Abstract
The increasing salinity of agricultural lands highlights the urgent need to improve salt tolerance in crops, a critical factor for ensuring food security. Epigenetic mechanisms are pivotal in plant adaptation to salt stress. This review elucidates the complex roles of DNA methylation, histone modifications, histone variants, and non-coding RNAs in the fine-tuning of gene expression in response to salt stress. It emphasizes how heritable changes, which do not alter the DNA sequence but significantly impact plant phenotype, contribute to this adaptation. DNA methylation is notably prevalent under high-salinity conditions and is associated with changes in gene expression that enhance plant resilience to salt. Modifications in histones, including both methylation and acetylation, are directly linked to the regulation of salt-tolerance genes. The presence of histone variants, such as H2A.Z, is altered under salt stress, promoting plant adaptation to high-salinity environments. Additionally, non-coding RNAs, such as miRNAs and lncRNAs, contribute to the intricate gene regulatory network under salt stress. This review also underscores the importance of understanding these epigenetic changes in developing plant stress memory and enhancing stress tolerance.
1. Introduction
Soil salinization is a critical issue in agriculture, with research by Negacz et al. indicating that approximately 17 million square kilometers of soil are affected by salinity [1]. This underscores the need to enhance crop salt tolerance to ensure global food security. Under salt stress, plants can regulate gene expression through epigenetic regulations, which involve alterations in chromatin architecture without changes in DNA sequences [2]. The well-known epigenetic regulatory strategies include DNA methylation, histone modifications, histone variants, and non-coding RNAs, all essential for plant adaptation to high-salt environments. DNA methylation, which adds a methyl group to the DNA sequence, is vital for gene expression regulation and environmental stress responses, particularly in plants with complex genomes [3]. There are many types of DNA methylation. We primarily focus on 5mC in this review. Histones, as proteins that package DNA, are regulated by post-translational modifications, such as methylation and acetylation, and histone variants, including H2A.Z and H2A.X, to modulate chromatin structure and accessibility [4]. Non-coding RNA (ncRNA), such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), do not encode proteins but contribute to the complex gene regulatory network under salt stress by modulating key genes involved in hormonal response pathways and stress signaling. These epigenetic regulations not only modulate gene expression and rapidly adjust the physiological responses, enhancing their salt tolerance, but also play a crucial role in establishing plant stress memory. This enables plants and their offspring to adapt more swiftly to recurring stress. In summary, epigenetic regulation is pivotal in the plant response to salt stress, affecting both immediate physiological reactions and long-term adaptability and evolution. This knowledge provides vital strategies for agricultural breeding, facilitating the development of salt-tolerant crop varieties and contributing to global food security.
2. DNA Methylation
2.1. DNA Methylation in Plants
There are various types of DNA methylation, with 6mA and 5mC being the most extensively studied [3]. Among these, 5mC is notably prevalent under salinity stress. The 5mC modification is commonly observed in all three contexts of plant DNA sequences: symmetrical CG and CHG, as well as asymmetrical CHH (where H represents A, T, or C). Distinct sequence contexts necessitate specific methylases for maintenance. For instance, MET1 (methyltransferase 1) is charged with responsible for maintaining CG methylation, while CMT2 (chromomethylase 2) and CMT3 (chromomethylase 3) are involved in maintaining methylation at CHG sites. CHH methylation is regulated by DRM2 or CMT2, depending on the genomic context [3,5]. DRM2 is known to catalyze CHH methylation at RdDM (RNA-directed DNA methylation) target regions [6], while CMT2 maintains CHH methylation at heterochromatin containing histone H1.
Studies indicate that alterations in DNA methylation are closely linked to promoters and transposable elements (TEs) [7,8]. DNA demethylation occurs through two main mechanisms: active demethylation and passive demethylation. The processes of these mechanisms differ between plants and animals [9]. In plants, passive demethylation occurs during DNA replication and is attributed to either reduced DNA methyltransferase activity or lacking methyl donors [10]. Active demethylation, on the other hand, involves the enzymatic removal of methylated cytosine, facilitated by DNA glycosylases such as Repressor of Silencing 1 (ROS1), Demeter (DME), Demeter-like 2 (DML2), and Demeter-like 3 (DML3) [11]. This process is subsequently completed through a basal excision repair (BER)-dependent mechanism [12]. Increasing evidence suggests that active DNA demethylation is crucial in various biological processes across plant species, including fruit ripening, auxin-mediated development, and responses to environmental stress [13].
S-Adenosyl-L-methionine (SAM) serves as a crucial methyl donor for cytosine and lysine methylation in plants [14]. The tomato synthase of S-Adenosyl-L-methionine is induced by salinity stress, which positively correlated with increased lignin deposition in the vascular tissues under such conditions [15]. Furthermore, overexpression of tomato SAM synthetase SlSAMS1 enhances salt tolerance by promoting gene body DNA methylation and the expression of the circadian rhythm core component SlGI [16,17]. Similarly, the downregulation of barley SAM synthetase HvSAMS3 significantly impairs plant tolerance to drought and salt stress [18]. These findings underscore the potential regulatory role of SAM in plants subjected to salt stress.
2.2. Global Alterations of DNA Methylation under Salt Stress
Salt stress has been demonstrated to globally impact DNA methylation across various plant species, leading to alterations in gene expression. In alfalfa (Medicago sativa), a genome-wide increase in DNA methylation was observed under salt stress, particularly under high-salinity conditions [19]. The high-salt environment may influence DNA methylation levels by affecting the activity of C5-methyltransferases (C5-MTases) or other DNA methyltransferases [20]. In Pyrus betulaefolia (a wild pear), methyltransferases (MTases) and salt-responsive genes were upregulated in response to a high-salt environment. The DNA methylation levels of salt-responsive genes were altered under salt stress, and the precise mechanisms by which methyltransferases regulate the salt response in P. betulaefolia remain unclear [21].
Salt stress induces changes in DNA methylation levels in plants, with salt-tolerant plants generally exhibiting increased methylation and salt-sensitive plants showing decreased methylation [22]. This suggests that DNA methylation is crucial for plant adaptation to salt stress. Recent studies indicate that hypermethylation occurs at a significantly higher rate in salt-tolerant accessions in comparison with salt-sensitive ones in sugar beet. Thus, hypomethylation is more commonly observed in salt-sensitive accessions of sugar beet [23,24]. In cotton, under salinity treatment, various types of DNA methylation were observed in three cultivars: salt-tolerant CCRI 35, Zhong 07, and salt-sensitive CCRI 12. Generally, the global DNA methylation levels of the salt-tolerant cultivars CCRI 35 and Zhong 07 significantly increased under salinity stress, while no significant change was noted in the salt-sensitive cultivar CCRI 12 [24]. Additionally, salt stress caused a notable decrease in DNA methylation in the salt-sensitive rice cultivar IR29, compared to the salt-tolerant cultivar FL478 [25]. These observations suggest that the salt-tolerance plant may possess a unique mechanism that enhances DNA methylation under high-salinity conditions. However, the specific mechanism by which salt stress affects DNA methylation remains unclear and represents a potential avenue for future research. Contrarily, in some cases, the level of 5mC methylation increased in the salt-sensitive wheat cultivar, whereas it decreased in the salt-tolerant wheat cultivar SR3 under salt stress [26].
Alterations of DNA methylation induced by salt stress exhibit both tissue and growth stage specificity. Differences in the DNA methylation of genes related to growth and development between two different rice varieties result in significant variations in their salt tolerance [25]. Konate et al. found that the frequency of DNA methylation increased in barley leaves compared to roots, indicating that salt-induced DNA methylation is tissue-specific [27]. Additionally, Chen et al. reported that under salt stress, 61.2% of CGs, 39.7% CHG, and 3.2% CHH sites Glycine roots were methylated, with these levels being significantly lower than those in control conditions [28].
2.3. Regulation of Key Stress-Responsive Genes by DNA Methylation
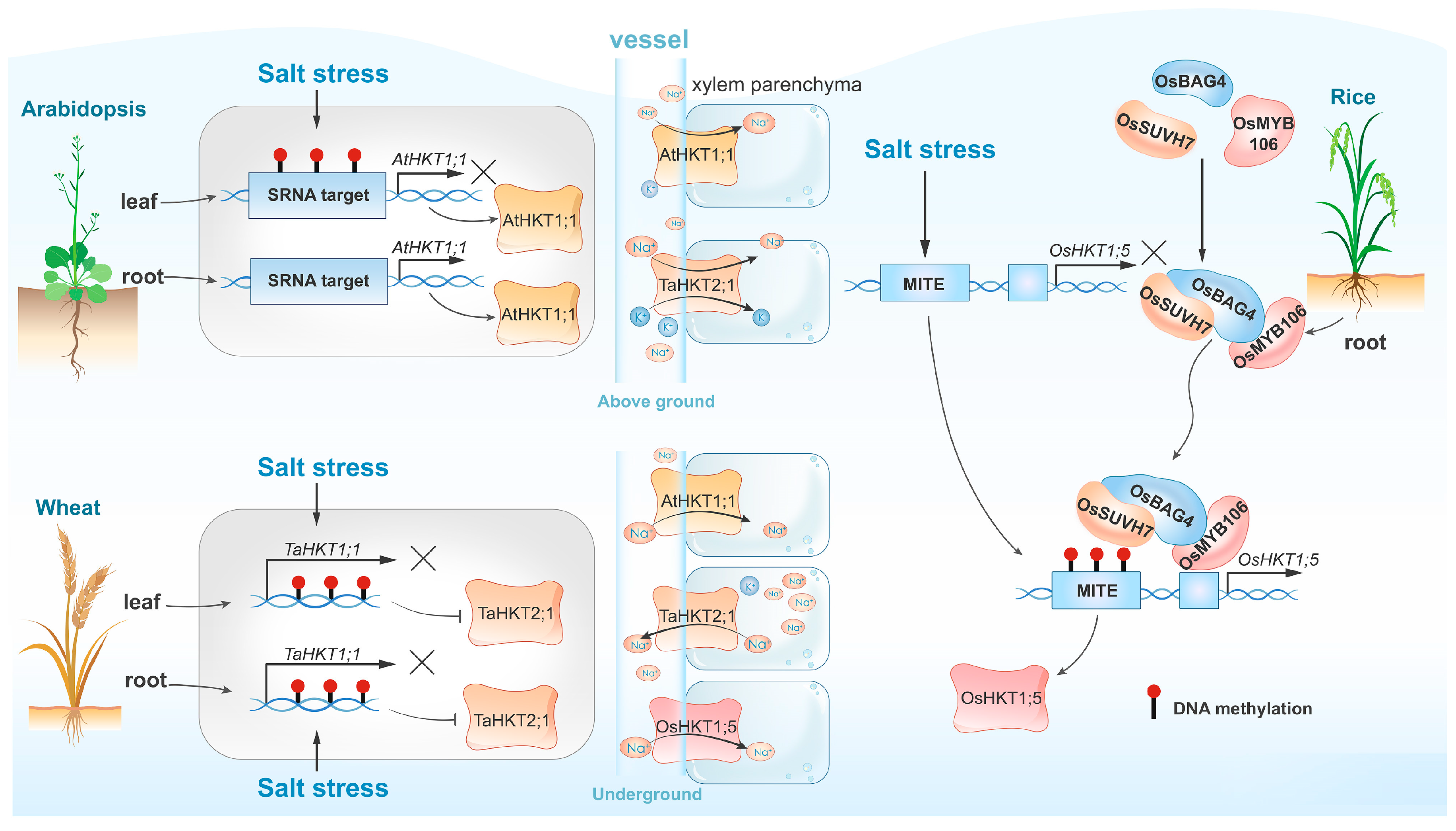
High-salinity environments induce genome-wide DNA hypermethylation of transposable elements (TEs), with cytosine methylation within TEs accounting for nearly one third of all cytosine methylation [29]. OsBAG4 is a key regulator associated with DNA methylation of TEs [30]. Previous research has demonstrated that OsBAG4 functions as a positive regulator of salt-stress tolerance by acting upstream of OsHKT1;5, which encodes the Na+ transporter essential for expelling Na+ from leaves and maintaining Na+/K+ homeostasis under salt stress [31]. OsMYB106, a MYB transcription factor, and OsSUVH7, a DNA methylation reader, both interact with OsBAG4, enhancing its DNA binding affinity and forming a stable complex. OsMYB106 binds to the cis-regulatory sequences of OsHKT1;5, while OsSUVH7 associates with the methylated miniature inverted-repeat transposable element (MITE) [32] (Figure 1). HKTs also play a crucial role in wheat. Kumar et al. have identified genotype- and tissue-specific increases in DNA methylation triggered by salt stress, leading to the downregulation of TaHKT2;1 and TaHKT2;3 expressions in both shoot and root tissues of the wheat cultivars Kharchia-65, therefore enhancing salt tolerance [33] (Figure 1). In Arabidopsis, heavy methylation of the HKT1 promoter across different sequence contexts may inhibit transcription in leaves and roots, whereas non-CG methylation may contribute to the fine regulation of HKT1 expression in leaves (Figure 1). This regulation is crucial for long-term adaptation to salt stress but is not essential for short-term salt tolerance [22]. Besides HKTs, other salt-responsive genes regulated by DNA methylation include the flavanol synthase genes TaFLS1 and TaWRS15 in wheat and barley (Hordeum vulgare) [26,34,35].
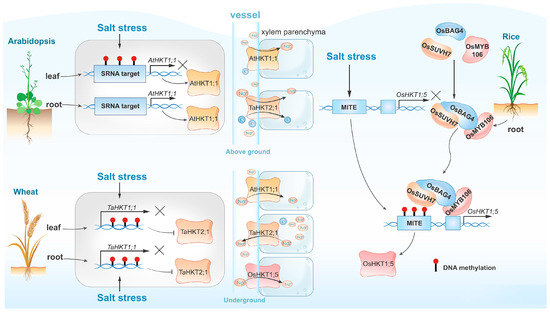
Figure 1.
Comparison of DNA methylation-dependent HKT expression and salt tolerance across various plant species. In Arabidopsis (top left), salt-induced DNA methylation exhibits organ heterogeneity, resulting in the specific expression of AtHKT1;1 in roots but not in leaves. In contrast, salt-induced DNA methylation suppresses the expression of the TaHKT2;1 gene in both wheat leaves and roots (bottom left). The rice regulatory complex consisting of OsBAG4, OsMYB106, and OsSUVH7 recognizes salt-induced methylation in MITE sequences, therefore activating the expression of OsHKT;5. Variations in the expression levels of HKT genes influence Na+ transport within the vascular system, consequently impacting the plant’s salt tolerance.
2.4. Plant Stress Memory by DNA Methylation
The MTases also play a crucial role in heritability, as they enable DNA methylation changes induced by environmental disturbances to be sustained over time or passed to subsequent generations. This allows both the original organism and its progeny to better adapt to recurring environmental conditions. The adaptive phenomenon, which enables plants to “remember” past experiences and recall mechanisms of responding to environmental stress, is referred to as plant stress memory (PSM) [36]. Using Arabidopsis thaliana as a model, approximately 75% of salinity stress-induced differential methylated cytosine positions are inherited, although some of these changes may be lost in future generations [37] (Table 1). In addition to salt stress, CHG demethylation observed in heavy metal-treated rice leaf tissues can be transmitted to the next generation, indicating a meiosis-based inheritance mechanism [38]. Similarly, heavy metal-induced methylation changes in TEs are also inherited in rice [39]. In the annual plant Polygonum persicaria, heritable DNA methylation changes have been observed as well. Longer root systems and greater biomass are exhibited in the progeny of drought-stressed parents in comparison with progeny from non-drought-treated parents of the same genetic line [26,40]. Moreover, drought-induced changes in DNA methylation are inherited in subsequent generations, which modulate the expression of drought-responsive genes [41,42]. Most memory studies have focused on 5mC modification, leaving the role of 6mA largely unexplored. Notably, the number of generations over which DNA-methylated memory can be inherited is also an interesting question. Further exploration is needed to elucidate the molecular mechanisms underlying the formation and maintenance of plant stress memory.

Table 1.
Memorized salt stress by DNA methylation and histone acetylation.
3. Histone Methylation
3.1. Histone Methylation in Plants
The stability of chromatin is enhanced through the interaction between the negatively charged phosphate groups of DNA molecules and the positively charged amino acids in histone proteins. Post-translational modifications (PTMs) of both histone tail regions and histone fold domains are essential to regulating chromatin structure and its accessibility for various biological processes [48]. Histone methylation influences the association between DNA and histones by altering local hydrophobicity [48,49]. Consequently, histone methylation is associated with either actively transcribed or repressed genes based on the specific methylated amino acid residue [50]. Although histone methylation does not directly cause transcriptional activation or repression, it modulates the transcriptional potential of genes [51]. The methylation marks are added to lysine or arginine residues in histone H3 or H4 by specific enzymes, i.e., histone lysine methyltransferases (HKMTs) or protein arginine methyltransferases (PRMTs) [52]. These methylation modifications can occur at different amino acids and involve various methylation states, including mono-, di-, or tri-methylation. Once established, the marks can be recognized by reader protein and removed by histone demethylases (HDMs) [52].
Different types of histone methylation have distinct biological functions [53,54]. In plants, the correlation between histone methylation and gene activation/repression depends on the specific methylation mark [55]. For instance, H3K4me3 and H3K36me3 are generally associated with active transcription, whereas genes marked with H3K27me3 or H3K9me3 often exhibit low transcript levels [56]. Significant differences exist between plants and animals. In animals, all forms of H3K4me modifications are associated with gene activation, whereas in plants, only H3K4me3 correlates with active transcription [57,58]. Regarding H3K9, about 40% of Arabidopsis coding genes are marked by H3K9me3, and only a minor fraction of the markers are found on TEs and pseudogenes [58,59]. Additionally, differences between plants and animals are also observed in H3K27 and H3K36 methylation patterns [60,61,62,63].
3.2. Global Alterations of Histone Methylation under Salt Stress
Recent studies have illustrated that changes in histone methylation are closely linked to activation or repression of gene expression [55,64,65]. The levels of H3K4me3 are increased while H3K27me3 levels are decreased, influencing downstream genes such as RD29A/RD29B, AtHKT1, and RSM1 in response to high-salinity stress [64,66,67]. In Arabidopsis, some salt-related genes, such as SUVH2/8 and MSH6, are downregulated with enhanced H3K9me2 under salt stress [68]. Moreover, in soybeans subjected to high-salinity stress, histone marks, including H3K4me2 and H3K4me3, are significantly upregulated, coordinating several key biological processes, such as stress response, cell wall modification, and ion homeostasis [69]. The upregulation of Glyma20g30840, Glyma08g41450, and Glyma11g02400 genes under high-salinity stress is likely mediated by increased H3K4me3 and decreased H3K9me2 levels [67]. Furthermore, histone methylation changes at the OsBZ8 gene locus are identified in both salt-tolerant and sensitive rice cultivars Nonabokra and IR64, respectively. Notably, Nonabokra rice obtains lower H3K27me3 and higher H3K4me3, while IR64 rice obtains higher H3K27me3 [70]. Additionally, in alfalfa, the transcription factor MsMYB4 plays a crucial role in salt stress response, with its activation correlated with increased levels of H3K4me3 and H3K9ac at specific promoter sites [71]. The changes in H3K4me3 and H3K9ac are primarily a result of the active gene expression rather than a cause, as MsMYB4 expression was altered at 3 h after stress exposure [72].
3.3. Regulatory Mechanisms of Histone Methylation under Salt Stress
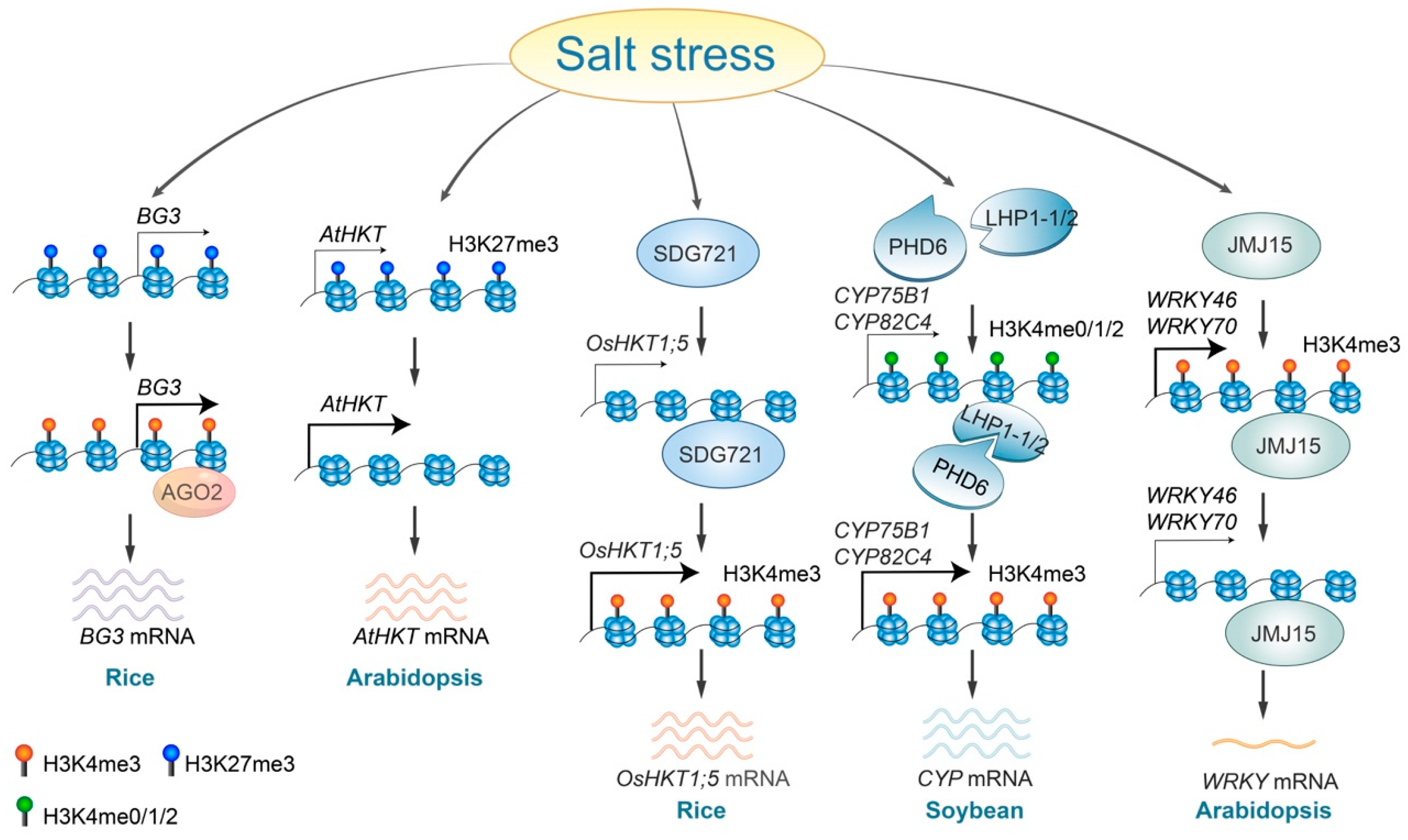
Salinity stress alters histone methylation levels through various regulatory pathways. In rice, the accumulation of AGO2 proteins at the BIG GRAINS3 (BG3) locus leads to enhanced BG3 expression. This is achieved by increasing H3K4me3 and decreasing H3K27me3 levels [73] (Figure 2). The histone demethylase JMJ15, which directly binds to and removes the H3K4me3 mark from the promoter and coding regions of WRKY46 and WRKY70, mediates the repression of these WRKY genes, therefore contributing to increased salt tolerance in plants [66,74] (Figure 2). Plant homeodomain (PHD) finger proteins function as histone code readers that identify and attach to H3K4 marks on the H3 tail [75]. The soybean GmPHD6 specifically recognizes low levels of H3K4 methylation (H3K4me0/1/2) through its N-terminal domain but does not recognize H3K4me3. The GmPHD6 protein engages with its coactivator, LHP1-1/2, via its PHD finger to assemble a transcriptional activation complex. Overexpression of two GmPHD6 target genes, CYP75B1 and CYP82C4, enhances stress tolerance of soybean [76] (Figure 2).
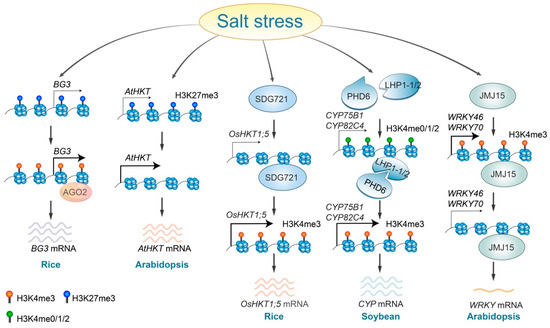
Figure 2.
Regulatory roles and mechanisms of histone methylation in response to salt stress. Salt stress triggers an AGO2-dependent increase in H3K4me3 levels at the BG3 locus, leading to the activation of BG3 expression. In Arabidopsis, salt stress removes H3K27me3 at the AtHKT locus, promoting its expression, whereas in rice, the SDG721 protein is necessary to elevate H3K4me3 levels, therefore enhancing OsHKT expression. Additionally, salt stress facilitates the binding of the PHD6-LHP1-1/2 complex, resulting in increased H3K4me3 levels at the CYP75B1 and CYP82C4 loci and subsequently enhancing their expression. Conversely, JMJ15 removes H3K4me3 from the WRKY46 and WRKY70 loci in response to salt stress, therefore reducing gene expression levels.
Under salt-stress conditions, SDG721, a SET DOMAIN GROUP protein possessing H3K4 methyltransferase activity, attaches to and adds the H3K4 mark within the promoter and coding regions of the OsHKT1;5, therefore regulating its expression levels [65,77]. During salt stress, removing H3K27me3 from AtHKT1, which is typically highly enriched, activates AtHKT1 gene expression in Arabidopsis [46] (Figure 2). Additionally, a recent study indicates that salt stress induced the accumulation of H3 methylglyoxalation at the genomic loci of some salt-stress-responsive genes, which subsequently enhanced chromatin accessibility and gene expression [78]. Although several core enzymes, such as PHD6, JMJ15, and SDG721, have been identified, the regulatory mechanisms by which salt stress alters histone methylation remain unsolved.
3.4. Memorized Stress by Histone Methylation
DNA methylation imparts heritable stress memory to plants; however, this effect does not extend in the same manner as histone methylation [43,44,45,46,79,80] (Table 1). H3K4me3 plays a role in transcriptional memory, but in drought stress, this memory is short-lived and cannot be transmitted to the progenies [81]. Specifically, H3K4me3 is crucial for the activation of drought memory genes GhP5CS1, GhNCED9, GhSnRK2, and GhPYL9-11A during repeated drought stress. The levels of H3K4me3 associated with drought stress memory are diminished by the fifth day of the recovery period [42]. In contrast, the transcriptional memory triggered by heat stress often persists for up to a week. This response is typically associated with increased levels of H3K4 methylation [82]. Ding et al. found increased levels of H3K4me3 modification at trainable gene loci under repeated stress treatment, which was maintained during the recovery phase [83]. Another classic example is cold stress-induced memory of vernalization. Prolonged exposure to cold conditions leads to the repression of FLC expression, which is subsequently restored when the temperature rises in spring. Histone marks H3K4me3, and H3K36me3 positively regulate FLC expression, while H3K27me3 exerts an opposing effect on FLC expression and stress memory [36,84]. Additionally, a recent study has demonstrated that light modulates the salt-induced transcriptional memory through HY5-mediated regulation of H3K4me3 mark at the memory gene P5CS1 [47].
4. Histone Acetylation
4.1. Histone Acetylation in Plants
The chromatin region with histone acetylation exhibits higher transcriptional activity. This is commonly attributed to the fact that the acetyl group neutralizes the positive charge of the histone, therefore altering the distance between nucleosomes and reducing the affinity between DNA and histone proteins [56,85]. Cells utilize two types of enzymes to modulate this dynamic process: histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs, which catalyze the acetylation of lysine residues, are further classified into four families: GNAT (GCN5-related N-acetyltransferase), MYST (Moz, Ybf2, Sas2, TIP60), p300/CBP, and TAFII250 [86]. Conversely, HDACs, which catalyze the removal of acetyl groups from lysine residues, are categorized into two classes: class I (including HDA 6/7/9/19) and class II (including 5/14/15/18) [87,88]. Despite some variations, HATs and HDACs exhibit conserved functions across a range of plant species, including Arabidopsis thaliana, Oryza sativa, foxtail millet [89], Gossypium arboreum, and kenaf [90]. These enzymes play essential roles in regulating various biological processes, such as ABA signaling transduction, SOS signaling transduction, ROS homeostasis maintenance, and LEA protein accumulation, all of which are critical for plant adaptation to salt stress [91,92,93]. Acetyl-coenzyme A (acetyl-CoA) is a pivotal metabolic intermediate that regulates essential cellular processes, including energy metabolism, mitosis, and autophagy. It functions as a crucial precursor for lipid synthesis and influences the acetylation profile of various proteins, notably histones [94,95]. The acetyl group from acetyl-CoA is transferred by histone acetyltransferases (HATs) to the ε-amino groups of lysine residues located at the N-terminal ends of histones. The acetyl-CoA utilized for histone acetylation is primarily generated by ATP-citrate lyase (ACL) in the tricarboxylic acid (TCA) cycle, occurring in either the mitochondria or the nucleus [96]. Consistently, mutations in the Arabidopsis ATP-citrate lyase subunit A (ACLA) result in decreased acetylation at H3K27 [97]. Nevertheless, direct evidence elucidating the acetyl-CoA promoted histone acetylation in response to salt stress remains to be unrevealed.
4.2. Global Alteration of Histone Acetylation under Salt Stress
Histone acetylation predominantly occurs at the K9, K14, and K27 residues of histone H3 and the K16 residue of histone H4. There is some acetylation in the plant salt regulatory gene region, and the acetylation pattern of the plant salt regulatory gene region will change greatly or slightly under salt stress, which will affect the plant’s tolerance to salt [70]. Recent studies have identified significant changes in the deacetylation of H3K9 and H3K14 under salt stress, resulting in the suppression of genes within the affected genomic regions. Notably, stress-induced acetylation of histone H3 is relatively rare compared to the deacetylation and subsequent gene repression observed under salt stress [98].
4.3. Regulatory Roles and Mechanisms of Histone Acetylation under Salt Stress
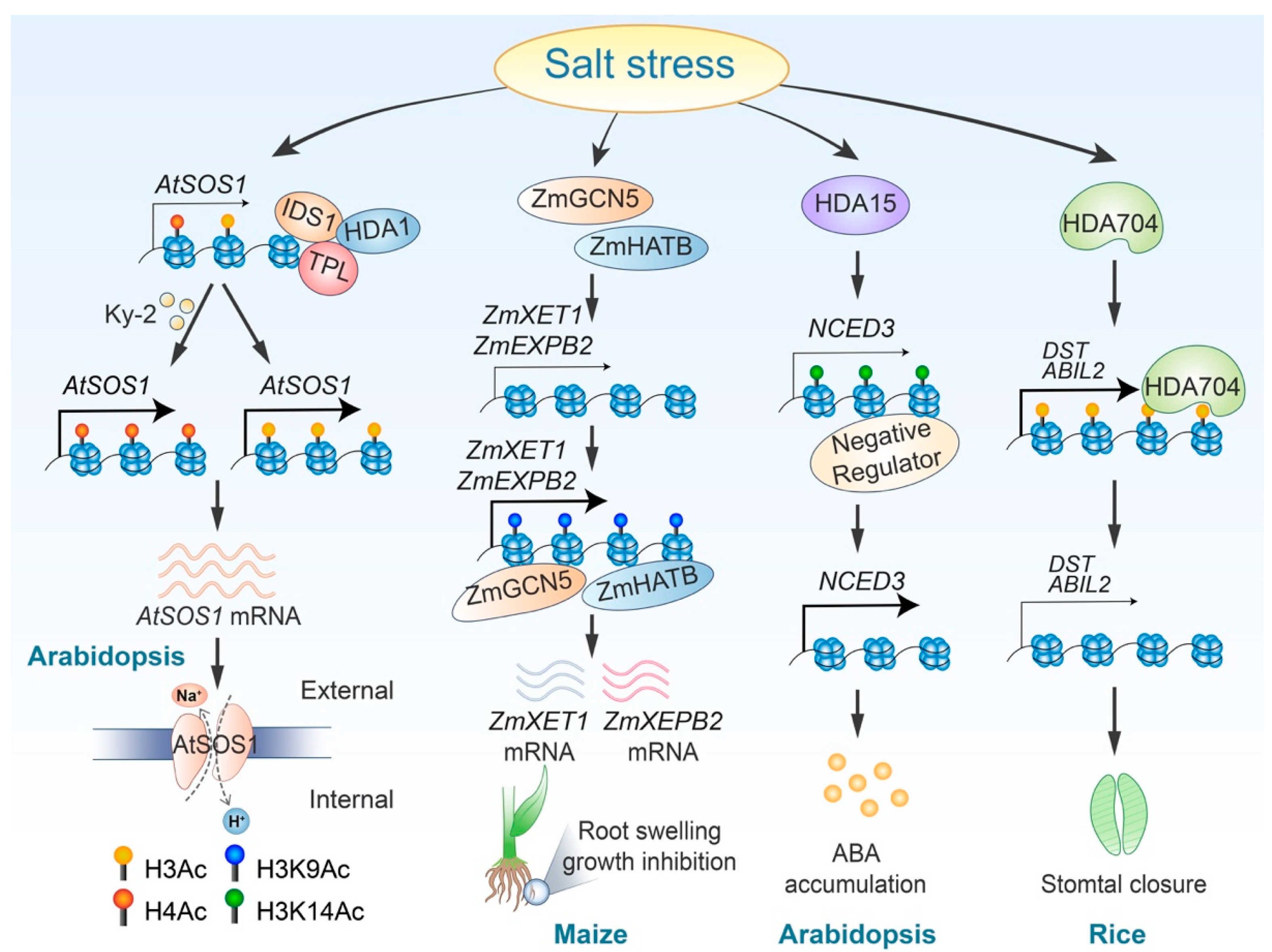
Histone acetylation plays a crucial role in regulating plant responses to salt stress by modulating various signaling pathways. Both histone acetylation and deacetylation serve as core regulators for ABA signaling pathways. For instance, Arabidopsis histone deacetylase HDA15 enhances deacetylation at the genomic region of NCED3, a gene involved in ABA biosynthesis. This process inhibits the binding of negative regulators to this genomic locus, ultimately promoting NCED3 expression and ABA synthesis [99]. Conversely, another histone deacetylase, HDA710, which is induced by high salt stress and phytohormones such as jasmonic acid (JA) and abscisic acid (ABA), catalyzes the deacetylation of histones H3 and H4, therefore negatively regulating ABA signaling [100]. Furthermore, a poplar RPD3/HDA1-type histone deacetylase, 84KHDA909, has been shown to increase ABA accumulation in plants and alter the transcript abundance of ABA response genes when transferred to Arabidopsis [101]. In addition to its role in ABA signaling, histone acetylation and deacetylation also influence the SOS signaling pathway. Acetylation of histone H4 is essential for the activation of the SOS1 gene [102]. Meanwhile, the transcription factor IDS1 (INDETERMINATE SPIKELET1), which belongs to the apetala2/ethylene response factor family, can collaborate with histone deacetylase HDA1 to repress SOS1 (SALT OVERLY SENSITIVE1) expression by modulating H3 histone acetylation [93] (Table 2 and Figure 3).

Table 2.
Regulatory roles of plant histone acetylation in response to salt stress.
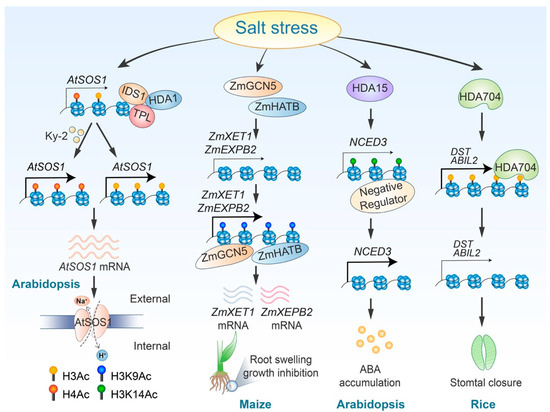
Figure 3.
Regulatory roles and mechanisms of histone acetylation in response to salt stress. Ky-2 treatment inhibits the deacetylation of AtSOS1, therefore enhancing plant salt tolerance. Additionally, salt stress triggers the removal of the HDA1-IDS1-TPL complex, resulting in elevated H3 acetylation levels at the AtSOS1 locus and increased AtSOS1 expression. ZmGCN5 and ZmHATB promote H3K9 acetylation at the ZmXET1 and ZmEXPB2 loci. HDA15 reduces the binding of a negative regulator to the NCED3 gene locus by removing H3K14 acetylation, consequently increasing NCED3 expression. Furthermore, salt stress induces deacetylation of DST and ABIL2 by HDA704, modulating stomatal closure.
In addition to their roles in signaling pathways, histone acetylation/deacetylation also regulates metabolites associated with salt stress. Plants utilize histone acetylation and deacetylation to manage reactive oxygen species (ROS). For instance, in wheat, TaHAG1 directly targets a subset of genes involved in hydrogen peroxide production, leading to H3 acetylation at these gene regions and, therefore, maintaining ROS homeostasis [108,112]. Similarly, in rice, analogous regulations have been observed [106]. Furthermore, the expression levels of beet POX genes, which are involved in ROS removal, are positively correlated with the levels of H3K9ac and H3K27ac [103]. In addition to regulating ROS removal, plants also modulate the synthesis of certain stress-responsive substances through histone acetylation and deacetylation to mitigate the effects of salt stress. Late embryogenesis abundant proteins (LEAs) are stress-induced proteins that enhance plant salt tolerance. The coordination of IDS1 and HDA1 negatively regulates LEA1 synthesis [93]. Similarly, another class I histone deacetylase, HDA19, exerts a comparable regulatory effect [87] (Table 2).
Under high-salinity conditions, root growth is inhibited, and root cells tend to swell, a process closely related to cell wall enlargement. In corn (Zea mays), the upregulation of the ZmEXPB2 and ZmXET1 genes, which are involved in cell wall modification under salt stress, is associated with increased H3K9 acetylation [105] (Figure 3). Wang et al. demonstrated that in Chrysanthemum morifolium, the heat shock factor A4 (CmHSFA4) recruits the corepressor TOPLESS (CmTPL) to inhibit the transcription of CmMYB121, a gene responsive to salt stress. This inhibition occurs through the reduction of H3 and H4 histone acetylation levels at the CmMYB121 locus [111]. Additionally, in rice, the circadian clock regulatory core component OsPRR73 interacts with the histone deacetylase HDA10 to suppress the transcription of the Na+ absorption transporter OsHKT2;1 in response to salt stress [107] (Table 2). Rice stomatal closure under salt stress is regulated by HDA704-mediated histone deacetylation of the DST and ABIL2 genes [110] (Figure 3). Altogether, the plant utilizes histone acetylation and deacetylation as “on” and “off” switches to regulate gene expression levels, therefore modulating various pathways in response to the salt stress.
Histone acetylation does not function in isolation but often coordinates with other DNA and histone modifications. For example, under salt stress, both tobacco and Arabidopsis cells exhibit rapid upregulation of histone H3 Ser-10 phosphorylation, which is followed by subsequent phosphorylation of H3 and acetylation of histone H4 [109] (Table 2). Additionally, activation of four DNA-methylated-controlled transcription factors has been found to correlate with increased levels of histone H3K4 trimethylation and H3K9 acetylation [67].
5. Histone Variants
5.1. Histone Variants in Plants
Histone variants are non-allelic protein isomers, such as H2A.X, H2A.Z, macroH2A, CENP-A, and H3.3, that play crucial roles in chromatin structural diversification and gene expression. While they share sequence homology and major structural similarity with core histones, histone variants possess unique distributions and functions. The differential expression of histone variants at specific tissue and developmental stages indicates their specialized roles in modifying the structural and functional properties of chromatin [113]. Among eukaryotes, H2A is the most diverse histone, with its variants performing specialized functions during nucleosome assembly and genome packaging [114]. Consequently, histone variants of H2A are more prevalent across organisms, whereas other histone variants exhibit less diversity. Notably, the H2A family shows the greatest sequence differentiation at their C-terminus.
5.2. Regulatory Roles and Mechanisms of Histone Variants under Salt Stress
H2A.Z is a highly conserved histone variant that plays a critical role in regulating plant growth and development. A recent study has shown that H2A.Z is essential for salt tolerance in Arabidopsis thaliana. Under salt stress, H2A.Z is deposited at the promoter region near transcriptional initiation site (TSS) sites, influencing transcriptional regulation. However, the accumulation of H2A.Z is often negatively correlated with gene expression under salt stress [115]. Notably, in rice, the deposition of H2A.Z at the TSS is modulated by several stress-responsive regulators. For instance, in osarp6 knockdown plants, the expression levels of stress-responsive genes, ABA INSENSITIVE 1 (ABI1) and ABA INSENSITIVE 2 (ABI2) are decreased [116]. The expression of the Arabidopsis transcription factor AtMYB44, which responds to salt stress, is regulated by H2A.Z deposition. Under salt stress, there is a marked reduction in H2A.Z deposition at the promoter of AtMYB44, which correlates with decreased occupancy of AtMYB44 in the same region [117]. This observation aligns with the general trend that gene expression levels are negatively correlated with H2A.Z enrichment under salt stress [118]. The rice H3 variant RH3.2A, which encodes the H3.2-type histone protein, shows upregulated expression in rice roots under both salt stress and ABA treatment [119].
Histone variants utilize specialized histone deposition mechanisms to ensure timely and site-specific binding to chromatin [120]. The regulation is partly mediated through the influence of histone modifications on nucleosomes and nucleosomes with specific deposition at relevant sites. ATP-dependent chromatin remodeling complexes modify nucleosome structure, therefore influencing the accessibility of packaged DNA sequences to trans-acting factors[121]. Histone variants further affect nucleosome dynamics following their deposition in conjunction with covalent post-translational modifications (PTMs) [119,122]. The precise mechanism by which histone variants mediate plant salt tolerance remains to be fully elucidated. However, transcriptomic analysis of histone variants in plants has provided valuable insights. For instance, Wang et al. conducted genome-wide characterization, phylogeny, and expression analysis of the histone gene family in cucumber [123].
6. Non-Coding RNAs
6.1. Non-Coding RNAs in Plants
MicroRNAs (miRNAs) are small, non-coding single-stranded RNAs, typically 21–24 nucleotides in length, that function as a gene regulator by modulating the abundance of their target genes. Plant miRNAs exhibit high complementarity to the specific sites of their target mRNAs, leading to the cleavage of most targeted mRNAs [124]. These miRNAs frequently target transcription factors involved in plant growth and development, therefore playing a crucial role in plant responses to abiotic stresses by regulating key transcription factors. Long non-coding RNAs (lncRNAs), which are non-coding RNAs longer than 200 nucleotides, are also vital in numerous biological processes, including dosage compensation, epigenetic regulation, cell cycle regulation, and cell differentiation. Both miRNAs and lncRNAs are significant regulatory elements in plants under salt stress, with many of these molecules showing altered expression in response to salt stress treatments.
6.2. Global Alterations of Non-Coding RNAs under Salt Stress
Under salt stress, the expression levels of miRNAs are significantly altered in a species-specific manner. In some plant species, the numbers of upregulated and downregulated miRNAs are comparable. For instance, in wheat, 49 miRNAs exhibited notable changes in expression levels under salt stress, with 25 showing significant upregulation and 24 showing significant downregulation [125]. A similar pattern was observed in the salt-sensitive broad bean Hassawi-3 [126]. Moreover, in contrast, most miRNAs are downregulated after salt stress treatment in some plant species. For example, in grapevine, 39 miRNAs were differentially expressed after salt stress, with 14 significantly upregulated and 25 significantly downregulated [127]. Comparable trends have been observed in the citrus root [128], rice [129], and the salt-tolerant cultivar Fraxinus velutina R7 [130]. Furthermore, in certain plant species, miRNA changes induced by salt stress are predominantly upregulated. In cotton, 51 miRNAs were upregulated, and 37 miRNAs were downregulated after 4 h of salt stress, while 48 miRNAs were significantly upregulated, and 27 miRNAs were downregulated after 5 days of long-term salt stress [131]. This pattern is similar to that observed in the Fraxinus velutina salt-sensitive cultivar S4 [130].
The salt-induced alterations of miRNA expression are time-dependent. For example, in fennel, six miRNAs were differentially expressed under salt stress; five were upregulated at 24 h, while one was downregulated. However, at 72 h post-salt stress, all studied miRNAs were upregulated [132]. A similar temporal pattern has been observed in members of the miR399 family in grapes [133]. Additionally, miRNA expression exhibits tissue specificity. A recent study reported the miRNA expression pattern in salt-tolerant Doc Phung (DP) rice under salt-stress conditions. Among 69 differentially expressed miRNAs, 50 miRNAs (five upregulated and 45 downregulated) were differentially expressed in shoot, while 28 miRNAs (13 upregulated and 15 downregulated) were differentially expressed in root tissue of the DP rice, respectively [129]. A similar pattern has been observed in carrot [134]. Notably, under salt stress, the expression pattern of a specific miRNA can vary across different plant species. For instance, miR156 is upregulated in Arabidopsis thaliana, Raphanus sativus, Saccharum spp., and Suaeda maritima but downregulated in Populus trichocarpa [124]. Similarly, miR159 is downregulated in Arabidopsis thaliana, Nicotiana tabacum, and Oryza sativa after salt stress, whereas it is upregulated in Panicum virgatum, Saccharum spp., and Suaeda maritima [135].
To regulate plant response to salt stress, the expression of long non-coding RNAs (lncRNAs) undergoes significant alteration. A study on tobacco has identified 2428 differentially expressed lncRNAs (DE-lncRNAs) in response to high-salinity treatment over time, with 2147 DE-lncRNAs detected in the roots and 495 in the leaves [136]. Functional predictions suggest that these DE-lncRNAs are involved in starch and sucrose metabolism pathways in roots and cysteine and methionine metabolism pathways in leaves. Additionally, under salt stress, 8724 lncRNA candidates were identified in the salt-tolerant rice species FL478, and 9235 lncRNA candidates were identified in the rice species IR19 [136]. In tomatoes, 154 and 137 lncRNAs exhibited differential expression in the M82 and S. pennellii varieties, respectively [137]. Functional analysis of target genes of these DE-lncRNAs in tomato indicates that some genes contribute to the salt-stress response by modulating the abscisic acid (ABA) signaling pathway [138].
6.3. Regulation of Core Stress-Responsive Genes by Non-Coding RNAs
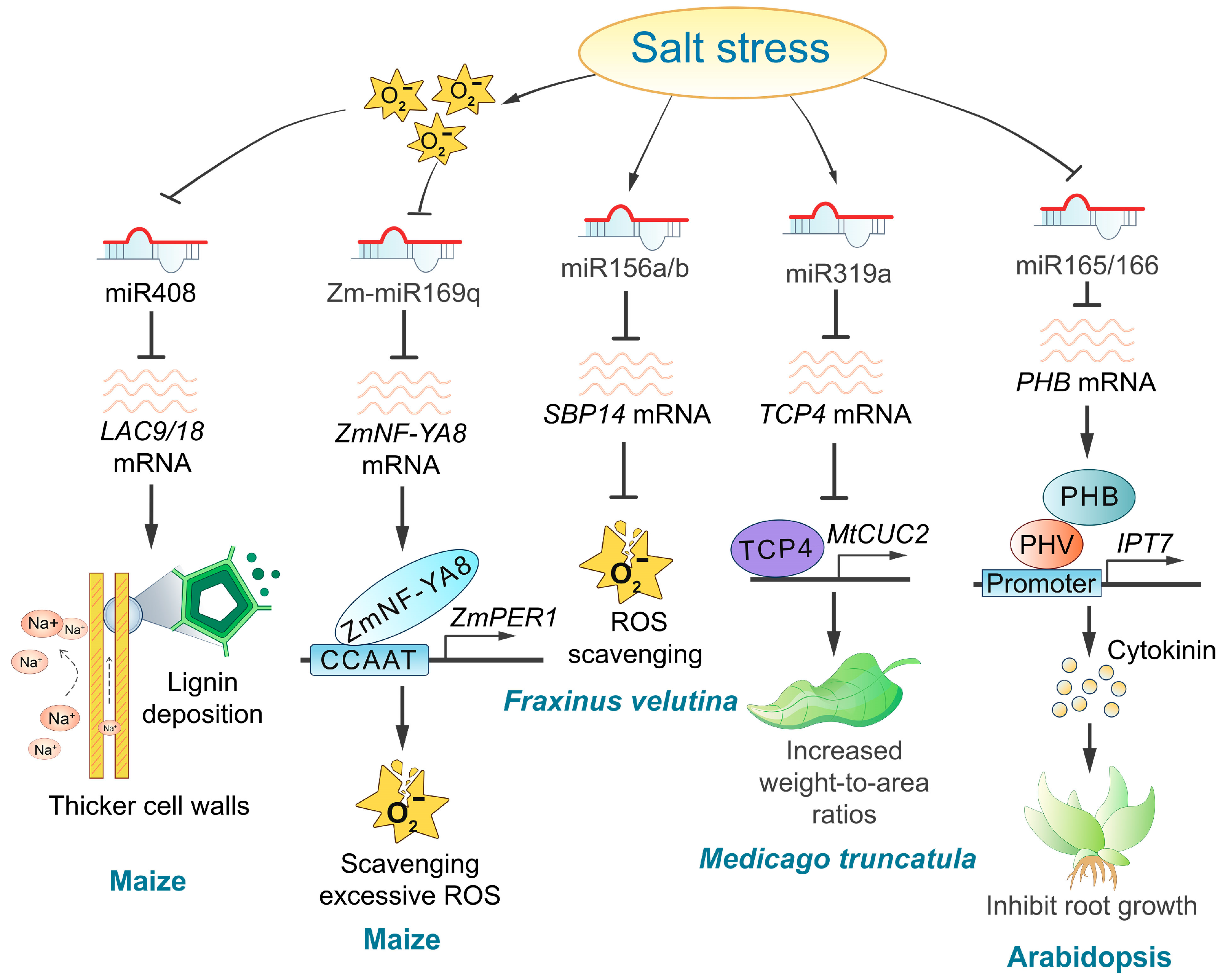
Several studies have demonstrated that miRNAs play a crucial role in plant responses to salt stress by regulating hormone response pathways [139,140,141]. Notable examples include Arabidopsis miR165/166, grape miR390/394, tomato miR164, and Fraxinus miR393a/TIR1 [127,130,142]. The salt-induced downregulation of miR166 causes an upregulation of PHB expression, therefore triggering a salt-dependent rise in cytokinin levels through IPT7 gene induction (Figure 4). Higher cytokinin levels at the transition zone activate the AHK3/ARR1/12 pathway, which promotes SHY2 expression, which triggers cell differentiation and inhibits root meristem activity in response to salts [143]. Oxidative stress, a secondary consequence of plant salt stress, significantly impacts miRNA expression levels [144,145]. During salt stress, the accumulation of reactive oxygen species (ROS) inhibits the transcription of pre-miR169q, leading to an increased abundance of its target, ZmNF-YA8. The upregulation of ZmNF-YA8, in turn, promotes the expression of ZmPERs, which enhances peroxidase (POD) enzyme activity and contributes to the plant’s response to salt stress [146]. Additionally, differentially expressed miRNAs influence ROS accumulation in plants. For example, miR156a/b targets SBP14 in Fraxinus velutina, downregulating SBP14 expression under salt stress and therefore facilitating ROS clearance [130] (Figure 4). Furthermore, the knockdown of miR164a in tomatoes has been shown to reduce ROS accumulation in transgenic plants [142]. Conversely, the role of ROS as an upstream regulator of miR408 in maize remains speculative, with the precise relationship between them yet to be clarified [147]. Overexpression of MIR408b in maize has been shown to reduce lignin deposition, decrease the thickness of the pavement cell wall, and lower the number of cells in vascular bundles under salt stress. This suggests that miR408 may influence the influx of high Na+ concentration by regulating maize cell wall lignification, therefore impacting salt tolerance [147] (Table 3 and Figure 4). Glutathione (GSH), a tripeptide, plays a multifaceted role in plants’ responses to environmental stresses [148,149]. It functions as an antioxidant by mitigating oxidative stress, preventing lipid peroxidation, and protecting the plasma membrane. These actions subsequently reduce passive Na+ influx, therefore enhancing salt tolerance in plants. Furthermore, GSH is essential for maintaining cellular redox homeostasis under salt stress [148]. In the context of salt stress, the salt-insensitive tomato species Lycopersicon pennellii exhibits upregulation of both GSH biosynthesis and the activity of metabolizing enzymes compared to salt-sensitive tomato varieties [150]. Additionally, in salt-tolerant carrot varieties, increased expression of miR266 downregulates its target gene gamma-glutamyl peptidase 1 (GGP1), resulting in elevated GSH levels and enhanced reactive oxygen species (ROS) scavenging efficiency [134]. Additionally, plants improve salt tolerance by regulating ion homeostasis, as demonstrated by the miR164d and the miR396a in Fraxinus velutina [130]. Notably, miR319 affects leaf phenotype and delays leaf senescence, therefore enhancing plant salt tolerance [151] (Figure 4).
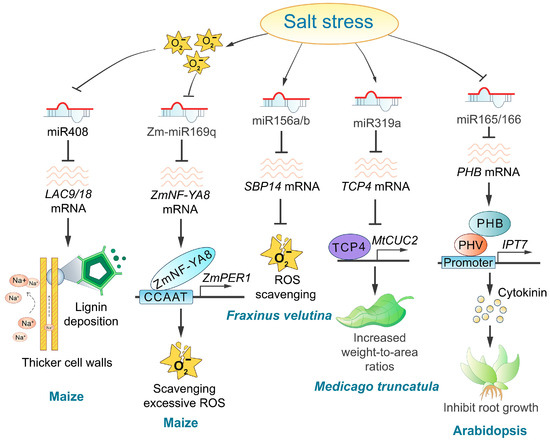
Figure 4.
Regulatory roles and mechanisms of miRNAs in response to salt stress. Several miRNAs, including miR408, Zm-miR169q, and miR156a/b, play crucial regulatory roles in response to oxidative stress under high-salinity conditions. Additionally, other miRNAs, such as miR319a and miR165/166, are involved in the regulation of plant leaf senescence and phytohormonal responses to salt stress.
Numerous lncRNAs have been identified as responsive to abiotic stress in plants, influencing ion transport and promoting signal transduction. Differentially expressed lncRNAs act as endogenous target mimics of certain miRNAs, such as rice miRNA osa-miR5809b, modulating their biological functions under salt stress [152]. In Medicago truncatula, the lncRNA MtCIR1 functions as a negative regulator of salt stress by enhancing abscisic acid (ABA) accumulation through the inhibition of the ABA catabolic enzyme CYP707A2, thus increasing seed germination sensitivity to salt stress. In Tribulus terrestris and Arabidopsis thaliana, MtCIR1 expression negatively regulates the salt-stress response by downregulating Na+ transporter genes, leading to higher Na+ accumulation in leaves and increased sensitivity of transgenic plants to salt-stress [153]. Differentially expressed poplar lncRNAs are observed between salt-tolerant and salt-sensitive cultivars [154] (Table 3). These lncRNAs may improve the adaptability of poplars to varying environmental conditions. Recent research has highlighted that poplar lncRNA.2-FL plays a crucial role in salt-stress tolerance in the FL478 cultivar by modulating 173 target genes in trans [155].
Both miRNA and lncRNA may play essential roles in plant stress memory. MiRNAs have been identified as key regulators of plant stress memory by mediating post-transcriptional silencing of target genes under stress conditions. Similarly, lncRNAs are involved in the formation of plant stress memories and contribute to enhanced stress tolerance [156]. Further research is needed to elucidate the unique regulatory mechanisms of miRNAs and lncRNAs in the formation of plant stress memory.

Table 3.
Regulatory roles of non-coding RNAs in response to salt stress.
Table 3.
Regulatory roles of non-coding RNAs in response to salt stress.
| Non-Coding RNA | Species | Changes under Salt Stress | Target Genes and Biological Functions | References |
|---|---|---|---|---|
| MiR156 | Malus domestica (apple) | Downregulation of MIR156a | Upregulation of MdSPL13. OE of MIR156a reduces salt tolerance | [157] |
| Zea mays | Downregulation of MIR156 | R2R3 Myb SBP-domain protein | [158] | |
| MiR164 | Solanum lycopersicum (tomato) | N.A. | KO of Sly-miR164a leads to reduced ROS and enhanced salt tolerance | [142] |
| Zea mays | Downregulation of MIR164 | NAC1, ARF8 | [158] | |
| MiR165/166 | Arabidopsis thaliana | Downregulation of MIR165A, MIR166A and MIR166B | Salt stress induces PHB expression and production of cytokinin. | [143] |
| MiR168 | Oryza sativa | N.A. | PINHEAD (OsAGO1). KD of miR168 leads to enhanced salt tolerance. | [159] |
| Zea mays | Upregulation of MIR168 | AGO1 | [158] | |
| MiR169 | Zea mays | Downregulation of zma-miR169 family members | ZmNF-YA1; ZmNF-YA4; ZmNF-YA6; ZmNF-YA7; ZmNF-YA11; ZmNF-YA13; ZmNF-YA14 | [160] |
| MiR172 | Glycine max (soybean) | Upregulation of gma-miR172a | SSAC1. OE of gma-miR172a leads to downregulation of SSAC1 and enhanced salt tolerance. | [161] |
| Glycine max (soybean) | Upregulation of miR172c | NNC1. OE of miR172c leads to enhanced salt tolerance. | [162] | |
| Oryza sativa | Upregulation of miR172a/b | IDS1. OE of miR172 leads to downregulation of IDS1 and enhanced salt tolerance. | [163] | |
| MiR319 | Arabidopsis thaliana | Upregulation of miR319 | [164] | |
| Medicago truncatula (model legume) | Downregulation of miR319 | TCP4. OE of Mtr-miR319a leads to the downregulation of TCP4 and enhanced salt tolerance. | [151] | |
| Solanum linnaeanum (eggplant) | Downregulation of miR319 | TCP family transcription factor | [165] | |
| Triticum aestivum | Upregulation of miR319a | [166] | ||
| Zea mays | Downregulation of miR319 | TCPs | [158] | |
| MiR390 | Populus spp. (poplar) | Upregulation of miR390 | TAS3. OE of miR390 leads to downregulation of ARFs (ARF3.1, ARF3.2, and ARF4) and enhanced salt tolerance. | [139] |
| MiR393 | Arabidopsis thaliana | Upregulation of MIR393A | TIR1, ABF2, ABF3. Loss of miR393ab leads to an increase of lateral root number under salt stress, whereas OE of miR393 leads to enhanced salt tolerance. | [140,167] |
| Oryza sativa | Upregulation of OsmiR393 | OsTIR1 and OsAFB2. OE of OsmiR393 leads to less tolerance to salt stress. | [168,169] | |
| MiR394 | Arabidopsis thaliana | Upregulation of miR394 | LCR. OE of miR394 leads to less tolerance to salt stress. | [141] |
| MiR395 | Zea mays | Upregulation of MIR395 | NADP-dependent malic protein, ATP sulfurylase | [158] |
| MiR396 | Chrysanthemum indicum | Upregulation of cin-miR396a | CiGRF1 and CiGRF5. OE of cin-miR396a leads to less tolerance to salt stress. | [170] |
| Oryza sativa | Upregulation of miR396b and downregulation of miR396c | GRF6. Loss of miR396 leads to enhanced salt tolerance. | [171,172] | |
| Zea mays | Upregulation of MIR396 | Cytochrome oxidase | [158] | |
| MiR397 | Arabidopsis thaliana | Upregulation of miR397 | LAC2, LAC4, and LAC17. OE of AtmiR397 leads to less tolerance to salt stress. | [173] |
| MiR399 | Arabidopsis thaliana | Upregulation of miR399f | CSP41b and ABF3. OE of miR399f leads to enhanced salt tolerance. | [174] |
| MiR408 | Zea mays | Downregulation of miR408 | ZmLAC9. OE of miR408a/b leads to enhanced salt tolerance. | [147,175] |
| Salvia miltiorrhiza | Upregulation of Sm-MIR408 | OE of Sm-miR408 leads to enhanced salt tolerance. | [144] | |
| MiR414 | Gossypium hirsutum (cotton) | Downregulation of ghr-miR414c | GhFSD1. OE of ghr-miR414c leads to less tolerance to salt stress. | [145] |
| MiR528 | Oryza sativa | Upregulation of miR528 | AO. OE of miR528 leads to enhanced salt tolerance. | [176] |
| MiR1118 | Triticum aestivum | Downregulation of miR1118 | PIP1;5. | [177] |
| MiR1848 | Oryza sativa | Upregulation of osa-miR1848 | OsCYP51G3. OE of osa-miR1848 leads to less tolerance to salt stress. | [178] |
| Lnc_388, Lnc_883, Lnc_973, Lnc_253 | Gossypium hirsutum (cotton) | Upregulation of Lnc_388, Lnc_883, Lnc_973, and Lnc_253 | LRR8 (Lnc_388), msD3 (Lnc_883), miR399 (Lnc_973), and miR156 (Lnc_253). Loss of Lnc_973 leads to less tolerance to salt stress. | [179,180] |
| LncRNA354 | Gossypium hirsutum (cotton) | Upregulation of LncRNA354 | CeRNA for miR160b. Loss of LncRNA354 leads to enhanced salt tolerance. | [181] |
| Ptlinc-NAC72 | Populus trichocarpa | Upregulation of Ptlinc-NAC72 | PtNAC72.A/B. OE of Ptlinc-NAC72 leads to less tolerance to salt stress. | [182] |
| PUPPIES | Arabidopsis thaliana | Upregulation of PUPPIES | DOG1. Loss of PUPPIES leads to reduced expression of DOG1. | [183] |
| LncRNA77580 | Glycine max (soybean) | N.A. | OE of LncRNA77580 leads to less tolerance to salt stress. | [184] |
| LncERF024 | Populus ssp. | Upregulation of LncERF024 | OE of LncERF024 leads to enhanced salt tolerance. | [154] |
| DRIR | Arabidopsis thaliana | Upregulation of DRIR | OE of DRIR leads to enhanced salt tolerance. | [185] |
7. Conclusions and Perspectives
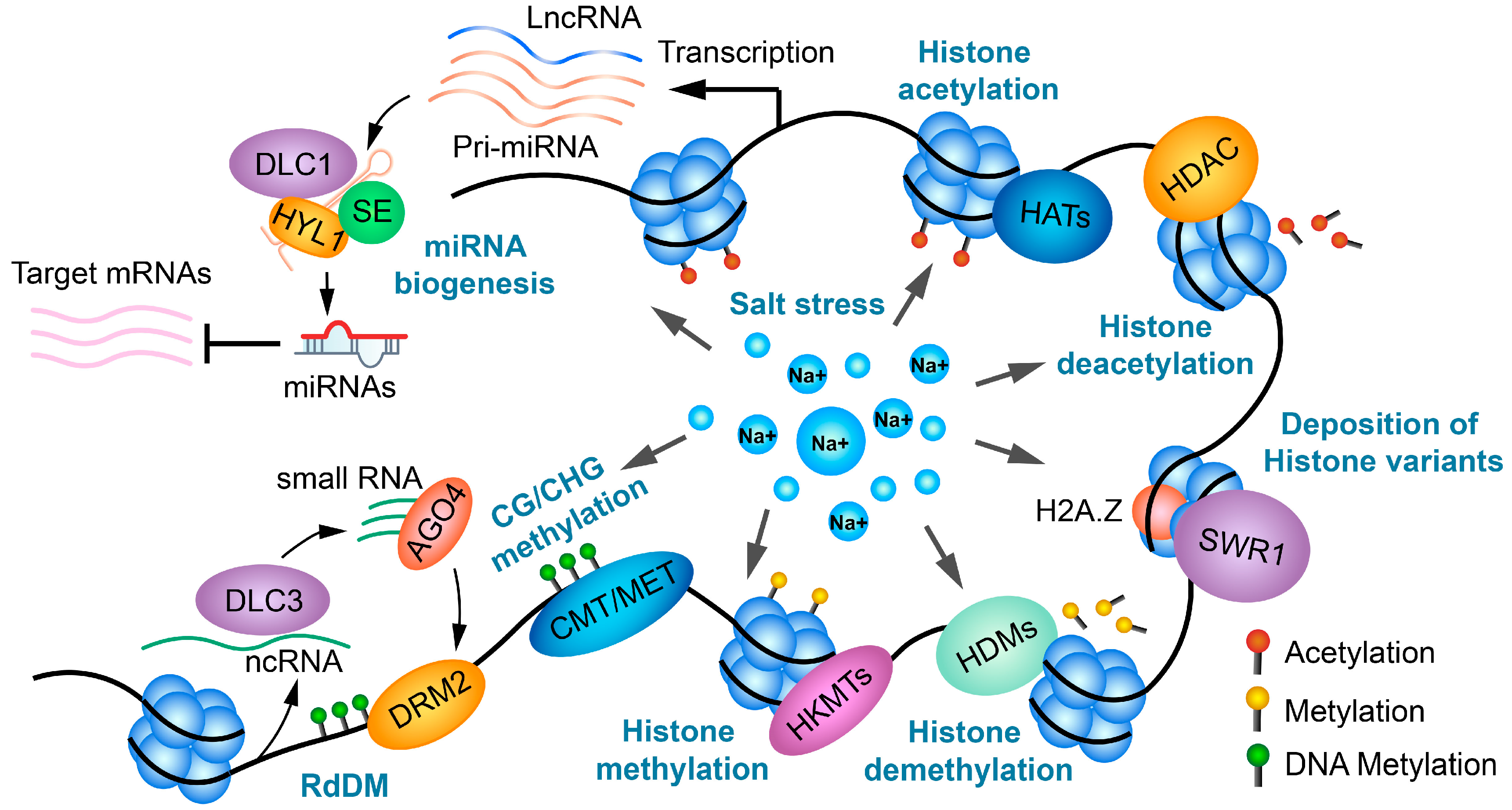
Since plants cannot proactively escape negative environments, they have evolved complex strategies to adapt to environmental stresses. Epigenetic regulation is a critical component of these adaptive mechanisms. This review collected and summarized recent studies on the overall state of DNA methylation, histone modifications, histone variants, and non-coding RNAs in plants, focusing on their global alterations in response to salt stress and their roles in enhancing salt tolerance (Figure 5). During salt stress, epigenetic regulators modulate genome architecture, reform the transcriptome, and establish a salt-specific regulatory network, therefore affecting physiological processes and plant phenotypes. Despite extensive research, several fundamental questions remain, such as how histone methyltransferases recognize their genomic targeting loci in response to salt stress, the mechanisms of DNA methyltransferase/demethylase recognition, and the deposition of H2A.Z. Most recent studies have focused on individual epigenetic processes, but further investigation is needed into the crosstalk and synergistic regulation between different epigenetic mechanisms in enhancing plant stress tolerance.
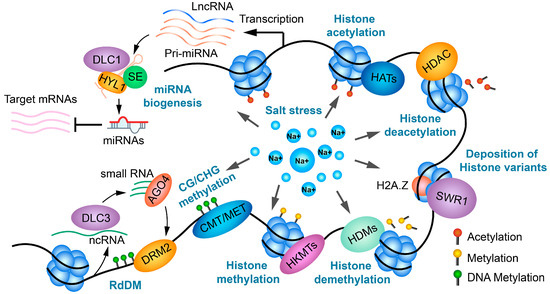
Figure 5.
A schematic diagram illustrating all epigenetic factors and non-coding RNAs in response to salt stress.
We also explore the emerging roles of epigenetic regulation in the establishment of plant stress memory (PSM), which is vital for plants to adapt to recurring stress. During stress, plants maintain a stress-responsive transcriptome. Post-stress, epigenetic marks may be inherited by subsequent generations through various mechanisms. However, the precise mechanisms and core factors involved in maintaining these epigenetic modifications to preserve genomic and transcriptional status remain unclear. Recent technological advancements, such as single-cell RNA sequencing (scRNA-seq), high-throughput chromosome conformation capture (Hi-C) [26], and CRISPR-Cas9 knockout tools [186], have advanced epigenetic research [186,187,188], potentially accelerating the systematic identification of PSM-associated factors. PSM and epigenetic engineering in crops could become crucial approaches for developing stress-resistant cultivars addressing severe global climate change [189]. A deeper understanding of the mechanisms underlying salt tolerance is essential for cultivating resilient crop varieties and ensuring food security.
Author Contributions
D.Z. (Dongyu Zhang) and J.L. conceived of the presented idea. D.Z. (Dongyu Zhang), D.Z. (Duoqian Zhang), Y.Z., G.L. and J.L. wrote the manuscript. J.L., B.Z. and D.S. helped supervise the project. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, and the grant number is 32170283.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Jiacai Chen for assisting with the creation of the models and for the aesthetic improvements. This work was supported by the “2115” project of China Agricultural University.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Negacz, K.; Malek, Ž.; de Vos, A.; Vellinga, P. Saline soils worldwide: Identifying the most promising areas for saline agriculture. J. Arid Environ. 2022, 203, 104775. [Google Scholar] [CrossRef]
- da Costa, G.S.; Cerqueira, A.F.; de Brito, C.R.; Mielke, M.S.; Gaiotto, F.A. Epigenetics Regulation in Responses to Abiotic Factors in Plant Species: A Systematic Review. Plants 2024, 13, 2082. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Kudapa, H.; Ramalingam, A.; Choudhary, D.; Sinha, P.; Garg, V.; Singh, V.K.; Patil, G.B.; Pandey, M.K.; Nguyen, H.T.; et al. Epigenetics and epigenomics: Underlying mechanisms, relevance, and implications in crop improvement. Funct. Integr. Genom. 2020, 20, 739–761. [Google Scholar] [CrossRef] [PubMed]
- Bender, J. DNA Methylation and Epigenetics. Annu. Rev. Plant Biol. 2004, 55, 41–68. [Google Scholar] [CrossRef]
- Matzke, M.A.; Kanno, T.; Matzke, A.J.M. RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu. Rev. Plant Biol. 2015, 66, 243–267. [Google Scholar] [CrossRef]
- Li, Y.; Guo, D. Transcriptome and DNA Methylome Analysis of Two Contrasting Rice Genotypes under Salt Stress during Germination. Int. J. Mol. Sci. 2023, 24, 3978. [Google Scholar] [CrossRef]
- Singroha, G.; Kumar, S.; Gupta, O.P.; Singh, G.P.; Sharma, P. Uncovering the Epigenetic Marks Involved in Mediating Salt Stress Tolerance in Plants. Front. Genet. 2022, 13, 811732. [Google Scholar] [CrossRef]
- He, X.-J.; Chen, T.; Zhu, J.-K. Regulation and function of DNA methylation in plants and animals. Cell Res. 2011, 21, 442–465. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of Modified DNA Bases: 5-Methylcytosine and Beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef]
- Lin, W.; Sun, L.; Huang, R.Z.; Liang, W.; Liu, X.; He, H.; Fukuda, H.; He, X.; Qian, W. Active DNA demethylation regulates tracheary element differentiation in Arabidopsis. Sci. Adv. 2020, 6, eaaz2963. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liang, W.; Liu, Y.; Ren, Z.; Ci, D.; Chang, J.; Qian, W. The Arabidopsis ATR-SOG1 signaling module regulates pleiotropic developmental adjustments in response to 3′-blocked DNA repair intermediates. Plant Cell 2022, 34, 852–866. [Google Scholar] [CrossRef]
- Li, Y.; Kumar, S.; Qian, W. Active DNA demethylation: Mechanism and role in plant development. Plant Cell Rep. 2017, 37, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Lindermayr, C.; Rudolf, E.E.; Durner, J.; Groth, M. Interactions between metabolism and chromatin in plant models. Mol. Metab. 2020, 38, 100951. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aguayo, I.; Rodríguez-Galán, J.M.; García, R.; Torreblanca, J.; Pardo, J.M. Salt stress enhances xylem development and expression of S-adenosyl-l-methionine synthase in lignifying tissues of tomato plants. Planta 2004, 220, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, G.; Guo, S.; Wang, Y.; Sun, J. SlSAMS1 enhances salt tolerance through regulation DNA methylation of SlGI in tomato. Plant Sci. 2023, 335, 111808. [Google Scholar] [CrossRef]
- Zhang, X.; Bao, Z.; Gong, B.; Shi, Q. S-adenosylmethionine synthetase 1 confers drought and salt tolerance in transgenic tomato. Environ. Exp. Bot. 2020, 179, 104226. [Google Scholar] [CrossRef]
- Ahmed, I.M.; Nadira, U.A.; Qiu, C.W.; Cao, F.; Chen, Z.H.; Vincze, E.; Wu, F. The Barley S-Adenosylmethionine Synthetase 3 Gene HvSAMS3 Positively Regulates the Tolerance to Combined Drought and Salinity Stress in Tibetan Wild Barley. Cells 2020, 9, 1530. [Google Scholar] [CrossRef]
- Al-Bahry, S.; Victor, R.; Al-Lawati, A.; Yaish, M. Salt stress alters DNA methylation levels in alfalfa (Medicago spp.). Genet. Mol. Res. 2016, 15, 15018299. [Google Scholar]
- Yang, X.; Bai, Z.; He, Y.; Wang, N.; Sun, L.; Li, Y.; Yin, Z.; Wang, X.; Zhang, B.; Han, M.; et al. Genome-wide characterization of DNA methyltransferase family genes implies GhDMT6 improving tolerance of salt and drought on cotton. BMC Plant Biol. 2024, 24, 312. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, C.; Xu, X.; Kan, J.; Li, H.; Lin, J.; Cheng, Z.; Chang, Y. Comprehensive Analysis of the DNA Methyltransferase Genes and Their Association with Salt Response in Pyrus betulaefolia. Forests 2023, 14, 1751. [Google Scholar] [CrossRef]
- Ashapkin, V.V.; Kutueva, L.I.; Aleksandrushkina, N.I.; Vanyushin, B.F. Epigenetic Mechanisms of Plant Adaptation to Biotic and Abiotic Stresses. Int. J. Mol. Sci. 2020, 21, 7457. [Google Scholar] [CrossRef] [PubMed]
- Gahlaut, V.; Skorupa, M.; Szczepanek, J.; Mazur, J.; Domagalski, K.; Tretyn, A.; Tyburski, J. Salt stress and salt shock differently affect DNA methylation in salt-responsive genes in sugar beet and its wild, halophytic ancestor. PLoS ONE 2021, 16, e0251675. [Google Scholar]
- Wang, B.; Fu, R.; Zhang, M.; Ding, Z.; Chang, L.; Zhu, X.; Wang, Y.; Fan, B.; Ye, W.; Yuan, Y. Analysis of methylation-sensitive amplified polymorphism in different cotton accessions under salt stress based on capillary electrophoresis. Genes Genom. 2015, 37, 713–724. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, X.; Pan, Y.; Zhu, L.; Fu, B.; Li, Z. DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stress. J. Genet. Genom. 2011, 38, 419–424. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yang, Z.; Liu, L.; Duan, L. DNA Methylation in Plant Responses and Adaption to Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 6910. [Google Scholar] [CrossRef]
- Konate, M.; Wilkinson, M.; Mayne, B.; Pederson, S.; Scott, E.; Berger, B.; Rodriguez Lopez, C. Salt Stress Induces Non-CG Methylation in Coding Regions of Barley Seedlings (Hordeum vulgare). Epigenomes 2018, 2, 12. [Google Scholar] [CrossRef]
- Chen, R.; Li, M.; Zhang, H.; Duan, L.; Sun, X.; Jiang, Q.; Zhang, H.; Hu, Z. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genom. 2019, 20, 730. [Google Scholar] [CrossRef]
- Lin, X.; Zhou, M.; Yao, J.; Li, Q.Q.; Zhang, Y.-Y. Phenotypic and Methylome Responses to Salt Stress in Arabidopsis thaliana Natural Accessions. Front. Plant Sci. 2022, 13, 841154. [Google Scholar] [CrossRef]
- Miryeganeh, M.; Marlétaz, F.; Gavriouchkina, D.; Saze, H. De novo genome assembly and in natura epigenomics reveal salinity-induced DNA methylation in the mangrove tree Bruguiera gymnorhiza. New Phytol. 2021, 233, 2094–2110. [Google Scholar] [CrossRef]
- Shahid, S. A DNA Methylation Reader with an Affinity for Salt Stress. Plant Cell 2020, 32, 3380–3381. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Nan, N.; Li, N.; Liu, Y.; Wang, T.-J.; Hwang, I.; Liu, B.; Xu, Z.-Y. A DNA Methylation Reader–Chaperone Regulator–Transcription Factor Complex Activates OsHKT1;5 Expression during Salinity Stress. Plant Cell 2020, 32, 3535–3558. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Beena, A.S.; Awana, M.; Singh, A. Salt-Induced Tissue-Specific Cytosine Methylation Downregulates Expression of HKT Genes in Contrasting Wheat (Triticum aestivum L.) Genotypes. DNA Cell Biol. 2017, 36, 283–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qin, L.; Xie, C.; Li, W.; Yuan, J.; Kong, L.; Yu, W.; Xia, G.; Liu, S. Induced and Constitutive DNA Methylation in a Salinity-Tolerant Wheat Introgression Line. Plant Cell Physiol. 2014, 55, 1354–1365. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Liu, Y.; Wang, X.; Chang, C. Insight into the Role of Epigenetic Processes in Abiotic and Biotic Stress Response in Wheat and Barley. Int. J. Mol. Sci. 2020, 21, 1480. [Google Scholar] [CrossRef]
- Siddique, A.B.; Parveen, S.; Rahman, M.Z.; Rahman, J. Revisiting plant stress memory: Mechanisms and contribution to stress adaptation. Physiol. Mol. Biol. Plants 2024, 30, 349–367. [Google Scholar] [CrossRef]
- Jiang, C.; Mithani, A.; Belfield, E.J.; Mott, R.; Hurst, L.D.; Harberd, N.P. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014, 24, 1821–1829. [Google Scholar] [CrossRef]
- Wu, K.; Ou, X.; Zhang, Y.; Xu, C.; Lin, X.; Zang, Q.; Zhuang, T.; Jiang, L.; von Wettstein, D.; Liu, B. Transgenerational Inheritance of Modified DNA Methylation Patterns and Enhanced Tolerance Induced by Heavy Metal Stress in Rice (Oryza sativa L.). PLoS ONE 2012, 7, e41143. [Google Scholar]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N.; et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef]
- Herman, J.J.; Sultan, S.E. DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160988. [Google Scholar] [CrossRef]
- Zheng, X.; Chen, L.; Xia, H.; Wei, H.; Lou, Q.; Li, M.; Li, T.; Luo, L. Transgenerational epimutations induced by multi-generation drought imposition mediate rice plant’s adaptation to drought condition. Sci. Rep. 2017, 7, 39843. [Google Scholar] [CrossRef]
- Kambona, C.M.; Koua, P.A.; Léon, J.; Ballvora, A. Stress memory and its regulation in plants experiencing recurrent drought conditions. Theor. Appl. Genet. 2023, 136, 26. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Blevins, T.; Yao, Y.; Golubov, A.; Bilichak, A.; Ilnytskyy, Y.; Hollunder, J.; Meins, F., Jr.; Kovalchuk, I. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 2010, 5, e9514. [Google Scholar] [CrossRef]
- Wibowo, A.; Becker, C.; Marconi, G.; Durr, J.; Price, J.; Hagmann, J.; Papareddy, R.; Putra, H.; Kageyama, J.; Becker, J.; et al. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 2016, 5, e13546. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Chang, N.; Zhao, Y.; Qin, X.; Lu, S.; Crabbe, M.J.C.; Guan, Y.; Zhang, T. Increased epigenetic diversity and transient epigenetic memory in response to salinity stress in Thlaspi arvense. Ecol. Evol. 2020, 10, 11622–11630. [Google Scholar] [CrossRef] [PubMed]
- Sani, E.; Herzyk, P.; Perrella, G.; Colot, V.; Amtmann, A. Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biol. 2013, 14, R59. [Google Scholar] [CrossRef]
- Feng, X.J.; Li, J.R.; Qi, S.L.; Lin, Q.F.; Jin, J.B.; Hua, X.J. Light affects salt stress-induced transcriptional memory of P5CS1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, E8335–E8343. [Google Scholar] [CrossRef]
- Nunez-Vazquez, R.; Desvoyes, B.; Gutierrez, C. Histone variants and modifications during abiotic stress response. Front. Plant Sci. 2022, 13, 984702. [Google Scholar] [CrossRef]
- Xiao, J.; Lee, U.-S.; Wagner, D. Tug of war: Adding and removing histone lysine methylation in arabidopsis. Curr. Opin. Plant Biol. 2016, 34, 41–53. [Google Scholar] [CrossRef]
- Yung, W.S.; Li, M.W.; Sze, C.C.; Wang, Q.; Lam, H.M. Histone modifications and chromatin remodelling in plants in response to salt stress. Physiol. Plant 2021, 173, 1495–1513. [Google Scholar] [CrossRef]
- Chua, Y.; Gray, J.C. Histone Modifications and Transcription in Plants; Blackwell Publishing Ltd.: Oxford, UK, 2006; pp. 79–111. [Google Scholar]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef]
- Zhang, X.; Clarenz, O.; Cokus, S.; Bernatavichute, Y.V.; Pellegrini, M.; Goodrich, J.; Jacobsen, S.E. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007, 5, e129. [Google Scholar] [CrossRef]
- Roudier, F.; Ahmed, I.; Berard, C.; Sarazin, A.; Mary-Huard, T.; Cortijo, S.; Bouyer, D.; Caillieux, E.; Duvernois-Berthet, E.; Al-Shikhley, L.; et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011, 30, 1928–1938. [Google Scholar] [CrossRef]
- Han, B.; Xu, W.; Ahmed, N.; Yu, A.; Wang, Z.; Liu, A. Changes and Associations of Genomic Transcription and Histone Methylation with Salt Stress in Castor Bean. Plant Cell Physiol. 2020, 61, 1120–1133. [Google Scholar] [CrossRef]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 106–122. [Google Scholar] [CrossRef]
- Jambhekar, A.; Dhall, A.; Shi, Y. Roles and regulation of histone methylation in animal development. Nat. Rev. Mol. Cell Biol. 2019, 20, 625–641. [Google Scholar] [CrossRef]
- Haider, S.; Farrona, S. Decoding histone 3 lysine methylation: Insights into seed germination and flowering. Curr. Opin. Plant Biol. 2024, 81, 102598. [Google Scholar] [CrossRef]
- Roudier, F.; Teixeira, F.K.; Colot, V. Chromatin indexing in Arabidopsis: An epigenomic tale of tails and more. Trends Genet. 2009, 25, 511–517. [Google Scholar] [CrossRef]
- Sequeira-Mendes, J.; Aragüez, I.; Peiró, R.; Mendez-Giraldez, R.; Zhang, X.; Jacobsen, S.E.; Bastolla, U.; Gutierrez, C. The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell 2014, 26, 2351–2366. [Google Scholar] [CrossRef]
- Nützmann, H.-W.; Doerr, D.; Ramírez-Colmenero, A.; Sotelo-Fonseca, J.E.; Wegel, E.; Di Stefano, M.; Wingett, S.W.; Fraser, P.; Hurst, L.; Fernandez-Valverde, S.L.; et al. Active and repressed biosynthetic gene clusters have spatially distinct chromosome states. Proc. Natl. Acad. Sci. USA 2020, 117, 13800–13809. [Google Scholar] [CrossRef]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Cao, X.; Deng, X. Histone methylation in epigenetic regulation and temperature responses. Curr. Opin. Plant Biol. 2021, 61, 102001. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Cui, X.; Shen, Y. The roles of histone methylation in the regulation of abiotic stress responses in plants. Plant Stress 2024, 11, 100303. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Liu, B.; Xu, Z.Y. Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants. J. Integr. Plant Biol. 2022, 64, 2252–2274. [Google Scholar] [CrossRef]
- Shen, Y.; Chi, Y.; Lu, S.; Lu, H.; Shi, L. Involvement of JMJ15 in the dynamic change of genome-wide H3K4me3 in response to salt stress. Front. Plant Sci. 2022, 13, 1009723. [Google Scholar] [CrossRef]
- Song, Y.; Ji, D.; Li, S.; Wang, P.; Li, Q.; Xiang, F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012, 7, e41274. [Google Scholar] [CrossRef]
- Bilichak, A.; Ilnystkyy, Y.; Hollunder, J.; Kovalchuk, I. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 2012, 7, e30515. [Google Scholar] [CrossRef] [PubMed]
- Yung, W.S.; Wang, Q.; Huang, M.; Wong, F.L.; Liu, A.; Ng, M.S.; Li, K.P.; Sze, C.C.; Li, M.W.; Lam, H.M. Priming-induced alterations in histone modifications modulate transcriptional responses in soybean under salt stress. Plant J. 2022, 109, 1575–1590. [Google Scholar] [CrossRef]
- Paul, A.; Dasgupta, P.; Roy, D.; Chaudhuri, S. Comparative analysis of Histone modifications and DNA methylation at OsBZ8 locus under salinity stress in IR64 and Nonabokra rice varieties. Plant Mol. Biol. 2017, 95, 63–88. [Google Scholar] [CrossRef]
- Dong, W.; Gao, T.; Wang, Q.; Chen, J.; Lv, J.; Song, Y. Salinity stress induces epigenetic alterations to the promoter of MsMYB4 encoding a salt-induced MYB transcription factor. Plant Physiol. Biochem. 2020, 155, 709–715. [Google Scholar] [CrossRef]
- Yung, W.S.; Huang, C.; Li, M.W.; Lam, H.M. Changes in epigenetic features in legumes under abiotic stresses. Plant Genome 2023, 16, e20237. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Xiao, Y.; Niu, M.; Meng, W.; Li, L.; Zhang, X.; Liu, D.; Zhang, G.; Qian, Y.; Sun, Z.; et al. ARGONAUTE2 Enhances Grain Length and Salt Tolerance by Activating BIG GRAIN3 to Modulate Cytokinin. Plant Cell 2020, 32, 2291–2306. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Conde, E.S.N.; Audonnet, L.; Servet, C.; Wei, W.; Zhou, D.X. Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 2014, 5, 290. [Google Scholar] [CrossRef]
- Mellor, J. It Takes a PHD to Read the Histone Code. Cell 2006, 126, 22–24. [Google Scholar] [CrossRef]
- Wei, W.; Tao, J.J.; Chen, H.W.; Li, Q.T.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; Chen, S.Y. A Histone Code Reader and a Transcriptional Activator Interact to Regulate Genes for Salt Tolerance. Plant Physiol. 2017, 175, 1304–1320. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Xue, S.; Quan, T.; Cui, D.; Han, L.; Cong, W.; Li, M.; Yun, D.J.; Liu, B.; et al. SET DOMAIN GROUP 721 protein functions in saline–alkaline stress tolerance in the model rice variety Kitaake. Plant Biotechnol. J. 2021, 19, 2576–2588. [Google Scholar] [CrossRef]
- Fu, Z.W.; Li, J.H.; Feng, Y.R.; Yuan, X.; Lu, Y.T. The metabolite methylglyoxal-mediated gene expression is associated with histone methylglyoxalation. Nucleic Acids Res. 2021, 49, 1886–1899. [Google Scholar] [CrossRef]
- QiZhi, F.; ChunWu, Y.; XiuYun, L.; JinMing, W.; XiuFang, O.; ChunYu, Z.; Yu, C.; Bao, L. Salt and alkaline stress induced transgenerational alteration in DNA methylation of rice (Oryza sativa). Aust. J. Crop Sci. 2012, 6, 877–883. [Google Scholar]
- Roychoudhury, A.; Banerjee, A.; Lahiri, V. Metabolic and molecular-genetic regulation of proline signaling and itscross-talk with major effectors mediates abiotic stress tolerance in plants. Turk. J. Bot. 2015, 39, 887–910. [Google Scholar] [CrossRef]
- Tian, Z.; Li, K.; Sun, Y.; Chen, B.; Pan, Z.; Wang, Z.; Pang, B.; He, S.; Miao, Y.; Du, X. Physiological and transcriptional analyses reveal formation of memory under recurring drought stresses in seedlings of cotton (Gossypium hirsutum). Plant Sci. 2024, 338, 111920. [Google Scholar] [CrossRef]
- Liu, H.c.; Lämke, J.; Lin, S.y.; Hung, M.J.; Liu, K.M.; Charng, Y.y.; Bäurle, I. Distinct heat shock factors and chromatin modifications mediate the organ-autonomous transcriptional memory of heat stress. Plant J. 2018, 95, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Fromm, M.; Avramova, Z. Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat. Commun. 2012, 3, 740. [Google Scholar] [CrossRef] [PubMed]
- Mozgova, I.; Hennig, L. The Polycomb Group Protein Regulatory Network. Annu. Rev. Plant Biol. 2015, 66, 269–296. [Google Scholar] [CrossRef] [PubMed]
- Onufriev, A.V.; Schiessel, H. The nucleosome: From structure to function through physics. Curr. Opin. Struct. Biol. 2019, 56, 119–130. [Google Scholar] [CrossRef]
- Earley, K.W.; Shook, M.S.; Brower-Toland, B.; Hicks, L.; Pikaard, C.S. In Vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J. 2007, 52, 615–626. [Google Scholar] [CrossRef]
- Ueda, M.; Matsui, A.; Tanaka, M.; Nakamura, T.; Abe, T.; Sako, K.; Sasaki, T.; Kim, J.M.; Ito, A.; Nishino, N.; et al. The Distinct Roles of Class I and II RPD3-Like Histone Deacetylases in Salinity Stress Response. Plant Physiol. 2017, 175, 1760–1773. [Google Scholar] [CrossRef]
- Zheng, Y.; Ding, Y.; Sun, X.; Xie, S.; Wang, D.; Liu, X.; Su, L.; Wei, W.; Pan, L.; Zhou, D.X. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 2016, 67, 1703–1713. [Google Scholar] [CrossRef]
- Xing, G.; Jin, M.; Qu, R.; Zhang, J.; Han, Y.; Han, Y.; Wang, X.; Li, X.; Ma, F.; Zhao, X. Genome-wide investigation of histone acetyltransferase gene family and its responses to biotic and abiotic stress in foxtail millet (Setaria italica [L.] P. Beauv). BMC Plant Biol. 2022, 22, 292. [Google Scholar] [CrossRef]
- Wei, F.; Tang, D.; Li, Z.; Kashif, M.H.; Khan, A.; Lu, H.; Jia, R.; Chen, P. Molecular cloning and subcellular localization of six HDACs and their roles in response to salt and drought stress in kenaf (Hibiscus cannabinus L.). Biol. Res. 2019, 52, 20. [Google Scholar] [CrossRef]
- Liu, C.T.; Mao, B.G.; Yuan, D.Y.; Chu, C.C.; Duan, M.J. Salt tolerance in rice: Physiological responses and molecular mechanisms. Crop J. 2022, 10, 13–25. [Google Scholar] [CrossRef]
- Su, P.; Yan, J.; Li, W.; Wang, L.; Zhao, J.; Ma, X.; Li, A.; Wang, H.; Kong, L. A member of wheat class III peroxidase gene family, TaPRX-2A, enhanced the tolerance of salt stress. BMC Plant Biol. 2020, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zhang, S.; Tao, W.; Zhang, X.; Liu, J.; Sun, J.; Zhang, H.; Pu, L.; Huang, R.; Chen, T. INDETERMINATE SPIKELET1 Recruits Histone Deacetylase and a Transcriptional Repression Complex to Regulate Rice Salt Tolerance. Plant Physiol. 2018, 178, 824–837. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Galluzzi, L.; Bravo-San Pedro, J.M.; Madeo, F.; Kroemer, G. Acetyl Coenzyme A: A Central Metabolite and Second Messenger. Cell Metab. 2015, 21, 805–821. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Lu, Y.; Zhao, Y.; Zhou, D.X. Histone Acetylation Dynamics Integrates Metabolic Activity to Regulate Plant Response to Stress. Front. Plant Sci. 2019, 10, 1236. [Google Scholar] [CrossRef]
- Chen, C.; Li, C.; Wang, Y.; Renaud, J.; Tian, G.; Kambhampati, S.; Saatian, B.; Nguyen, V.; Hannoufa, A.; Marsolais, F.; et al. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants 2017, 3, 814–824. [Google Scholar] [CrossRef]
- Lorena, M.-P.; Vicent, P.; María, D.C.; Vicente, T. Dynamic remodeling of histone modifications in response to osmotic stress in Saccharomyces cerevisiae. BMC Genom. 2014, 15, 247. [Google Scholar]
- Truong, H.A.; Lee, S.; Trinh, C.S.; Lee, W.J.; Chung, E.H.; Hong, S.W.; Lee, H. Overexpression of the HDA15 Gene Confers Resistance to Salt Stress by the Induction of NCED3, an ABA Biosynthesis Enzyme. Front. Plant Sci. 2021, 12, 640443. [Google Scholar] [CrossRef]
- Ullah, F.; Xu, Q.; Zhao, Y.; Zhou, D.X. Histone deacetylase HDA710 controls salt tolerance by regulating ABA signaling in rice. J. Integr. Plant Biol. 2020, 63, 451–467. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, H.; Zhang, B.; Cheng, Y.; Ma, X. Overexpression of histone deacetylase gene 84KHDA909 from poplar confers enhanced tolerance to drought and salt stresses in Arabidopsis. Plant Sci. 2022, 324, 111434. [Google Scholar] [CrossRef]
- Sako, K.; Kim, J.M.; Matsui, A.; Nakamura, K.; Tanaka, M.; Kobayashi, M.; Saito, K.; Nishino, N.; Kusano, M.; Taji, T.; et al. Ky-2, a Histone Deacetylase Inhibitor, Enhances High-Salinity Stress Tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2016, 57, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Yolcu, S.; Ozdemir, F.; Guler, A.; Bor, M. Histone acetylation influences the transcriptional activation of POX in Beta vulgaris L. and Beta maritima L. under salt stress. Plant Physiol. Biochem. 2016, 100, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-T.; Luo, M.; Wang, Y.-Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, S.; Zhao, L.; Tan, J.; Zhang, Q.; Gao, F.; Wang, P.; Hou, H.; Li, L. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014, 14, 105. [Google Scholar] [CrossRef]
- Zhu, N.; Cheng, S.; Liu, X.; Du, H.; Dai, M.; Zhou, D.-X.; Yang, W.; Zhao, Y. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci. 2015, 236, 146–156. [Google Scholar] [CrossRef]
- Wei, H.; Wang, X.; He, Y.; Xu, H.; Wang, L. Clock component OsPRR73 positively regulates rice salt tolerance by modulating OsHKT2;1-mediated sodium homeostasis. EMBO J. 2021, 40, e105086. [Google Scholar] [CrossRef]
- Zheng, M.; Lin, J.; Liu, X.; Chu, W.; Li, J.; Gao, Y.; An, K.; Song, W.; Xin, M.; Yao, Y.; et al. Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol. 2021, 186, 1951–1969. [Google Scholar] [CrossRef]
- Sokol, A.; Kwiatkowska, A.; Jerzmanowski, A.; Prymakowska-Bosak, M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 2007, 227, 245–254. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, W.; da Silva, J.A.T.; Liu, X.; Duan, J. Rice histone deacetylase HDA704 positively regulates drought and salt tolerance by controlling stomatal aperture and density. Planta 2021, 254, 79. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Jiang, Y.; Wang, H.; Zhou, L.; Li, F.; Wang, L.; Jiang, J.; Chen, F.; Chen, S. Transcription factor CmHSFA4-CmMYBS3 complex enhances salt tolerance in chrysanthemum by repressing CmMYB121 expression. Plant Physiol. 2024, 195, 3119–3135. [Google Scholar] [CrossRef]
- Zheng, L.; Ma, S.; Shen, D.; Fu, H.; Wang, Y.; Liu, Y.; Shah, K.; Yue, C.; Huang, J. Genome-wide identification of Gramineae histone modification genes and their potential roles in regulating wheat and maize growth and stress responses. BMC Plant Biol. 2021, 21, 543. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.V.; Wigge, P.A. H2A.Z-Containing Nucleosomes Mediate the Thermosensory Response in Arabidopsis. Cell 2010, 140, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, A.; Molaro, A. Histone renegades: Unusual H2A histone variants in plants and animals. Semin. Cell Dev. Biol. 2023, 135, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Zhang, Y.; Liu, X.; Yuan, Y.; Zang, W.; Li, Z.; Yan, X.; Pang, Q.; Zhang, A. Histone variant H2A.Z is required for plant salt response by regulating gene transcription. Plant Cell Environ. 2024, 47, 2691–2707. [Google Scholar] [CrossRef]
- Do, B.H.; Hiep, N.T.; Lao, T.D.; Nguyen, N.H. Loss-of-Function Mutation of ACTIN-RELATED PROTEIN 6 (ARP6) Impairs Root Growth in Response to Salinity Stress. Mol. Biotechnol. 2023, 65, 1414–1420. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Cheong, J.-J. H2A.Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress. Biochem. Biophys. Res. Commun. 2018, 499, 1039–1043. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.-J. Overexpression of AtMYB44 Enhances Stomatal Closure to Confer Abiotic Stress Tolerance in Transgenic Arabidopsis. Plant Physiol. 2008, 146, 323–324. [Google Scholar] [CrossRef]
- Qiu, S.-P.; Huang, J.; Pan, L.-J.; Wang, M.-M.; Zhang, H.-S. Salt Induces Expression of RH3.2A, Encoding an H3.2-type Histone H3 Protein in Rice (Oryza sativa L.). Acta Genet. Sin. 2006, 33, 833–840. [Google Scholar] [CrossRef]
- Probst, A.V.; Desvoyes, B.; Gutierrez, C.; Parry, G. Similar yet critically different: The distribution, dynamics and function of histone variants. J. Exp. Bot. 2020, 71, 5191–5204. [Google Scholar] [CrossRef]
- Hargreaves, D.C.; Crabtree, G.R. ATP-dependent chromatin remodeling: Genetics, genomics and mechanisms. Cell Res. 2011, 21, 396–420. [Google Scholar] [CrossRef]
- Xu, W.; Li, Y.; Cheng, Z.; Xia, G.; Wang, M. A wheat histone variant gene TaH2A.7 enhances drought tolerance and promotes stomatal closure in Arabidopsis. Plant Cell Rep. 2016, 35, 1853–1862. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Zhou, F.; Zhang, L.; Gong, J.; Cheng, C.; Chen, J.; Lou, Q. Genome-wide characterization, phylogenetic and expression analysis of Histone gene family in cucumber (Cucumis sativus L.). Int. J. Biol. Macromol. 2023, 230, 123401. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Ma, C.; Zheng, C.; Yao, Y.; Du, Y. Advances in the regulation of plant salt-stress tolerance by miRNA. Mol. Biol. Rep. 2022, 49, 5041–5055. [Google Scholar] [CrossRef]
- Qiao, H.; Jiao, B.; Wang, J.; Yang, Y.; Yang, F.; Geng, Z.; Zhao, G.; Liu, Y.; Dong, F.; Wang, Y.; et al. Comparative Analysis of miRNA Expression Profiles under Salt Stress in Wheat. Genes 2023, 14, 1586. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, S.M.; Alaraidh, I.A.; Khan, M.A.; Migdadi, H.M.; Alghamdi, S.S.; Alsahli, A.A. Identification and Characterization of Salt-Responsive MicroRNAs in Vicia faba by High-Throughput Sequencing. Genes 2019, 10, 303. [Google Scholar] [CrossRef]
- Wei, L.; Du, Y.; Xiang, J.; Zheng, T.; Cheng, J.; Wu, J. Integrated mRNA and miRNA transcriptome analysis of grape in responses to salt stress. Front. Plant Sci. 2023, 14, 1173857. [Google Scholar] [CrossRef]
- Xie, R.; Zhang, J.; Ma, Y.; Pan, X.; Dong, C.; Pang, S.; He, S.; Deng, L.; Yi, S.; Zheng, Y.; et al. Combined analysis of mRNA and miRNA identifies dehydration and salinity responsive key molecular players in citrus roots. Sci. Rep. 2017, 7, 42094. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Nguyen, N.L.; Nguyen, V.T.; Tran, T.H.G.; Nguyen, T.H.; Nguyen, T.K.L.; Nguyen, H.H. Comparative analysis of microRNA expression profiles in shoot and root tissues of contrasting rice cultivars (Oryza sativa L.) with different salt stress tolerance. PLoS ONE 2023, 18, e0286140. [Google Scholar] [CrossRef]
- Liu, J.N.; Ma, X.; Yan, L.; Liang, Q.; Fang, H.; Wang, C.; Dong, Y.; Chai, Z.; Zhou, R.; Bao, Y.; et al. MicroRNA and Degradome Profiling Uncover Defense Response of Fraxinus velutina Torr. to Salt Stress. Front. Plant Sci. 2022, 13, 847853. [Google Scholar] [CrossRef]
- Yin, Z.; Han, X.; Li, Y.; Wang, J.; Wang, D.; Wang, S.; Fu, X.; Ye, W. Comparative Analysis of Cotton Small RNAs and Their Target Genes in Response to Salt Stress. Genes 2017, 8, 369. [Google Scholar] [CrossRef]
- Bravo-Vázquez, L.A.; García-Ortega, M.; Medina-Feria, S.; Srivastava, A.; Paul, S. Identification and expression profiling of microRNAs in leaf tissues of Foeniculum vulgare Mill. under salinity stress. Plant Signal Behav. 2024, 19, 2361174. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ren, Y.; Sun, Y.; Yin, Y.; Han, B.; Zhang, L.; Song, Y.; Zhang, Z.; Xu, Y.; Fan, D.; et al. Identification and Analysis of the MIR399 Gene Family in Grapevine Reveal Their Potential Functions in Abiotic Stress. Int. J. Mol. Sci. 2024, 25, 2979. [Google Scholar] [CrossRef] [PubMed]
- Szymonik, K.; Klimek-Chodacka, M.; Lukasiewicz, A.; Macko-Podgórni, A.; Grzebelus, D.; Baranski, R. Comparative analysis of the carrot miRNAome in response to salt stress. Sci. Rep. 2023, 13, 21506. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Khare, T.; Shriram, V.; Wani, S.H. Plant small RNAs: The essential epigenetic regulators of gene expression for salt-stress responses and tolerance. Plant Cell Rep. 2018, 37, 61–75. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, H.; Xu, G.; Zhang, P.; Zhai, N.; Zheng, Q.; Liu, P.; Jin, L.; Bai, G.; Zhang, H. Genome-wide analysis of long noncoding RNAs in response to salt stress in Nicotiana tabacum. BMC Plant Biol. 2023, 23, 646. [Google Scholar] [CrossRef]
- Li, N.; Wang, Z.; Wang, B.; Wang, J.; Xu, R.; Yang, T.; Huang, S.; Wang, H.; Yu, Q. Identification and Characterization of Long Non-coding RNA in Tomato Roots Under Salt Stress. Front. Plant Sci. 2022, 13, 834027. [Google Scholar] [CrossRef]
- Gu, Y.; Li, G.; Wang, P.; Guo, Y.; Li, J. A simple and precise method (Y2H-in-frame-seq) improves yeast two-hybrid screening with cDNA libraries. J. Genet. Genom. 2022, 49, 595–598. [Google Scholar] [CrossRef]
- He, F.; Xu, C.; Fu, X.; Shen, Y.; Guo, L.; Leng, M.; Luo, K. The MicroRNA390/TRANS-ACTING SHORT INTERFERING RNA3 Module Mediates Lateral Root Growth under Salt Stress via the Auxin Pathway. Plant Physiol. 2018, 177, 775–791. [Google Scholar] [CrossRef]
- Iglesias, M.J.; Terrile, M.C.; Windels, D.; Lombardo, M.C.; Bartoli, C.G.; Vazquez, F.; Estelle, M.; Casalongué, C.A. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS ONE 2014, 9, e107678. [Google Scholar] [CrossRef]
- Song, J.B.; Gao, S.; Sun, D.; Li, H.; Shu, X.X.; Yang, Z.M. miR394 and LCR are involved in Arabidopsis salt and drought stress responses in an abscisic acid-dependent manner. BMC Plant Biol. 2013, 13, 210. [Google Scholar] [CrossRef]
- Wan, X.; Wang, Z.; Duan, W.; Huang, T.; Song, H.; Xu, X. Knockdown of Sly-miR164a Enhanced Plant Salt Tolerance and Improved Preharvest and Postharvest Fruit Nutrition of Tomato. Int. J. Mol. Sci. 2023, 24, 4639. [Google Scholar] [CrossRef] [PubMed]
- Scintu, D.; Scacchi, E.; Cazzaniga, F.; Vinciarelli, F.; De Vivo, M.; Shtin, M.; Svolacchia, N.; Bertolotti, G.; Unterholzner, S.J.; Del Bianco, M.; et al. microRNA165 and 166 modulate response of the Arabidopsis root apical meristem to salt stress. Commun. Biol. 2023, 6, 834. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Niu, J.; Cao, X. Heterologous Expression of Salvia miltiorrhiza MicroRNA408 Enhances Tolerance to Salt Stress in Nicotiana benthamiana. Int. J. Mol. Sci. 2018, 19, 3985. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liu, D.; Chen, D.; Cheng, Y.; Zhang, X.; Song, L.; Hu, M.; Dong, J.; Shen, F. MicroRNA414c affects salt tolerance of cotton by regulating reactive oxygen species metabolism under salinity stress. RNA Biol. 2019, 16, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Zhu, M.; Luan, M.; Zhang, M.; Jin, L.; Liu, Y.; Zou, J.; Wang, L.; Xu, M. miR169q and NUCLEAR FACTOR YA8 enhance salt tolerance by activating PEROXIDASE1 expression in response to ROS. Plant Physiol. 2022, 188, 608–623. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Hu, Y.; Chen, H.; Du, Q.; Yang, J.; Li, W.X. MicroRNA408 negatively regulates salt tolerance by affecting secondary cell wall development in maize. Plant Physiol. 2023, 192, 1569–1583. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Fujita, M. Glutathione in plants: Biosynthesis and physiological role in environmental stress tolerance. Physiol. Mol. Biol. Plants 2017, 23, 249–268. [Google Scholar] [CrossRef]
- Lushchak, V.I. Glutathione Homeostasis and Functions: Potential Targets for Medical Interventions. J. Amino Acids 2012, 2012, 736837. [Google Scholar] [CrossRef]
- Mittova, V.; Theodoulou, F.L.; Kiddle, G.; Gómez, L.; Volokita, M.; Tal, M.; Foyer, C.H.; Guy, M. Coordinate induction of glutathione biosynthesis and glutathione-metabolizing enzymes is correlated with salt tolerance in tomato. FEBS Lett. 2003, 554, 417–421. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xu, L.; Zhang, L.; Li, X.; Cao, C.; Chen, L.; Kang, J.; Yang, Q.; Liu, Y.; Sod, B.; et al. Overexpression of Mtr-miR319a Contributes to Leaf Curl and Salt Stress Adaptation in Arabidopsis thaliana and Medicago truncatula. Int. J. Mol. Sci. 2022, 24, 429. [Google Scholar] [CrossRef]
- Rehman, O.U.; Uzair, M.; Farooq, M.S.; Saleem, B.; Attacha, S.; Attia, K.A.; Farooq, U.; Fiaz, S.; El-Kallawy, W.H.; Kimiko, I.; et al. Comprehensive insights into the regulatory mechanisms of lncRNA in alkaline-salt stress tolerance in rice. Mol. Biol. Rep. 2023, 50, 7381–7392. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Sun, X.; Liu, C.; Chu, J.; Zhao, M.; Zhang, W.H. A Medicago truncatula lncRNA MtCIR1 negatively regulates response to salt stress. Planta 2023, 257, 32. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, Q.; Bai, Q.; Feng, Y.; Mao, K.; Yang, M.; He, L.; Liu, M.; Liu, J.; Wan, D. LncRNA expression analysis by comparative transcriptomics among closely related poplars and their regulatory roles in response to salt stress. Tree Physiol. 2023, 43, 1233–1249. [Google Scholar] [CrossRef] [PubMed]
- Mirdar Mansuri, R.; Azizi, A.H.; Sadri, A.H.; Shobbar, Z.S. Long non-coding RNAs as the regulatory hubs in rice response to salt stress. Sci. Rep. 2022, 12, 21696. [Google Scholar] [CrossRef]
- Xu, W.B.; Cao, F.; Liu, P.; Yan, K.; Guo, Q.H. The multifaceted role of RNA-based regulation in plant stress memory. Front. Plant Sci. 2024, 15, 1387575. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef]
- Ding, D.; Zhang, L.; Wang, H.; Liu, Z.; Zhang, Z.; Zheng, Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann. Bot. 2009, 103, 29–38. [Google Scholar] [CrossRef]
- Wan, J.; Meng, S.; Wang, Q.; Zhao, J.; Qiu, X.; Wang, L.; Li, J.; Lin, Y.; Mu, L.; Dang, K.; et al. Suppression of microRNA168 enhances salt tolerance in rice (Oryza sativa L.). BMC Plant Biol. 2022, 22, 563. [Google Scholar] [CrossRef]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, Q.; Zhang, L.; Zhang, C.; Fan, Y.; Lang, Z.; Wang, L. Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS ONE 2014, 9, e91369. [Google Scholar] [CrossRef]
- Pan, W.J.; Tao, J.J.; Cheng, T.; Bian, X.H.; Wei, W.; Zhang, W.K.; Ma, B.; Chen, S.Y.; Zhang, J.S. Soybean miR172a Improves Salt Tolerance and Can Function as a Long-Distance Signal. Mol. Plant 2016, 9, 1337–1340. [Google Scholar] [CrossRef]
- Sahito, Z.A.; Wang, L.; Sun, Z.; Yan, Q.; Zhang, X.; Jiang, Q.; Ullah, I.; Tong, Y.; Li, X. The miR172c-NNC1 module modulates root plastic development in response to salt in soybean. BMC Plant Biol. 2017, 17, 229. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; He, Q.; Tang, S.; Wang, H.; Zhang, X.; Lv, M.; Liu, H.; Gao, Q.; Zhou, Y.; Wang, Q.; et al. The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New Phytol. 2021, 230, 1017–1033. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Zhou, X.H.; Liu, J. Conserved miRNAs and their response to salt stress in wild eggplant Solanum linnaeanum roots. Int. J. Mol. Sci. 2014, 15, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Sun, Y.F.; Song, N.; Wei, J.P.; Wang, X.J.; Feng, H.; Yin, Z.Y.; Kang, Z.S. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol. Biochem. 2014, 80, 90–96. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 73–83. [Google Scholar] [CrossRef]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. osa-MIR393: A salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2011, 38, 237–242. [Google Scholar] [CrossRef]
- Liu, X.; Xia, B.; Purente, N.; Chen, B.; Zhou, Y.; He, M. Transgenic Chrysanthemum indicum overexpressing cin-miR396a exhibits altered plant development and reduced salt and drought tolerance. Plant Physiol. Biochem. 2021, 168, 17–26. [Google Scholar] [CrossRef]
- Yuan, H.; Cheng, M.; Wang, R.; Wang, Z.; Fan, F.; Wang, W.; Si, F.; Gao, F.; Li, S. miR396b/GRF6 module contributes to salt tolerance in rice. Plant Biotechnol. J. 2024, 22, 2079–2092. [Google Scholar] [CrossRef]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Li, Y.; Cai, H.; Ji, W.; Guo, D.; Zhu, Y. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 2010, 231, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.Q.; Brown, C.W.; Pegler, J.L.; Eamens, A.L.; Grof, C.P.L. Molecular Manipulation of MicroRNA397 Abundance Influences the Development and Salt Stress Response of Arabidopsis thaliana. Int. J. Mol. Sci. 2020, 21, 7879. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Chun, H.J.; Kang, S.; Shin, G.; Park, S.J.; Hong, H.; Kim, C.; Kim, D.H.; Lee, S.Y.; Kim, M.C.; et al. A Role for Arabidopsis miR399f in Salt, Drought, and ABA Signaling. Mol. Cells 2016, 39, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Tuteja, N. microRNAs targeting DEAD-box helicases are involved in salinity stress response in rice (Oryza sativa L.). BMC Plant Biol. 2012, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Guo, W.; Li, J.; Pan, X.; Pan, L.; Zhao, J.; Zhang, Y.; Cai, S.; Huang, X.; Wang, A.; et al. The miR528-AO Module Confers Enhanced Salt Tolerance in Rice by Modulating the Ascorbic Acid and Abscisic Acid Metabolism and ROS Scavenging. J. Agric. Food Chem. 2021, 69, 8634–8648. [Google Scholar] [CrossRef] [PubMed]
- Shamloo-Dashtpagerdi, R.; Sisakht, J.N.; Tahmasebi, A. MicroRNA miR1118 contributes to wheat (Triticum aestivum L.) salinity tolerance by regulating the Plasma Membrane Intrinsic Proteins1;5 (PIP1;5) gene. J. Plant Physiol. 2022, 278, 153827. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Ou, X.; Tang, H.; Wang, R.; Wu, P.; Jia, Y.; Wei, X.; Xu, X.; Kang, S.H.; Kim, S.K.; et al. Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 2015, 208, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zhang, X.; Wang, W.; Yuan, R.; Shen, F. Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol. 2018, 18, 23. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Cheng, Y.; Song, L.; Hu, M.; Shen, J.; Xu, Q.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 459. [Google Scholar] [CrossRef]
- Zhang, X.; Shen, J.; Xu, Q.; Dong, J.; Song, L.; Wang, W.; Shen, F. Long noncoding RNA lncRNA354 functions as a competing endogenous RNA of miR160b to regulate ARF genes in response to salt stress in upland cotton. Plant Cell Environ. 2021, 44, 3302–3321. [Google Scholar] [CrossRef]
- Ye, X.; Wang, S.; Zhao, X.; Gao, N.; Wang, Y.; Yang, Y.; Wu, E.; Jiang, C.; Cheng, Y.; Wu, W.; et al. Role of lncRNAs in cis- and trans-regulatory responses to salt in Populus trichocarpa. Plant J. 2022, 110, 978–993. [Google Scholar] [CrossRef]
- Montez, M.; Majchrowska, M.; Krzyszton, M.; Bokota, G.; Sacharowski, S.; Wrona, M.; Yatusevich, R.; Massana, F.; Plewczynski, D.; Swiezewski, S. Promoter-pervasive transcription causes RNA polymerase II pausing to boost DOG1 expression in response to salt. EMBO J. 2023, 42, e112443. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, X.; Niu, F.; Sun, X.; Hu, Z.; Gao, F.; Zhang, H.; Jiang, Q. Overexpression of lncRNA77580 Regulates Drought and Salinity Stress Responses in Soybean. Plants 2023, 12, 181. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A Nucleus-Localized Long Non-Coding RNA Enhances Drought and Salt Stress Tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Patil, D.P.; Xia, D.F.; Paine, C.B.; Fauser, F.; Richards, H.W.; Shivak, D.A.; Bendaña, Y.R.; Hinkley, S.J.; Scarlott, N.A.; et al. Enhancing gene editing specificity by attenuating DNA cleavage kinetics. Nat. Biotechnol. 2019, 37, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Watanabe, S.; Ando, N.; Ishihara, M.; Sato, M. Transplacental Gene Delivery (TPGD) as a Noninvasive Tool for Fetal Gene Manipulation in Mice. Int. J. Mol. Sci. 2019, 20, 5926. [Google Scholar] [CrossRef] [PubMed]
- Paschon, D.E.; Lussier, S.; Wangzor, T.; Xia, D.F.; Li, P.W.; Hinkley, S.J.; Scarlott, N.A.; Lam, S.C.; Waite, A.J.; Truong, L.N.; et al. Diversifying the structure of zinc finger nucleases for high-precision genome editing. Nat. Commun. 2019, 10, 1133. [Google Scholar] [CrossRef]
- Kim, E.Y.; Kim, K.D.; Cho, J. Harnessing epigenetic variability for crop improvement: Current status and future prospects. Genes Genom. 2021, 44, 259–266. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).