an elevation gradient on a southeast-facing mountain slope in the semi-arid eastern Qaidam Basin, northeastern Tibetan Plateau. Trees-Struct. Funct. 2016, 30, 1095–1109. [Google Scholar] [CrossRef]
- Guo, Q.; Li, H.; Luo, D.; Quan, H.; Bianba, D.; Zhang, W. Comparative drought tolerance of six native deciduous and broad-leaved woody plant species seedlings in the Qinghai-Tibet Plateau. Acta Physiol. Plant. 2016, 38, 14. [Google Scholar] [CrossRef]
- Bu, H.; Du, G.; Chen, X.; Xu, X.; Liu, K.; Wen, S. Community-wide germination strategies in an alpine meadow on the eastern Qinghai-Tibet plateau: Phylogenetic and life-history correlates. Plant Ecol. 2008, 195, 87–98. [Google Scholar] [CrossRef]
- Li, H.Q.; Zhang, F.W.; Li, Y.N.; Cao, G.M.; Zhao, L.; Zhao, X.Q. Seasonal and interannual variations of ecosystem photosynthetic features in an alpine dwarf shrubland on the Qinghai-Tibetan Plateau, China. Photosynthetica 2014, 52, 321–331. [Google Scholar] [CrossRef]
- Alexander, J.M.; Chalmandrier, L.; Lenoir, J.; Burgess, T.I.; Essl, F.; Haider, S.; Kueffer, C.; McDougall, K.; Milbau, A.; Nuñez, M.A.; et al. Lags in the response of mountain plant communities to climate change. Glob. Chang. Biol. 2018, 24, 563–579. [Google Scholar] [CrossRef]
- Rahbek, C.; Borregaard, M.K.; Antonelli, A.; Colwell, R.K.; Holt, B.G.; Nogues-Bravo, D.; Rasmussen, C.M.Ø.; Richardson, K.; Rosing, M.T.; Whittaker, R.J.; et al. Building mountain biodiversity: Geological and evolutionary processes. Science 2019, 365, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Albuja-Quintana, M.; Pozo, G.; Gordillo-Romero, M.; Armijos, C.E.; Torres, M.D.L. Genome report: First reference genome of Vaccinium floribundum Kunth, an emblematic Andean species. G3 Genes|Genomes|Genet. 2024, 14, jkae136. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Lee, T.M. Altitude dependence of alpine grassland ecosystem multifunctionality across the Tibetan Plateau. J. Environ. Manag. 2023, 332, 117358. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, P.; Bhargava, B.; Sharma, R.; Irfan, M.; Chandora, R. Transcriptomic and Metabolomic Reprogramming to Explore the High-Altitude Adaptation of Medicinal Plants: A Review. J. Plant Growth Regul. 2023, 42, 7315–7329. [Google Scholar] [CrossRef]
- Li, B.-Z.; Liu, R.-N.; Zhang, H.; Tian, Y.-N.; Chen, T.-T.; Li, J.-X.; Jiao, F.-H.; Jia, T.-F.; Li, Y.-X.; Zhang, X.-Y.; et al. ZmMYB56 regulates stomatal closure and drought tolerance in maize seedlings through the transcriptional regulation of ZmTOM7. New Crops. 2024, 1, 100012. [Google Scholar] [CrossRef]
- Tsegaw, M.; Zegeye, W.A.; Jiang, B.; Sun, S.; Yuan, S.; Han, T.; Wu, T. Progress and Prospects of the Molecular Basis of Soybean Cold Tolerance. Plants 2023, 12, 459. [Google Scholar] [CrossRef]
- Vega-Gálvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martínez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef]
- Vélez-Sánchez, J.; Casierra-Posada, F.; Fischer, G. Effect of Regulated Deficit Irrigation (RDI) on the Growth and Development of Pear Fruit (Pyrus communis L.), var. Triunfo de Viena. Sustainability 2023, 15, 13392. [Google Scholar] [CrossRef]
- Flores-Marquez, R.; Vera-Vílchez, J.; Verástegui-Martínez, P.; Lastra, S.; Solórzano-Acosta, R. An Evaluation of Dryland Ulluco Cultivation Yields in the Face of Climate Change Scenarios in the Central Andes of Peru by Using the AquaCrop Model. Sustainability 2024, 16, 5428. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, J. Response and Adaptive Mechanism of Flavonoids in Pigmented Potatoes at Different Altitudes. Plant Cell Physiol. 2024, 65, 1184–1196. [Google Scholar] [CrossRef]
- Aitken, S.N.; Yeaman, S.; Holliday, J.A.; Wang, T.; Curtis-McLane, S. Adaptation, migration or extirpation: Climate change outcomes for tree populations. Evol. Appl. 2008, 1, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Jump, A.S.; Penuelas, J. Running to stand still: Adaptation and the response of plants to rapid climate change. Ecol. Lett. 2005, 8, 1010–1020. [Google Scholar] [CrossRef]
- Latreille, A.C.; Pichot, C. Local-scale diversity and adaptation along elevational gradients assessed by reciprocal transplant experiments: Lack of local adaptation in silver fir populations. Ann. For. Sci. 2017, 74, 77. [Google Scholar] [CrossRef]
- Chen, M.X.; Tian, Y.; Zhu, F.Y.; Fan, T.; Yan, H.X.; Sun, P.C.; Li, M.; Hou, X.X.; Lin, P.; Song, Y.C.; et al. Alternative splicing of VRF1 acts as a molecular switch to regulate stress-induced early flowering. Cell Rep. 2024, 11, 114918. [Google Scholar] [CrossRef] [PubMed]
- Hoban, S.; Kelley, J.L.; Lotterhos, K.E.; Antolin, M.F.; Bradburd, G.; Lowry, D.B.; Poss, M.L.; Reed, L.K.; Storfer, A.; Whitlock, M.C. Finding the Genomic Basis of Local Adaptation: Pitfalls, Practical Solutions, and Future Directions. Am. Nat. 2016, 188, 379–397. [Google Scholar] [CrossRef] [PubMed]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Leimu, R.; Fischer, M. A Meta-Analysis of Local Adaptation in Plants. PLoS ONE 2008, 3, e4010. [Google Scholar] [CrossRef]
- Sork, V.L.; Aitken, S.N.; Dyer, R.J.; Eckert, A.J.; Legendre, P.; Neale, D.B. Putting the landscape into the genomics of trees: Approaches for understanding local adaptation and population responses to changing climate. Tree Genet. Genomes 2013, 9, 901–911. [Google Scholar] [CrossRef]
- Savolainen, O. The Genomic Basis of Local Climatic Adaptation. Science 2011, 334, 49–50. [Google Scholar] [CrossRef]
- Savolainen, V.; Pyhajarvi, T.; Knurr, T. Gene flow and local adaptation in tees. Annu. Rev. Ecol. Evol. Syst. 2007, 38, 595–619. [Google Scholar] [CrossRef]
- de Villemereuil, P.; Gaggiotti, O.E.; Mouterde, M.; Till-Bottraud, I. Common garden experiments in the genomic era: New perspectives and opportunities. Heredity 2016, 116, 249–254. [Google Scholar] [CrossRef] [PubMed]
- McKown, A.D.; Guy, R.D.; Klapste, J.; Geraldes, A.; Friedmann, M.; Cronk, Q.C.B.; El-Kassaby, Y.A.; Mansfield, S.D.; Douglas, C.J. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in Populus trichocarpa. New Phytol. 2014, 201, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Alberto, F.; Bouffier, L.; Louvet, J.M.; Lamy, J.B.; Delzon, S.; Kremer, A. Adaptive responses for seed and leaf phenology in natural populations of sessile oak along an altitudinal gradient. J. Evol. Biol. 2011, 24, 1442–1454. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.; Morgan, T.J.; Nippert, J.B.; Ocheltree, T.W.; Keith, R.; Dhakal, P.; Ungerer, M.C. Natural selection drives clinal life history patterns in the perennial sunflower species, Helianthus maximiliani. Mol. Ecol. 2011, 20, 2318–2328. [Google Scholar] [CrossRef] [PubMed]
- Lind, B.M.; Menon, M.; Bolte, C.E.; Faske, T.M.; Eckert, A.J. The genomics of local adaptation in trees: Are we out of the woods yet? Tree Genet. Genomes 2018, 14, 29. [Google Scholar] [CrossRef]
- Paschoa, R.P.D.; Pinto, V.B.; Pereira, J.P.; Cavatte, P.C.; Garbin, M.L.; Godinho, T.; Xavier, L.R.; Carrijo, T.T.; Silveira, V. Proteomic and physiological signatures of altitude adaptation in a Myrsine coriacea population under common garden conditions. J. Proteom. 2024, 299, 105156. [Google Scholar] [CrossRef]
- Aitken, S.N.; Whitlock, M.C. Assisted Gene Flow to Facilitate Local Adaptation to Climate Change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367. [Google Scholar] [CrossRef]
- Wang, X.; Sun, H.; Tan, C.; Wang, X.; Xia, M. Effects of Film Mulching on Plant Growth and Nutrients in Artificial Soil: A Case Study on High Altitude Slopes. Sustainability 2021, 13, 11026. [Google Scholar] [CrossRef]
- Han, T.-S.; Hu, Z.-Y.; Du, Z.-Q.; Zheng, Q.-J.; Liu, J.; Mitchell-Olds, T.; Xing, Y.-W. Adaptive responses drive the success of polyploid yellowcresses (Rorippa, Brassicaceae) in the Hengduan Mountains, a temperate biodiversity hotspot. Plant Divers. 2022, 44, 455–467. [Google Scholar] [CrossRef]
- Lajoie, G.; Vellend, M. Characterizing the contribution of plasticity and genetic differentiation to community-level trait responses to environmental change. Ecol. Evol. 2018, 8, 3895–3907. [Google Scholar] [CrossRef]
- Halbritter, A.H.; Fior, S.; Keller, I.; Billeter, R.; Edwards, P.J.; Holderegger, R.; Karrenberg, S.; Pluess, A.R.; Widmer, A.; Alexander, J.M. Trait differentiation and adaptation of plants along elevation gradients. J. Evol. Biol. 2018, 31, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Tiscar, P.A.; Lucas-Borja, M.E.; Candel-Perez, D. Lack of local adaptation to the establishment conditions limits assisted migration to adapt drought-prone Pinus nigra populations to climate change. For. Ecol. Manag. 2018, 409, 719–728. [Google Scholar] [CrossRef]
- Singh, A.; Roy, S. High altitude population of Arabidopsis thaliana is more plastic and adaptive under common garden than controlled condition. BMC Ecol. 2017, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.D.; Jiang, L.L.; Zhang, Z.H.; Cui, S.J.; Duan, J.C.; Wang, S.P.; Luo, C.Y.; Wang, Q.; Zhou, Y.; Li, X.E.; et al. Changes in flowering functional group affect responses of-community phenological sequences to temperature change. Ecology 2017, 98, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, W.; Zhou, M. Lack of local adaptation of invasive crofton weed (Ageratina adenophora) in different climatic areas of Yunnan Province, China. J. Plant Ecol. 2013, 6, 316–322. [Google Scholar] [CrossRef]
- Abdulridha, J.; Min, A.; Rouse, M.N.; Kianian, S.; Isler, V.; Yang, C. Evaluation of Stem Rust Disease in Wheat Fields by Drone Hyperspectral Imaging. Sensors 2023, 23, 4154. [Google Scholar] [CrossRef]
- Kai, P.M.; de Oliveira, B.M.; da Costa, R.M. Deep Learning-Based Method for Classification of Sugarcane Varieties. Agronomy 2022, 12, 2722. [Google Scholar] [CrossRef]
- Ding, R.; Luo, J.; Wang, C.; Yu, L.; Yang, J.; Wang, M.; Zhong, S.; Gu, R. Identifying and mapping individual medicinal plant Lamiophlomis rotata at high elevations by using unmanned aerial vehicles and deep learning. Plant Methods 2023, 19, 38. [Google Scholar] [CrossRef]
- Pieruschka, R.; Schurr, U. Plant Phenotyping: Past, Present, and Future. Plant Phenomics 2019, 2019, 7507131. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, G.; Liang, C.; Tian, Z. Omics-based interdisciplinarity is accelerating plant breeding. Curr. Opin. Plant Biol. 2022, 66, 102167. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Tabata, S.; Kaneko, T.; Nakamura, Y.; Kotani, H.; Kato, T.; Asamizu, E.; Miyajima, N.; Sasamoto, S.; Kimura, T.; Hosouchi, T.; et al. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 2000, 408, 823–826. [Google Scholar] [PubMed]
- Li, Y.; Cao, K.; Li, N.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Guo, J.; Wang, Q.; Ding, T.; et al. Genomic analyses provide insights into peach local adaptation and responses to climate change. Genome Res. 2021, 31, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Qiao, Q.; Novikova, P.Y.; Wang, Q.; Yue, J.; Guan, Y.; Ming, S.; Liu, T.; De, J.; Liu, Y.; et al. Qiong, Genome of Crucihimalaya himalaica, a close relative of Arabidopsis, shows ecological adaptation to high altitude. Proc. Natl. Acad. Sci. USA 2019, 116, 7137–7146. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.-Y.; Zhou, X.-L.; Wang, S.-Q.; Yang, G.-M.; Sun, W.-G.; Zhang, J.-Y.; Zhang, R.; Shen, S.-K. The first high-altitude autotetraploid haplotype-resolved genome assembled (Rhododendron nivale subsp. boreale) provides new insights into mountaintop adaptation. GigaScience 2024, 13, giae052. [Google Scholar] [CrossRef]
- Guo, X.; Hu, Q.; Hao, G.; Wang, X.; Zhang, D.; Ma, T.; Liu, J. The genomes of two Eutrema species provide insight into plant adaptation to high altitudes. DNA Res. 2018, 25, 307–315. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Zuo, H.; Zheng, W.; Zhang, S.; Huang, Y.; Pingcuo, G.; Ying, H.; Zhao, F.; Li, Y.; et al. Genomic basis of high-altitude adaptation in Tibetan Prunus fruit trees. Curr. Biol. 2021, 31, 3848–3860. [Google Scholar] [CrossRef]
- Zhang, X.; Kuang, T.; Dong, W.; Qian, Z.; Zhang, H.; Landis, J.B.; Feng, T.; Li, L.; Sun, Y.; Huang, J.; et al. Genomic convergence underlying high-altitude adaptation in alpine plants. J. Integr. Plant Biol. 2023, 65, 1620–1635. [Google Scholar] [CrossRef]
- Zheng, C.; Tan, L.; Sang, M.; Ye, M.; Wu, R. Genetic adaptation of Tibetan poplar (Populus szechuanica var. tibetica) to high altitudes on the Qinghai-Tibetan Plateau. Ecol. Evol. 2020, 10, 10974–10985. [Google Scholar] [CrossRef]
- Weigel, D.; Nordborg, M. Population Genomics for Understanding Adaptation in Wild Plant Species Annu. Rev. Genet. 2015, 492015, 315–338. [Google Scholar] [CrossRef]
- Luikart, G.; England, P.R.; Tallmon, D.; Jordan, S.; Taberlet, P. The power and promise of population genomics: From genotyping to genome typing. Nat. Rev. Genet. 2003, 4, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Tiffin, P.; Ross-Ibarra, J. Advances and limits of using population genetics to understand local adaptation. Trends Ecol. Evol. 2014, 29, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Lu, Z.; Song, Y.; Hu, X.; Corlett, R.T. A chromosome-scale genome assembly for the holly (Ilex polyneura) provides insights into genomic adaptations to elevation in Southwest China. Hortic. Res. 2022, 9, uhab049. [Google Scholar] [CrossRef] [PubMed]
- Manel, S.; Holderegger, R. Ten years of landscape genetics. Trends Ecol. Evol. 2013, 28, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Manel, S.; Joost, S.; Epperson, B.K.; Holderegger, R.; Storfer, A.; Rosenberg, M.S.; Scribner, K.T.; Bonin, A.; Fortin, M.J. Perspectives on the use of landscape genetics to detect genetic adaptive variation in the field. Mol. Ecol. 2010, 19, 3760–3772. [Google Scholar] [CrossRef]
- Holderegger, R.; Buehler, D.; Gugerli, F.; Manel, S. Landscape genetics of plants. Trends Plant Sci. 2010, 15, 675–683. [Google Scholar] [CrossRef]
- Leempoel, K.; Parisod, C.; Geiser, C.; Joost, S. Multiscale landscape genomic models to detect signatures of selection in the alpine plant Biscutella laevigata. Ecol. Evol. 2018, 8, 1794–1806. [Google Scholar] [CrossRef]
- Lou, S.; Guo, X.; Liu, L.; Song, Y.; Zhang, L.; Jiang, Y.; Zhang, L.; Sun, P.; Liu, B.; Tong, S.; et al. Allelic shift in cis-elements of the transcription factor RAP2.12 underlies adaptation associated with humidity in Arabidopsis thaliana. Sci. Adv. 2022, 8, eabn8281. [Google Scholar] [CrossRef]
- Song, Y.-C.; Chen, M.-X.; Zhang, K.-L.; Reddy, A.S.N.; Cao, F.-L.; Zhu, F.-Y. QuantAS: A comprehensive pipeline to study alternative splicing by absolute quantification of splice isoforms. New Phytol. 2023, 240, 928–939. [Google Scholar] [CrossRef]
- Lv, Z.; Jiang, R.; Chen, J.; Chen, W. Nanoparticle-mediated gene transformation strategies for plant genetic engineering. Plant J. 2020, 104, 880–891. [Google Scholar] [CrossRef]
- Kwak, S.-Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.-H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, Y.; Landis, J.B.; Shen, J.; Zhang, H.; Kuang, T.; Sun, W.; Sun, J.; Tiamiyu, B.B.; Deng, T.; et al. Transcriptomes of Saussurea (Asteraceae) Provide Insights into High-Altitude Adaptation. Plants 2021, 10, 1715. [Google Scholar] [CrossRef]
- Jia, Y.; Liu, M.-L.; Yue, M.; Zhao, Z.; Zhao, G.-F.; Li, Z.-H. Comparative Transcriptome Analysis Reveals Adaptive Evolution of Notopterygium incisum and Notopterygium franchetii, Two High-Alpine Herbal Species Endemic to China. Molecules 2017, 22, 1158. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Do, S.; Huynh, H.; Li, J.X.; Liu, Y.G.; Du, Z.Y.; Chen, M.X. Importance of pre-mRNA splicing and its study tools in plants. Adv. Biotechnol. 2024, 2, 4. [Google Scholar] [CrossRef]
- Tang, X.; Li, J.; Liu, L.; Jing, H.; Zuo, W.; Zeng, Y. Transcriptome Analysis Provides Insights into Potentilla bifurca Adaptation to High Altitude. Life 2022, 12, 1337. [Google Scholar] [CrossRef]
- Liu, X.-W.; Wang, Y.-H.; Shen, S.-K. Transcriptomic and metabolomic analyses reveal the altitude adaptability and evolution of different-colored flowers in alpine Rhododendron species. Tree Physiol. 2022, 42, 1100–1113. [Google Scholar] [CrossRef]
- Zhang, Z.-H.; Chang, X.; Su, D.-Y.; Yao, R.; Liu, X.-D.; Zhu, H.; Liu, G.-X.; Zhong, B.-J. Comprehensive transcriptome analyses of two Oocystis algae provide insights into the adaptation to Qinghai-Tibet Plateau. J. Syst. Evol. 2021, 59, 1209–1219. [Google Scholar] [CrossRef]
- Duruflé, H.; Hervé, V.; Ranocha, P.; Balliau, T.; Zivy, M.; Chourré, J.; Clemente, H.S.; Burlat, V.; Albenne, C.; Déjean, S.; et al. Dunand, Cell wall modifications of two Arabidopsis thaliana ecotypes, Col and Sha, in response to sub-optimal growth conditions: An integrative study. Plant Sci. 2017, 263, 183–193. [Google Scholar] [CrossRef]
- Zhang, F.; Gao, Q.; Khan, G.; Luo, K.; Chen, S. Comparative transcriptome analysis of aboveground and underground tissues of Rhodiola algida, an important ethno-medicinal herb endemic to the Qinghai-Tibetan Plateau. Gene 2014, 553, 90–97. [Google Scholar] [CrossRef]
- Rao, A.; Barkley, D.; França, G.S.; Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 2021, 596, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhang, H.; Xia, Y. Harnessing spatial transcriptomics for advancing plant regeneration research. Trends Plant Sci. 2024, 29, 718–720. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Kumari, M. Adaptive mechanisms of medicinal plants along altitude gradient: Contribution of proteomics. Biol. Plant. 2018, 62, 630–640. [Google Scholar] [CrossRef]
- Hu, J.; Rampitsch, C.; Bykova, N.V. Advances in plant proteomics toward improvement of crop productivity and stress resistance. Front. Plant Sci. 2015, 6, 209. [Google Scholar] [CrossRef]
- Diz, A.P.; Martinez-Fernandez, M.; Rolan-Alvarez, E. Proteomics in evolutionary ecology: Linking the genotype with the phenotype. Mol. Ecol. 2012, 21, 1060–1080. [Google Scholar] [CrossRef]
- Kumari, M.; Kumar, R. Functional trait correlation network and proteomic analysis reveal multifactorial adaptation mechanisms to a climatic gradient associated with high altitude in the Himalayan region. Plant Cell Environ. 2024, 47, 1556–1574. [Google Scholar] [CrossRef]
- Ma, L.; Yang, L.; Zhao, J.; Wei, J.; Kong, X.; Wang, C.; Zhang, X.; Yang, Y.; Hu, X. Comparative proteomic analysis reveals the role of hydrogen sulfide in the adaptation of the alpine plant Lamiophlomis rotata to altitude gradient in the Northern Tibetan Plateau. Planta 2015, 241, 887–906. [Google Scholar] [CrossRef]
- Ma, L.; Sun, X.; Kong, X.; Galvan, J.V.; Li, X.; Yang, S.; Yang, Y.; Yang, Y.; Hu, X. Physiological, biochemical and proteomics analysis reveals the adaptation strategies of the alpine plant Potentilla saundersiana at altitude gradient of the Northwestern Tibetan Plateau. J. Proteom. 2015, 112, 63–82. [Google Scholar] [CrossRef]
- De Vos, R.C.H.; Moco, S.; Lommen, A.; Keurentjes, J.J.B.; Bino, R.J.; Hall, R.D. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2007, 2, 778–791. [Google Scholar] [CrossRef]
- Yuan, H.; Cao, G.; Hou, X.; Huang, M.; Du, P.; Tan, T.; Zhang, Y.; Zhou, H.; Liu, X.; Liu, L.; et al. Development of a widely targeted volatilomics method for profiling volatilomes in plants. Mol. Plant 2022, 15, 189–202. [Google Scholar] [CrossRef]
- Kumari, M.; Joshi, R.; Kumar, R. Metabolic signatures provide novel insights to Picrorhiza kurroa adaptation along the altitude in Himalayan region. Metabolomics 2020, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Giupponi, L.; Leoni, V.; Pavlovic, R.; Giorgi, A. Influence of Altitude on Phytochemical Composition of Hemp Inflorescence: A Metabolomic Approach. Molecules 2020, 25, 1381. [Google Scholar] [CrossRef]
- Lei, L.; Yuan, X.; Fu, K.; Chen, Y.; Lu, Y.; Shou, N.; Wu, D.; Chen, X.; Shi, J.; Zhang, M.; et al. Pseudotargeted metabolomics revealed the adaptive mechanism of Draba oreades Schrenk at high altitude. Front. Plant Sci. 2022, 13, 1052640. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Dong, M.; Li, M.; Jin, L.; Pare, P.W. Light-Induced Flavonoid Biosynthesis in Sinopodophyllum hexandrum with High-Altitude Adaptation. Plants 2023, 12, 575. [Google Scholar] [CrossRef] [PubMed]
- He, Z.H.; Zhang, P.; Jia, H.T.; Zhang, S.L.; Nishawy, E.; Sun, X.P.; Dai, M.Q. Regulatory Mechanisms and Breeding Strategies for Crop Drought Resistance. New Crops. 2024, 1, 100029. [Google Scholar] [CrossRef]
- Giménez-Benavides, L.; Escudero, A.; Iriondo, J.M. Reproductive limits of a late-flowering high-mountain Mediterranean plant along an elevational climate gradient. New Phytol. 2007, 173, 367–382. [Google Scholar] [CrossRef]
- Gelvin, S.B. Plant Proteins Involved in Agrobacterium-Mediated Genetic Transformation. Annu. Rev. Phytopathol. 2010, 482010, 45–68. [Google Scholar] [CrossRef]
- Rhodes, C.A.; Pierce, D.A.; Mettler, I.J.; Mascarenhas, D.; Detmer, J.J. Genetically transformed maize plants from protoplasts. Science 1988, 240, 204–207. [Google Scholar] [CrossRef]
- Ahmed, S.; Gao, X.; Jahan, M.A.; Adams, M.; Wu, N.; Kovinich, N. Nanoparticle-based genetic transformation of Cannabis sativa. J. Biotechnol. 2021, 326, 48–51. [Google Scholar] [CrossRef]
- Demirer, G.S.; Zhang, H.; Matos, J.L.; Goh, N.S.; Cunningham, F.J.; Sung, Y.; Chang, R.; Aditham, A.J.; Chio, L.; Cho, M.-J.; et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019, 14, 456–464. [Google Scholar] [CrossRef]
- Cunningham, F.J.; Goh, N.S.; Demirer, G.S.; Matos, J.L.; Landry, M.P. Nanoparticle-Mediated Delivery towards Advancing Plant Genetic Engineering. Trends Biotechnol. 2018, 36, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut–dip–budding delivery system enables genetic modifications in plants without tissue culture. Innovation 2023, 4, 100345. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, S.; Deng, S.; Wang, M.; Wu, Y.; Li, M.; Dong, J.; Lu, S.; Su, C.; Li, G.; et al. A method of genetic transformation and gene editing of succulents without tissue culture. Plant Biotechnol. J. 2024, 22, 1981–1988. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.C.; Peng, M.; Liu, S.; Wang, Y.; Li, Z.; Zhao, S.; Li, S.; Quan, H.; Luo, Q.; Meng, F. Comparative proteomic analysis reveals the adaptation of Herpetospermum pedunculosum to an altitudinal gradient in the Tibetan Plateau. Biochem. Syst. Ecol. 2018, 80, 1–10. [Google Scholar] [CrossRef]
- Du, Z.; Lin, W.; Yu, B.; Zhu, J.; Li, J. Integrated Metabolomic and Transcriptomic Analysis of the Flavonoid Accumulation in the Leaves of Cyclocarya paliurus at Different Altitudes. Front. Plant Sci. 2022, 12, 794137. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.-L.; Leng, Y.-N.; Hao, R.-R.; Zhang, W.-Y.; Li, H.-F.; Chen, M.-X.; Zhu, F.-Y. Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques. Int. J. Mol. Sci. 2024, 25, 12666. https://doi.org/10.3390/ijms252312666
Zhang K-L, Leng Y-N, Hao R-R, Zhang W-Y, Li H-F, Chen M-X, Zhu F-Y. Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques. International Journal of Molecular Sciences. 2024; 25(23):12666. https://doi.org/10.3390/ijms252312666
Chicago/Turabian StyleZhang, Kai-Lu, Ya-Nan Leng, Rui-Rui Hao, Wen-Yao Zhang, Hong-Fei Li, Mo-Xian Chen, and Fu-Yuan Zhu. 2024. "Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques" International Journal of Molecular Sciences 25, no. 23: 12666. https://doi.org/10.3390/ijms252312666
APA StyleZhang, K.-L., Leng, Y.-N., Hao, R.-R., Zhang, W.-Y., Li, H.-F., Chen, M.-X., & Zhu, F.-Y. (2024). Adaptation of High-Altitude Plants to Harsh Environments: Application of Phenotypic-Variation-Related Methods and Multi-Omics Techniques. International Journal of Molecular Sciences, 25(23), 12666. https://doi.org/10.3390/ijms252312666







