Biological Insights and Recent Advances in Plant Long Non-Coding RNA
Abstract
:1. Introduction
2. LncRNA Genomic Regulation and Plant Biology
2.1. Generation and Classification of LncRNAs
2.2. Structure and Characteristics of Long Non-Coding RNA
2.3. Multi-Omics for LncRNA Functional Annotation
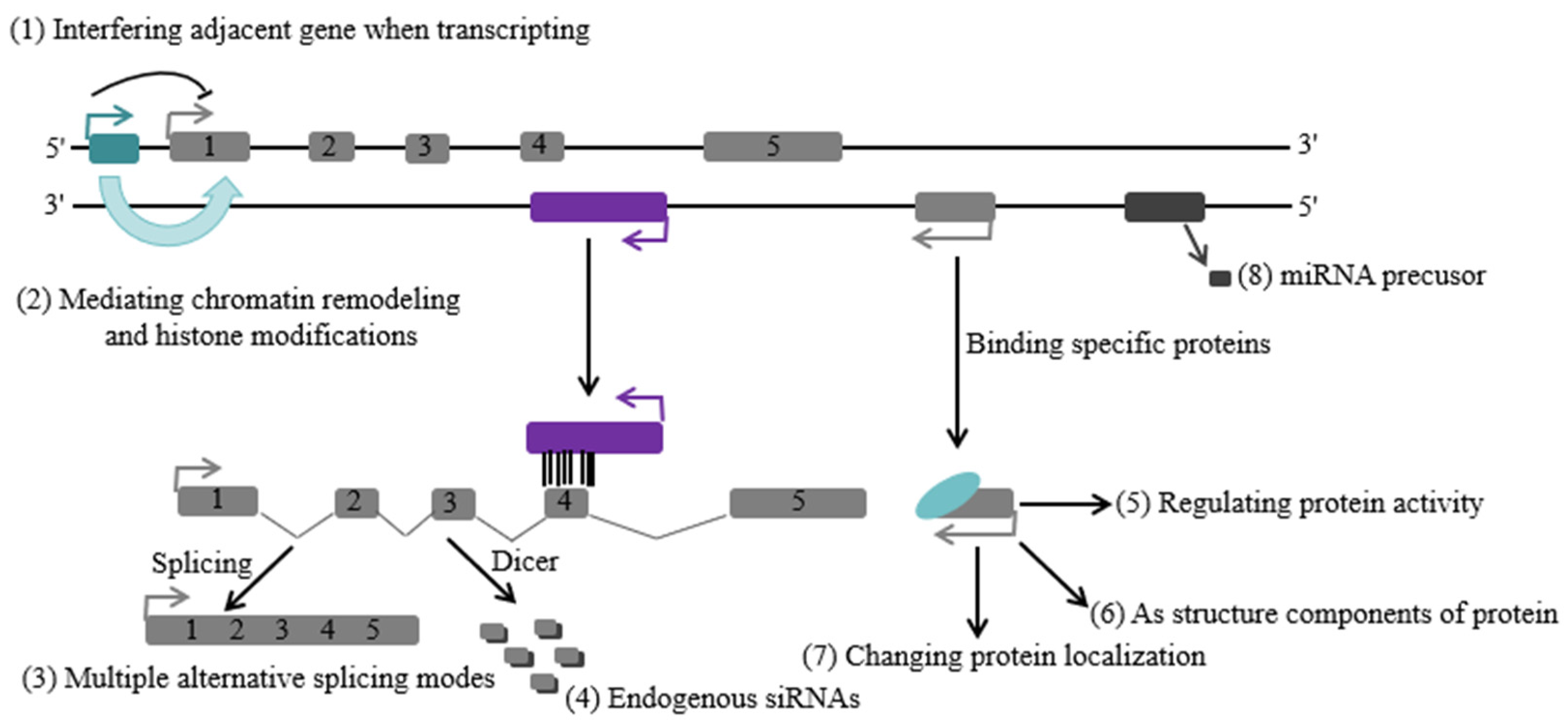
2.4. Functional Roles of LncRNAs in the Regulation of Gene Expression
- (1)
- Bait: LncRNAs can function as decoys to inhibit the binding of regulatory proteins to DNA. For instance, lncRNA Gas5 features a hairpin sequence that resembles the DNA binding domain of the glucocorticoid receptor. Starvation can induce the expression of Gas5, which then functions as a decoy to facilitate the release of the glucocorticoid receptor from its DNA binding site, thereby inhibiting the transcription of genes associated with metabolism [40]. Similarly, lncRNA PANDA can prevent apoptosis mediated by p53 by binding with the transcription factor NFYA through a similar mechanism [41];
- (2)
- Scaffolding or bridging: LncRNAs serve as regulators that guide two or more proteins into specific networks. As a scaffold, telomerase RNA TERC participates in the assembly of the telomerase complex [42]. Similarly, HOTAIR operates as a structural framework by combining the PRC2 and LSD1/CoREST complexes within a particular domain;
- (3)
- Guide: The binding of specific protein complexes to specific DNA regions requires the guidance of lncRNAs. Long non-coding RNAs, which play a guiding role, have two main functions—binding specific proteins and selectively acting on certain regions of the genome by some mechanism. Additionally, lncRNAs with a guiding function can exert their biological roles through either cis or trans action [43,44].
2.5. The Databases of Plant Long Non-Coding RNA
3. Expression of LncRNAs in Plants and Their Biological Functions
3.1. Functions of LncRNAs in Plant Progression
3.2. LncRNAs Participate in Plant Reproductive Growth
3.3. NA Participates in Abiotic Stress Responsive Regulation
3.3.1. Drought Stress
3.3.2. Cold Stress
3.3.3. Salt Stress
3.4. LncRNA Participates in Biotic Stresses
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Niu, X.L.; Feng, W.J.; Ma, J.H.; Xing, G.F. Research progress on biological functions of long non-coding RNA in plants. Biotechnol. Bulletin. 2015, 31, 1–7. [Google Scholar]
- Cho, J. Transposon-derived non-coding RNAs and their function in plants. Front. Plant Sci. 2018, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Romero-Barrios, N.; Jégu, T. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Dai, X.; Harrison, A.P.; Chen, M. RNA regulatory networks in animals and plants: A long noncoding RNA perspective. Brief. Funct. Genom. 2015, 14, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.S. Non-coding RNA. Progress. Biochem. Biophys. 2013, 40, 591–592. [Google Scholar]
- Zhang, X.P.; Wang, W.; Zhu, W.D.; Dong, J.; Cheng, Y.Y.; Yin, Z.J.; Shen, F.F. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int. J. Mol. Sci. 2019, 20, 573. [Google Scholar] [CrossRef]
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long non-coding RNAs: Mechanism of action and functional utility. Noncoding RNA Res. 2016, 1, 43–50. [Google Scholar] [CrossRef]
- Hsiao, L.V.; Wang, J.L.; Chekanova, A. Long noncoding RNAs in plants. Adv. Exp. Med. Biol. 2017, 1008, 133–154. [Google Scholar]
- Jampala, P.; Garhewal, A.; Lodha, M. Functions of long non-coding RNA in Arabidopsis thaliana. Plant Signal Behav. 2021, 16, 1925440. [Google Scholar] [CrossRef]
- Qin, S.W.; Lu, M.T.; Zhou, Y.Y. The latest research progress of long non-coding RNA in animals and plants. J. Tarim. Univ. 2016, 28, 103–112. [Google Scholar]
- Sun, X.; Zheng, H.X.; Sui, N. Regulation mechanism of long non-coding RNA in plant response to stress. Bio-Chem Biophys. Res. Commun. 2018, 503, 402–407. [Google Scholar] [CrossRef]
- Kaashyap, M.; Kaur, S.; Ford, R.; Edwards, D.; Siddique, K.H.M.; Varshney, R.K.; Mantri, N. Comprehensive transcriptomic analysis of two RIL parents with contrasting salt responsiveness identifies polyadenylated and non-polyadenylated flower lncRNAs in chickpea. Plant Biotechnol. J. 2022, 20, 1402–1416. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, Y.; He, R.; Jiang, L.; Qu, Z.; Gu, J.; Yang, J.; Legascue, M.F.; Wang, Z.Y.; Ariel, F.; et al. LncRNA DANA1 promotes drought tolerance and histone deacetylation of drought responsive genes in Arabidopsis. EMBO Rep. 2024, 25, 796–812. [Google Scholar] [CrossRef]
- Nejat, N.; Mantri, N. Emerging roles of long non-coding RNAs in plant response to biotic and abiotic stresses. Crit. Rev. Biotechnol. 2018, 38, 93–105. [Google Scholar] [CrossRef]
- Mishra, A.; Bohra, A. Non-coding RNAs and plant male sterility: Current knowledge and future prospects. Plant Cell Rep. 2018, 37, 177–191. [Google Scholar] [CrossRef]
- Tseng, K.C.; Wu, N.Y.; Chow, C.N.; Zheng, H.Q.; Chou, C.Y.; Yang, C.W.; Wang, M.J.; Chang, S.B.; Chang, W.C. JustRNA: A database of plant long noncoding RNA expression profiles and functional network. J. Exp. Bot. 2023, 74, 4949–4958. [Google Scholar] [CrossRef]
- Snijders, C.; Bassil, K.C.; Nijs, L.D. Methodologies of neuroepigenetic research: Background, challenges and future perspectives. Prog. Mol. Biol. Transl. Sci. 2018, 158, 15–27. [Google Scholar]
- Gloss, B.S.; Dinger, M.E. The specificity of long non-coding RNA expression. Biochim. Biophys. Acta 2016, 1859, 16–22. [Google Scholar] [CrossRef]
- Guo, X.L.; Gao, L.; Wang, Y.; David, K.Y.C.; Wang, T.; Deng, Y. Advances in long non-coding RNAs: Identification, structure prediction and function annotation. Brief. Funct. Genom. 2016, 15, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Florian, K.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNA. Cell 2018, 172, 393–407. [Google Scholar]
- Kornienko, A.E.; Dotter, C.P.; Guenzl, P.M.; Gisslinger, H.; Giaalinger, B.; Cleary, C.; Kralovics, R.; Pauler, F.M.; Barlow, D.P. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.R.; Li, S.P.; Cui, X.M.; Zheng, Y. A comprehensive review of web-based resources of non-coding RNAs for plant science research. Int. J. Biol. Sci. 2018, 14, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Paulo, P.; John, A.; Mattick, S. Noncoding RNA in development. Mamm. Genome 2008, 19, 454–492. [Google Scholar]
- Chris, P.P.; Peter, L.O.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Molecular Cell. 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Li, R.; Wang, T.; Zhu, H.L. Biological function and research methods of long noncoding RNA. China Biotechnol. 2015, 35, 66–70. [Google Scholar]
- Li, P.; Yang, H.; Wang, L.; Liu, H.J.; Huo, H.Q.; Zhang, C.J.; Liu, A.Z.; Zhu, A.D.; Hu, J.Y.; Lin, Y.J.; et al. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 2019, 10, 55. [Google Scholar] [CrossRef]
- Mercer, T. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef]
- Platt, E.J.; Smith, L.; Thayer, M.J. L1 retrotransposon antisense RNA within ASAR lncRNAs controls chromosome-wide replication timing. J. Cell Biol. 2018, 217, 541–553. [Google Scholar] [CrossRef]
- Kung, J.T.; Colognori, D.; Lee, J.T. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–659. [Google Scholar] [CrossRef]
- Akhade, V.S.; Pal, D.; Kanduri, C. Long noncoding RNA: Genome organization and mechanism of action. Adv. Exp. Med. Biol. 2017, 1008, 47–74. [Google Scholar]
- Wang, J.; Zhang, J.; Zheng, H. Mouse transcriptome: Neutral evolution of ‘non-coding’ complementary DNAs. Nature 2004, 431, 757. [Google Scholar] [CrossRef]
- Butcher, S.E.; Pyle, A.M. The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks. Acc. Chem. Res. 2011, 44, 1302–1311. [Google Scholar] [CrossRef]
- Holmes, Z.E.; Hamilton, D.J.; Hwang, T.; Parsonnet, N.V.; Rinn, J.L.; Wuttke, D.S.; Batey, R.T. The Sox2 transcription factor binds RNA. Nat. Commun. 2020, 14, 1805. [Google Scholar] [CrossRef]
- Liu, X.; Hong, C.; Jiang, Y.; Li, W.; Chen, Y.; Ma, Y.; Zhao, P.; Li, T.; Chen, H.; Liu, X.; et al. Co-expression module analysis reveals high expression homogeneity for both coding and non-coding genes in sepsis. BMC Genom. 2023, 24, 418. [Google Scholar] [CrossRef]
- Wen, J.; Wu, Y.; Luo, Q. DNA methyltransferases-associated long non-coding RNA PRKCQ-AS1 regulate DNA methylation in myelodysplastic syndrome. Int. J. Lab. Hematol. 2024, 28, 1102. [Google Scholar]
- Wu, S.C.; Kallin, E.M.; Zhang, Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010, 20, 1109–1116. [Google Scholar] [CrossRef]
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647. [Google Scholar] [CrossRef]
- Herman, A.B.; Tsitsipatis, D.; Gorospe, M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell. 2022, 82, 2252–2266. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 2011, 43, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Spitale, R.C.; Tsai, M.C.; Chang, H.Y. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics 2011, 6, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. Linc RNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Severing, E.; Faino, L.; Jamge, S.; Busscher, M.; Kuijer-Zhang, Y.; Bellinazzo, F.; Busscher-Lange, J.; Angenent, G.C.; Immink, R.G.H.; Pajoro, A. Arabidopsis thaliana ambient temperature responsive lncRNAs. BMC Plant Biol. 2018, 18, 145. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, C.; Xu, Z.; Wang, Y.; Liu, X.; Kang, Z.; Ji, W. Long non-coding genes implicated in response to stripe rust pathogen stress in wheat (Triticum aestivum L.). Mol. Biol. Rep. 2013, 40, 6245–6253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hu, W.; Hao, J.; Lv, S.; Wang, C.; Tong, W.; Wang, Y.; Wang, Y.; Liu, X.; Ji, W. Genome-wide identification and functional prediction of novel and fungi-responsive lincRNAs in Triticum aestivum. BMC Genom. 2016, 17, 238. [Google Scholar] [CrossRef]
- Yuan, J.; Li, J.; Yang, Y.; Tan, C.; Zhu, Y.; Hu, L.; Qi, Y.; Lu, Z.J. Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 2018, 93, 814–827. [Google Scholar] [CrossRef]
- Shin, S.Y.; Jeong, J.S.; Lim, J.Y.; Kim, T.; Park, J.H.; Kim, J.K.; Shin, C. Transcriptomic Analyses of Rice (Oryza Sativa) Genes and Non-Coding RNAs Under Nitrogen Starvation Using Multiple Omics Technologies. BMC Genom. 2018, 19, 532. [Google Scholar] [CrossRef]
- Wang, T.Z.; Liu, M.; Zhao, M.G.; Chen, R.J.; Zhang, W.H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef]
- Wekesa, J.S.; Luan, Y.; Chen, M.; Meng, J. A hybrid prediction method for plant lncRNA-protein interaction. Cells 2019, 8, 521. [Google Scholar] [CrossRef]
- Chao, Y.; Yuan, J.; Guo, T.; Xu, L.; Mu, Z.; Han, L. Analysis of transcripts and splice isoforms in Medicago sativa L. by single-molecule long-read sequencing. Plant Mol. Biol. 2019, 99, 219–235. [Google Scholar] [CrossRef]
- Yang, Y.W.; Liu, T.L.; Shen, D.Y.; Wang, J.Y.; Ling, X.T.; Hu, Z.Z.; Chen, T.Z.; Hu, J.L.; Zhang, B.L. Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathog. 2019, 15, e1007534. [Google Scholar] [CrossRef] [PubMed]
- Szcześniak, M.W.; Rosikiewicz, W.; Makałowska, I. CANTATAdb: A collection of plant long non-coding RNAs. Plant Cell Physiol. 2016, 57, e8. [Google Scholar] [CrossRef]
- Szcześniak, M.W.; Bryzghalov, O.; Ciomborowska-Basheer, J.; Makałowska, I. CANTATAdb 2.0: Expanding the collection of plant long noncoding RNAs. Methods Mol. Biol. 2019, 1933, 415–429. [Google Scholar]
- Li, X.Y.; Bu, D.C.; Sun, L.; Wu, Y.; Fang, S.S.; Li, H.; Luo, H.T.; Luo, C.L.; Fang, W.Z.; Chen, R.Z.; et al. Using the NONCODE database resource. Curr. Protoc. Bioinform. 2017, 58, 12161–121619. [Google Scholar]
- Paytuví Gallart, A.; Hermoso Pulido, A.; Anzar Martínez de Lagrán, I.; Sanseverino, W.; Aiese Cigliano, R. GREENC: A Wiki-based database of plant lncRNAs. Nucleic Acids Res. 2016, 44, D1161–D1166. [Google Scholar] [CrossRef] [PubMed]
- Quek, X.C.; Thomson, D.W.; Maag, J.L.; Bartonicek, N.; Signal, B.; Clark, M.B.; Gloss, B.S.; Dinger, M.E. lncRNAdb v2.0: Expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015, 43, D168–D173. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, Z.H.; Ling, Y. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef]
- Ogawa, Y.; Sun, B.K.; Lee, J.T. Intersection of the RNA interference and X-inactivation pathways. Science 2008, 320, 1336–1341. [Google Scholar] [CrossRef]
- Xuan, H.D.; Zhang, L.Z.; Liu, X.S.; Han, G.M.; Li, J.; Li, X.; Liu, A.G.; Liao, M.Z.; Zhang, S.H. PLNlncRbase: A resource for experimentally identified lncRNAs in plants. Gene 2015, 573, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell. 2014, 55, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Henriques, R.; Wang, H.; Liu, J.; Boix, M.; Huang, L.F.; Chua, N.H. The anti-phasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 2017, 216, 854–867. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of miR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 5351. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.-S.; Chen, R.; Harrison, M.J. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006, 45, 712–726. [Google Scholar] [CrossRef]
- Li, R.; Fu, D.Q.; Zhu, B.Z.; Luo, Y.B.; Zhu, H.L. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018, 94, 513–524. [Google Scholar] [CrossRef]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell. 2014, 30, 166–176. [Google Scholar] [CrossRef]
- Babaei, S.; Singh, M.B.; Bhalla, P.L. Role of long non-coding RNAs in rice reproductive development. Front. Plant Sci. 2022, 13, 1040366. [Google Scholar] [CrossRef]
- Ding, J.; Lu, Q.; Ouyang, Y.; Mao, H.; Zhang, P.; Yao, J.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. USA 2012, 109, 2654–2659. [Google Scholar] [CrossRef]
- Itoh, H.; Izawa, T. The coincidence of critical day length recognition for florigen gene expression and floral transition under long-day conditions in rice. Mol. Plant 2013, 6, 635–649. [Google Scholar] [CrossRef]
- Amasino, R.M.; Michaels, S.D. The timing of flowering. Plant Physiol 2010, 154, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Xi, Y.P.; Sung, S.B. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Li, J.R.; Lian, B.; Gu, H.Q.; Li, Y.; Qi, Y.J. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide screening and functional analysis identify a large number of long non-coding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Wang, R.H.; Mao, B.G.; Zhao, B.G.; Wang, J.B. Identification of lncRNAs involved in rice ovule development and female gametophyte abortion by genome-wide screening and functional analysis. BMC Genom. 2019, 20, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zhao, H.Y.; Cui, P.; Albesher, N.; Xiong, L.M. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef]
- Di, C.; Yuan, J.P.; Wu, Y.; Li, J.G.; Lin, H.X.; Hu, L.; Zhang, T.; Qi, Y.J.; Gerstein, M.B.; Guo, Y.; et al. Characterization of stress-responsive lncRNAs in Arabidopsis thaliana by integrating expression, epigenetic and structural features. Plant J. 2014, 80, 848–861. [Google Scholar] [CrossRef]
- Magar, N.D.; Shah, P.; Barbadikar, K.M.; Bosamia, T.C.; Madhav, M.S.; Mangrauthia, S.K.; Pandey, M.K.; Sharma, S.; Shanker, A.K.; Neeraja, C.N.; et al. Long non-coding RNA-mediated epigenetic response for abiotic stress tolerance in plants. Plant Physiol. Biochem. 2024, 206, 108165. [Google Scholar] [CrossRef]
- Patra, G.K.; Gupta, D.; Rout, G.R.; Panda, S.K. Role of long non coding RNA in plants under abiotic and biotic stresses. Plant Physiol. Biochem. 2023, 194, 96–110. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.Y.; Xu, J.; Wang, H.; Deng, S.L.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012, 24, 4333–4345. [Google Scholar] [CrossRef]
- Wu, R.N.; Wang, H.; Yang, C.C.; Wang, Z.Y.; Wu, Y.J. Construction of lncRNA-At5NC056820 overexpression vector in Arabidopsis thaliana and study on drought resistance of transgenic plants. Acta Bot. Boreal-Occident. Sin. 2017, 37, 1904–1909. [Google Scholar]
- Li, N.; Wang, B.K.; Wang, J.; Huang, S.Y.; Dai, L.; Pa, T.G.; Gao, J.; Yu, Q.H. Advances in functional research of long non-coding RNAs in plants. Plant Physiol. J. 2019, 55, 1427–1435. [Google Scholar]
- Wang, P.F.; Dai, L.M.; Ai, J.; Wang, Y.M.; Ren, F.S. Identification and functional prediction of cold-related long noncoding RNA (lncRNA) in grapevine. Sci. Rep. 2019, 9, 6638. [Google Scholar]
- Lu, X.K.; Chen, X.G.; Mu, M.; Wang, J.J.; Wang, X.G.; Wang, D.L.; Yin, Z.J.; Fan, W.L.; Wang, S.A.; Guo, L.X.; et al. Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0156723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; He, R.; Yang, J.; Cai, J.; Qu, Z.; Yang, R.; Gu, J.; Wang, Z.Y.; Adelson, D.L.; Zhu, Y.; et al. The long non-coding RNA DANA2 positively regulates drought tolerance by recruiting ERF84 to promote JMJ29-mediated histone demethylation. Mol. Plant 2023, 16, 1339–1353. [Google Scholar] [CrossRef]
- Liu, W.H.; Cheng, C.Z.; Lin, Y.L.; Xu, X.H.; Lai, Z.X. Genome-wide identification and characterization of mRNAs and lncRNAs involved in cold stress in the wild banana (Musa itinerans). PLoS ONE 2018, 13, E0200002. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, Y.; Liu, H. MIR398 and plant stress responses. Physiol. Plant. 2011, 143, 1–9. [Google Scholar] [CrossRef]
- Lu, Q.W.; Guo, F.Y.; Xu, Q.H.; Cang, J. LncRNA improves cold resistance of winter wheat by interacting with miR398. Funct Plant Biol. 2020, 47, 544–557. [Google Scholar] [CrossRef]
- Yang, H.; Cui, Y.; Feng, Y.; Hu, Y.; Liu, L.; Duan, L. Long Non-Coding RNAs of Plants in Response to Abiotic Stresses and Their Regulating Roles in Promoting Environmental Adaption. Cells 2023, 12, 729. [Google Scholar] [CrossRef]
- Wang, H.; Niu, Q.W.; Wu, H.W.; Liu, J.; Ye, J.; Yu, N.; Chua, N.H. Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 2015, 84, 404–416. [Google Scholar] [CrossRef]
- Huanca-Mamani, A.; Arias-Carrasco, R.; Cárdenas-Ninasivincha, S.; Rojas-Herrera, M.; Sepúlveda-Hermosilla, G.; Maracaja-Coutinho, V. Long non-coding RNAs responsive to salt and boron stress in the hyper-arid Lluteño maize from atacama desert. Genes 2018, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.N.; Zhang, X.P.; Wang, W.; Yuan, R.; Shen, F.F. Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol. 2018, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.J.; Xiao, W.F.; Qiu, J.R.; Xin, Y.; Liu, Q.P.; Chai, W.G.; Ruan, S.L. Advances in small RNA regulating plant immune response. Plant Physiol. J. 2018, 54, 539–548. [Google Scholar]
- Kwenda, S.; Birch, P.R.J.; Moleleki, L.N. Genome-wide identification of potato long intergenic noncoding RNAs responsive to Pectobacterium carotovorum subspecies brasiliense infection. BMC Genom. 2016, 17, 614. [Google Scholar] [CrossRef]
- Cui, J.; Luan, Y.S.; Jiang, N.; Bao, H.; Meng, J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017, 89, 577–589. [Google Scholar] [CrossRef]
- Zhang, L.Z.; Wang, M.J.; Li, N.N.; Wang, H.L.; Qiu, P.; Pei, L.L.; Xu, Z.; Wang, T.Y.; Gao, E.L.; Liu, J.X.; et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol. J. 2018, 16, 1172–1185. [Google Scholar] [CrossRef]
- Wang, J.Y.; Yang, Y.W.; Jin, L.M.; Ling, X.T.; Liu, T.L.; Chen, T.Z.; Ji, Y.H.; Yu, W.G.; Zhang, B.L. Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Biol. 2018, 18, 104. [Google Scholar] [CrossRef]
- Yu, Y.; Zhou, Y.-F.; Feng, Y.-Z.; He, H.; Lian, J.-P.; Yang, Y.-W.; Lei, M.-Q.; Zhang, Y.-C.; Chen, Y.-Q. Transcriptional landscape of pathogen-responsive lncRNAs in rice unveils the role of ALEX1 in jasmonate pathway and disease resistance. Plant Biotechnol. J. 2020, 18, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zang, S.; Zou, W.; Pan, Y.B.; Yao, W.; You, C.; Que, Y. Long Non-Coding RNAs: New Players in Plants. Int. J. Mol. Sci. 2022, 23, 9301. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Böhmdorfer, G.; Rowley, M.J.; Kuciński, J.; Zhu, Y.; Amies, I.; Wierzbicki, A.T. RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J. 2014, 79, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Spannagl, M.; Nussbaumer, T.; Bader, K.C.; Martis, M.M.; Seidel, M.; Kugler, K.G. PGSB PlantDB: Updates to the database framework for comparative plant genome research. Nucleic Acids Res. 2016, 44, 1141–1147. [Google Scholar] [CrossRef]
- Roulé, T.; Crespi, M.; Blein, T. Regulatory long non-coding RNAs in root growth and development. Biochem. Soc. Trans. 2022, 50, 403–412. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; He, R.; Qu, Z.; Gu, J.; Jiang, L.; Zhan, X.; Gao, Y.; Adelson, D.L.; Li, S.; Wang, Z.Y.; et al. Long noncoding RNA ARTA controls ABA response through MYB7 nuclear trafficking in Arabidopsis. Dev. Cell. 2023, 58, 1206–1217. [Google Scholar] [CrossRef]
- Li, N.; Liu, T.; Guo, F.; Yang, J.; Shi, Y.; Wang, S.; Sun, D. Identification of long non-coding RNA-microRNA-mRNA regulatory modules and their potential roles in drought stress response in wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 1011064. [Google Scholar] [CrossRef]
- Shin, W.J.; Nam, A.H.; Kim, J.Y.; Kwak, J.S.; Song, J.T.; Seo, H.S. Intronic long noncoding RNA, rice flowering associated (RIFLA), regulates OsMADS56-mediated flowering in rice. Plant Sci. 2022, 320, 111278. [Google Scholar] [CrossRef] [PubMed]
- Fedak, H.; Palusinska, M.; Krzyczmonik, K.; Brzezniak, L.; Yatusevich, R.; Pietras, Z.; Kaczanowski, S.; Swiezewski, S. Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci. USA 2016, 113, E7846–E7855. [Google Scholar] [CrossRef]
- Wu, H.W.; Deng, S.; Xu, H.; Mao, H.Z.; Liu, J.; Niu, Q.W.; Wang, H.; Chua, N.H. A noncoding RNA transcribed from the AGAMOUS (AG) second intron binds to CURLY LEAF and represses AG expression in leaves. New Phytol. 2018, 4, 1480–1491. [Google Scholar] [CrossRef]
- Allmer, J. Noncoding RNA Databases. Curr. Pharm. Biotechnol. 2023, 24, 825–831. [Google Scholar] [CrossRef]
- Budak, H.; Kaya, S.B.; Cagirici, H.B. Long Non-coding RNA in Plants in the Era of Reference Sequences. Front. Plant Sci. 2020, 11, 276. [Google Scholar] [CrossRef] [PubMed]
| Classification | LncRNA Type | References |
|---|---|---|
| lncRNA function | Decoy archetype | [25,26,27] |
| Scaffold archetype | ||
| Guide archetype | ||
| Signal archetype | ||
| Position on genome | Enhancer lncRNA | [28,29] |
| Co-transcriptional lncRNA Anti-sense lncRNA | ||
| Intronic transcript lncRNA | ||
| Large intergenic ncRNA |
| Database | Website | References |
|---|---|---|
| CANTATA | http://yeti.amu.edu.pl/CANTATA/ accessed on 12 September 2024 | [54,55] |
| NONCODE | http://www.noncode.org/ accessed on 12 September 2024 | [56] |
| GreeNC | http://greenc.sequentiabiotech.com/wiki2/Main_Page accessed on 5 November 2024 | [57] |
| PLncDB | https://bis.zju.edu.cn/PlncRNADB/index.php accessed on 21 September 2024 | [58] |
| JustRNA | http://JustRNA.itps.ncku.edu.tw accessed on 19 October 2024 | [16] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Z.; Yang, Y.; Iqbal, A.; Wu, Q.; Zhou, L. Biological Insights and Recent Advances in Plant Long Non-Coding RNA. Int. J. Mol. Sci. 2024, 25, 11964. https://doi.org/10.3390/ijms252211964
Zhao Z, Yang Y, Iqbal A, Wu Q, Zhou L. Biological Insights and Recent Advances in Plant Long Non-Coding RNA. International Journal of Molecular Sciences. 2024; 25(22):11964. https://doi.org/10.3390/ijms252211964
Chicago/Turabian StyleZhao, Zhihao, Yaodong Yang, Amjad Iqbal, Qiufei Wu, and Lixia Zhou. 2024. "Biological Insights and Recent Advances in Plant Long Non-Coding RNA" International Journal of Molecular Sciences 25, no. 22: 11964. https://doi.org/10.3390/ijms252211964
APA StyleZhao, Z., Yang, Y., Iqbal, A., Wu, Q., & Zhou, L. (2024). Biological Insights and Recent Advances in Plant Long Non-Coding RNA. International Journal of Molecular Sciences, 25(22), 11964. https://doi.org/10.3390/ijms252211964





