Raman Spectroscopic Algorithms for Assessing Virulence in Oral Candidiasis: The Fight-or-Flight Response
Abstract
1. Introduction
2. Results
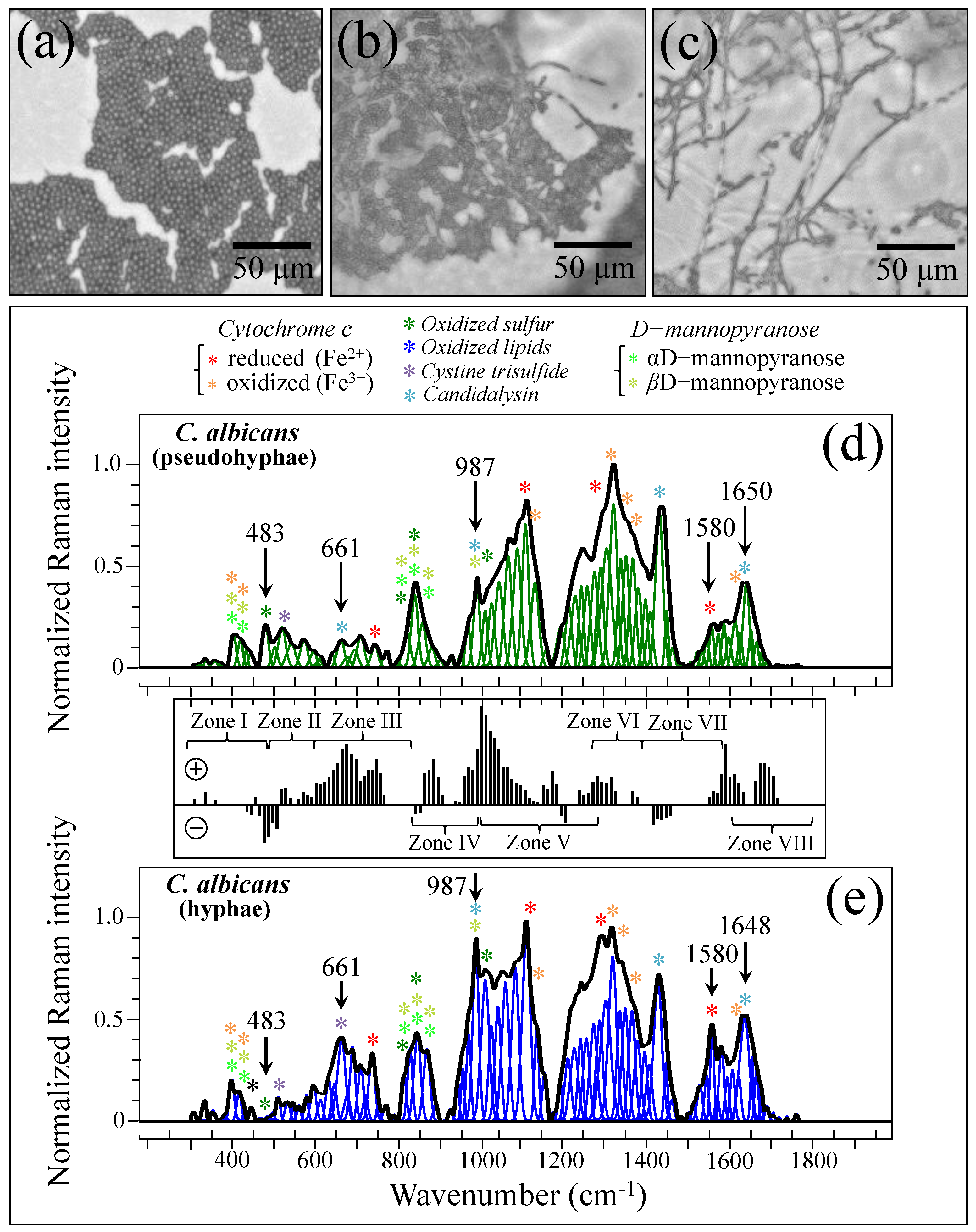
2.1. Microscopy Observations and Clinical Characterizations
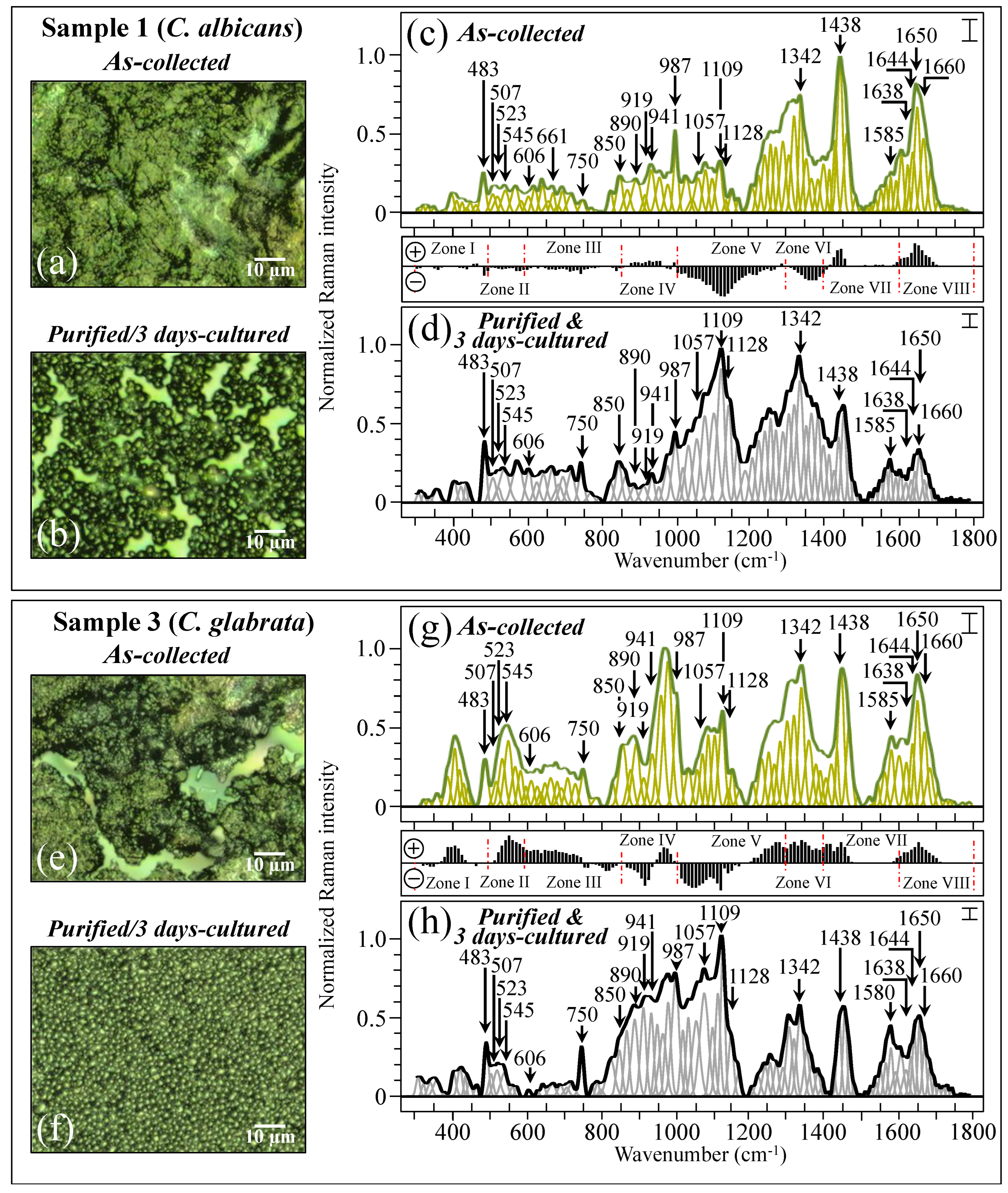
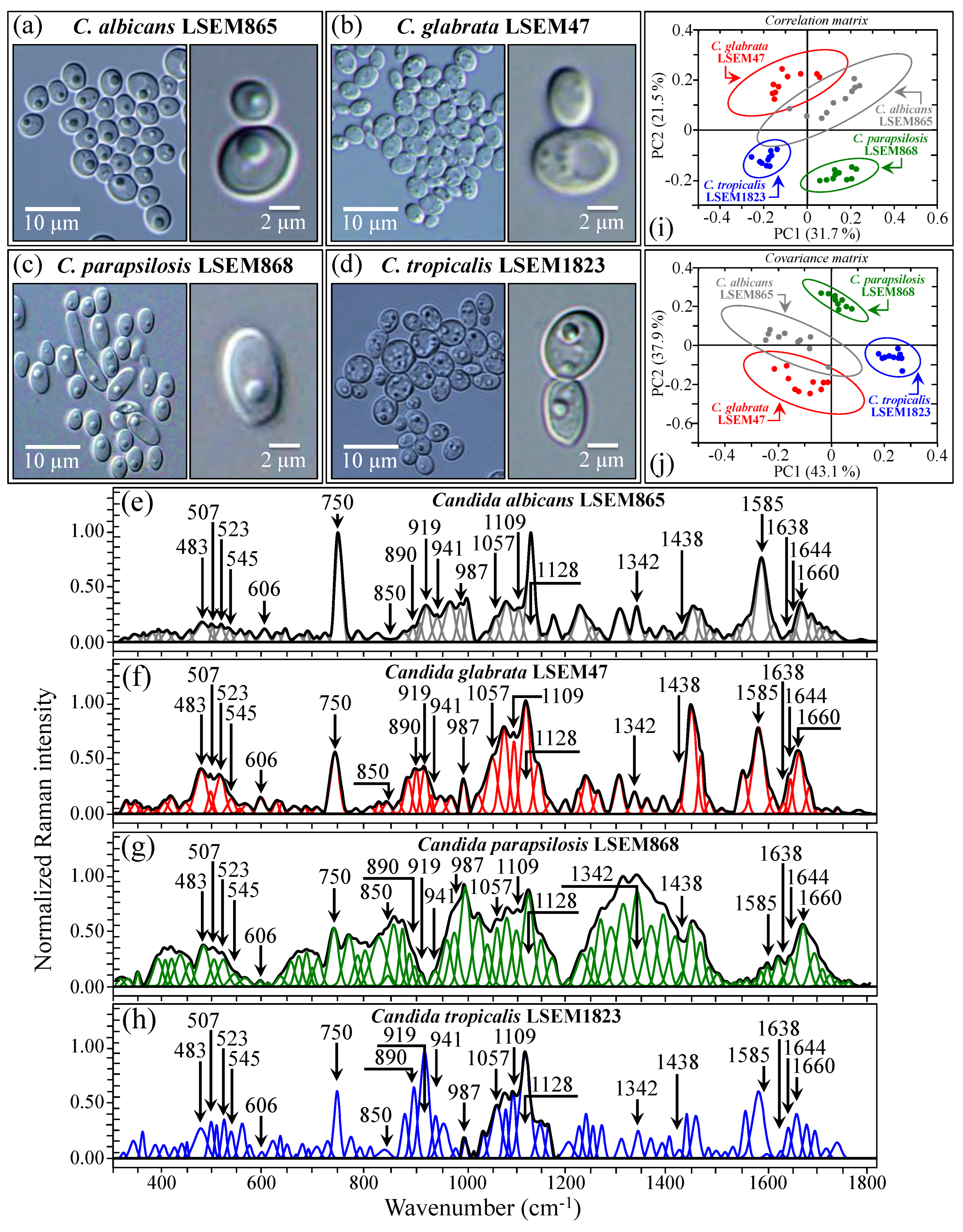
2.2. Raman Spectroscopic Measurements
2.3. Principal Component Analysis on Reference and Clinical Samples
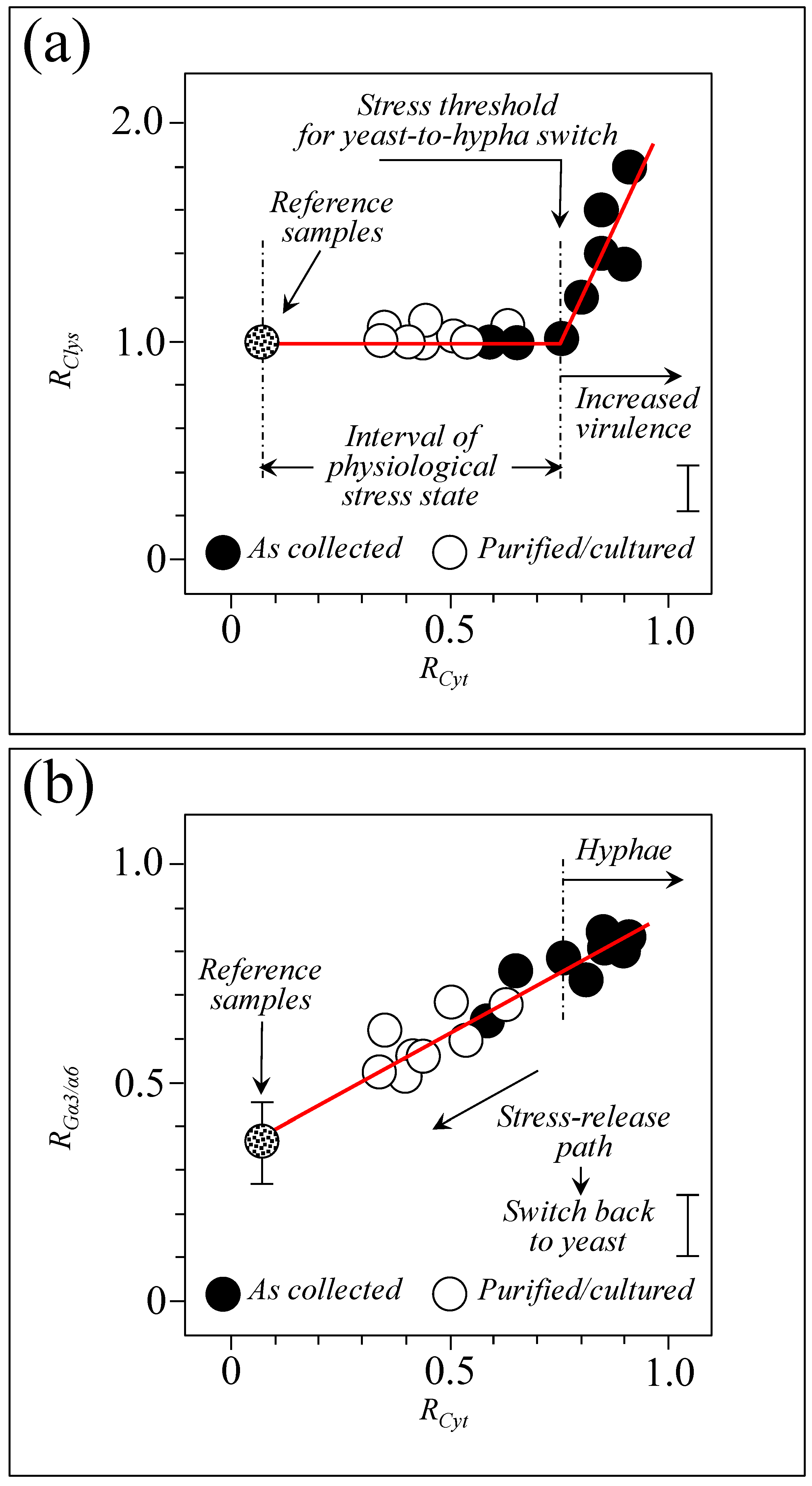
3. Discussion
3.1. The Multifactorial Origin of Raman Complexity in Clinical Samples
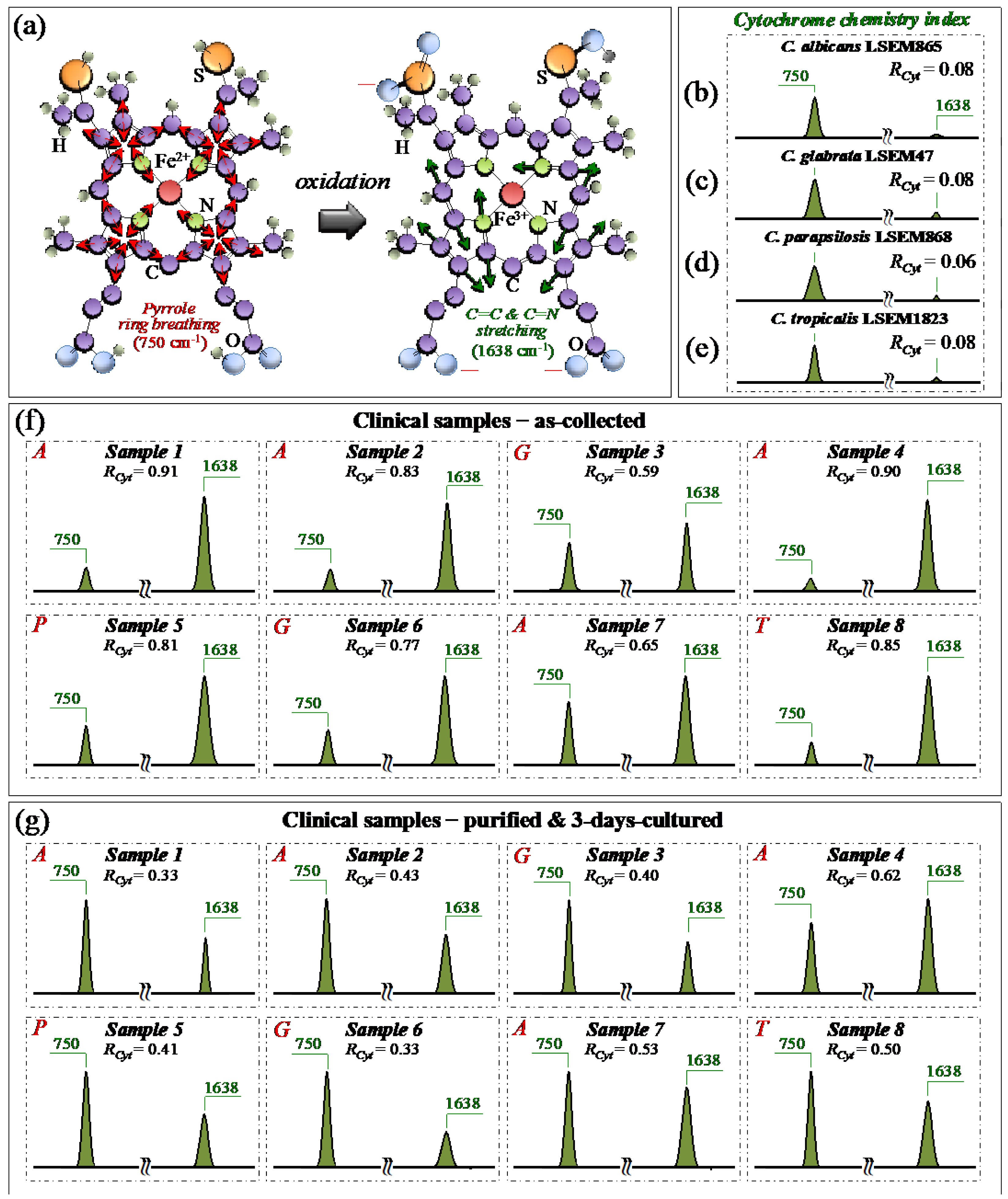
3.2. Raman Fingerprints for Oxidation of Sulfur-Containing Amino Acids
3.3. Oxidative Stress Assessments by Monitoring Cytochrome C Redox State
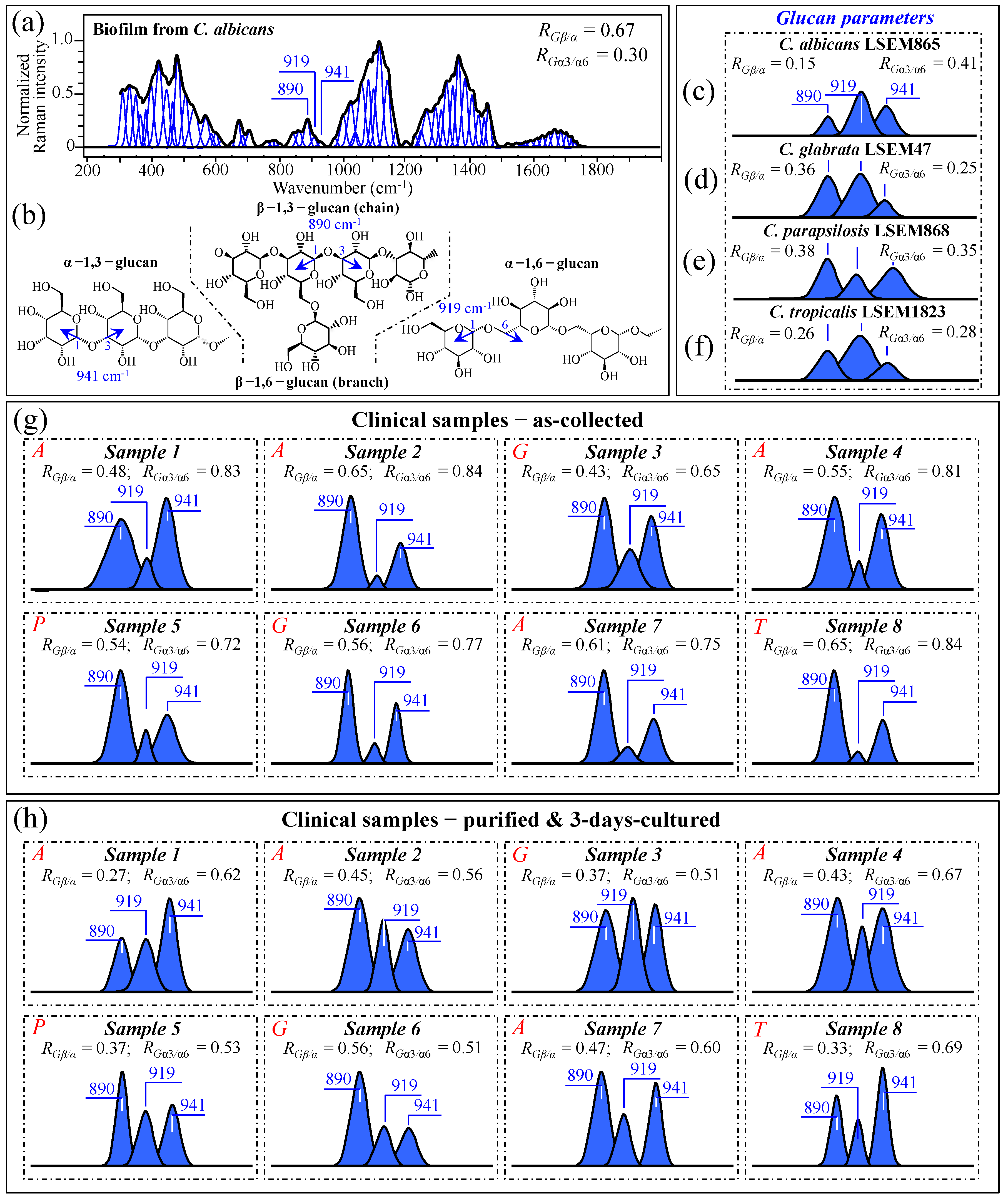
3.4. Raman Characterization of Glucan Structures in Biofilm and Cell Walls
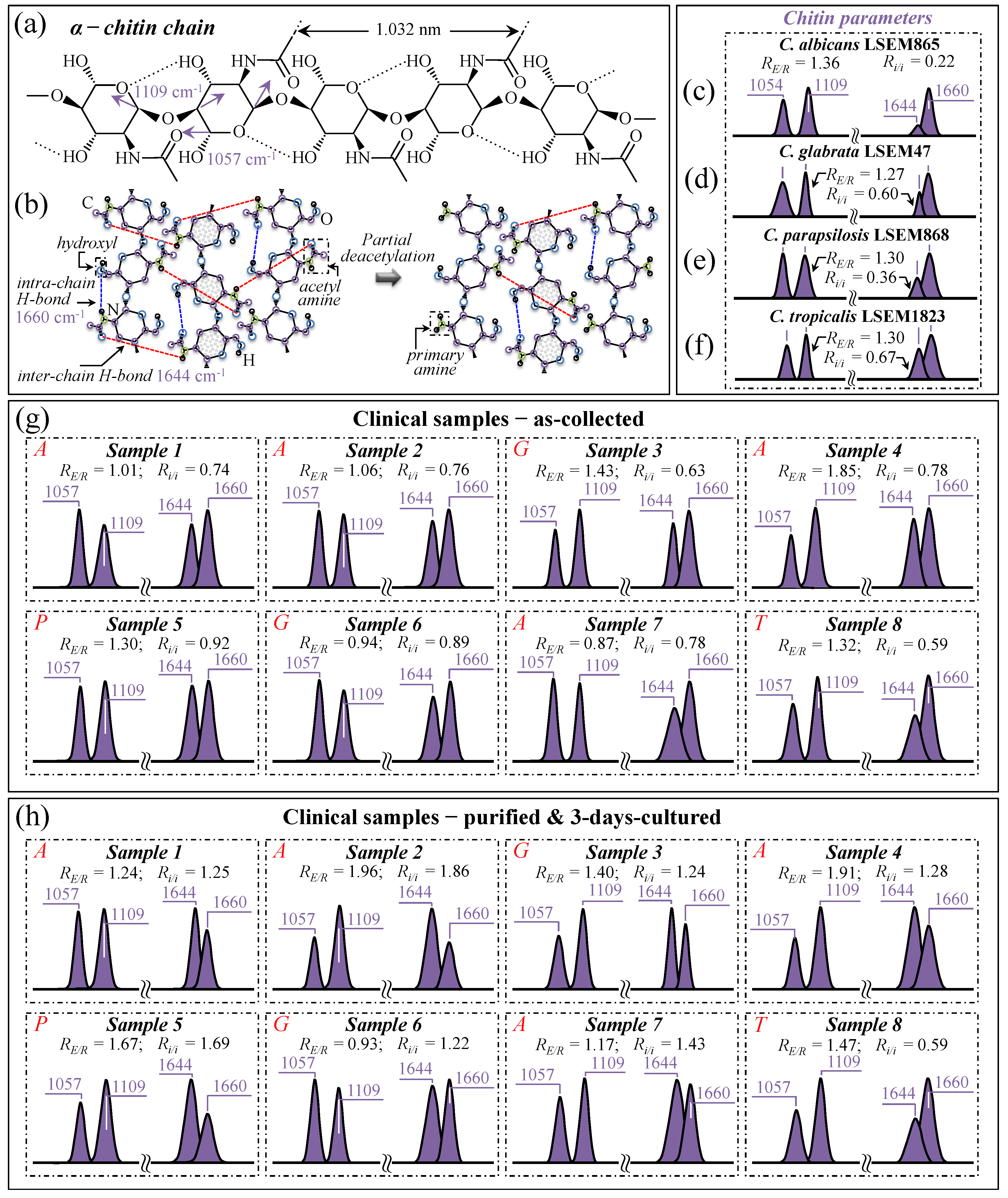
3.5. Raman Characterization of Chitin Structures in Biofilm and Cell Walls
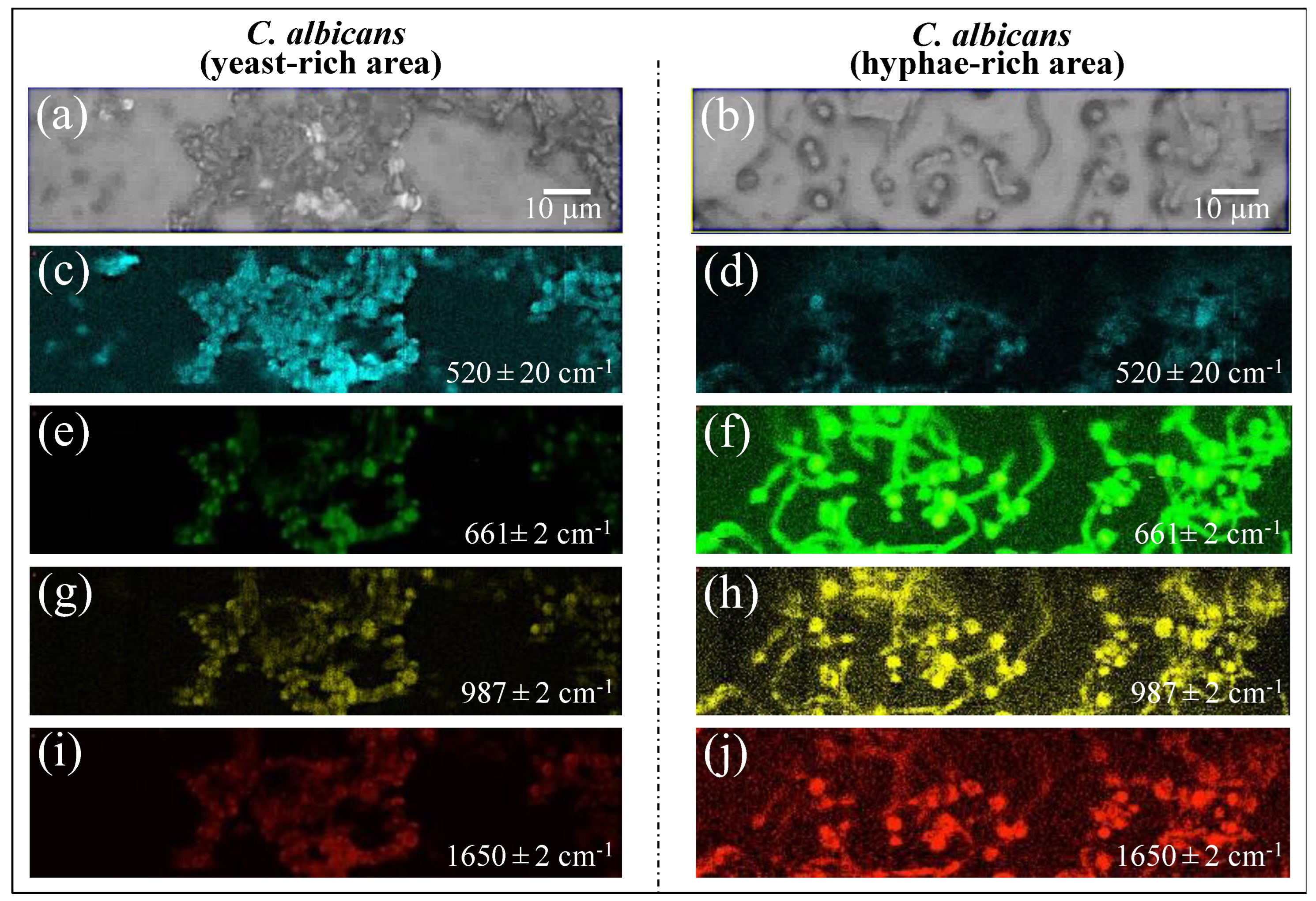
3.6. Spectroscopic Fingerprints of Environmentally Driven Morphogenesis
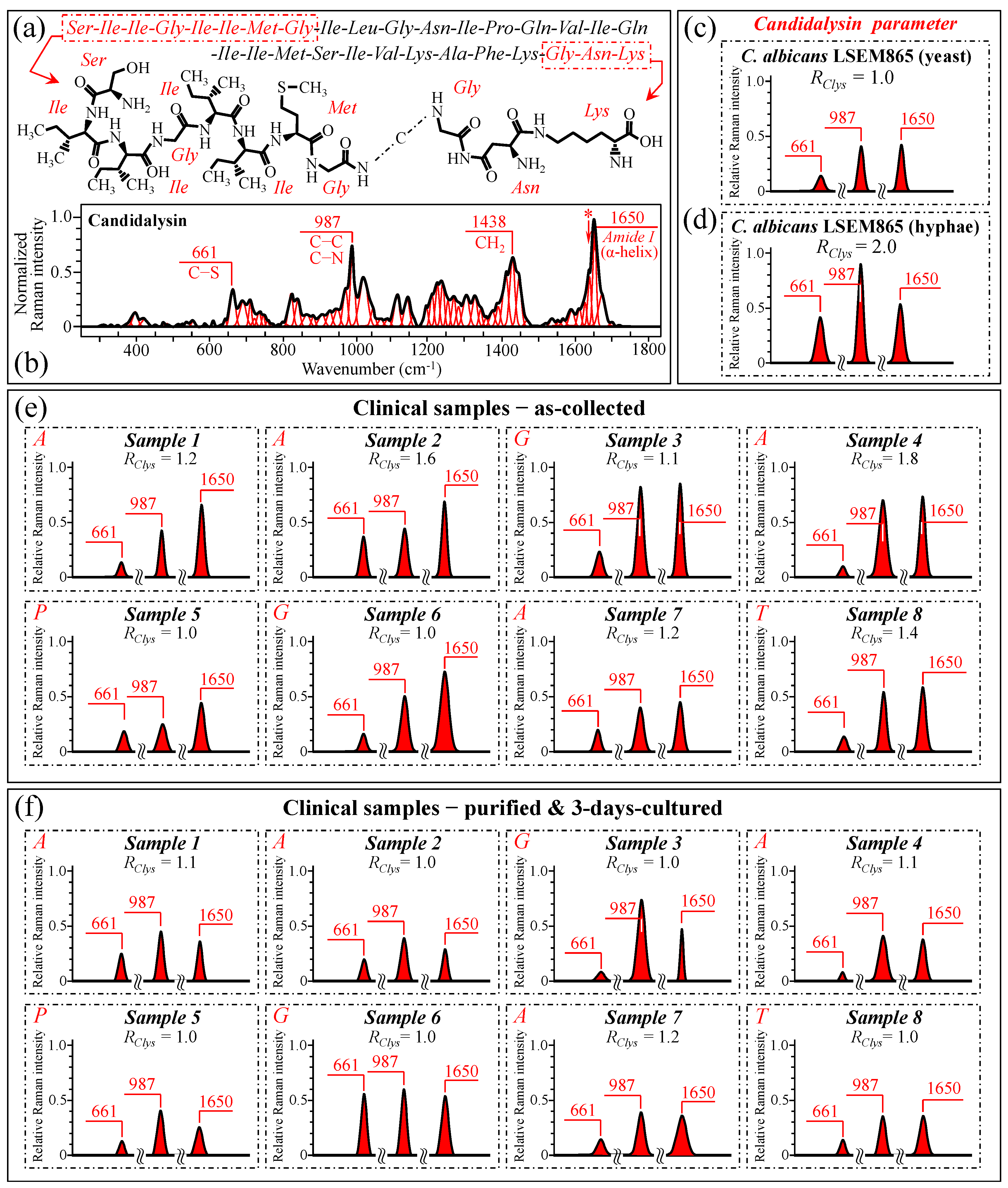
3.7. Spectroscopic Fingerprints of Peptide Toxins
3.8. Statistical Validation by Means of Raman Imaging
3.9. Other Overlapping Signals Characteristic of Bacterial Biofilms
3.10. Raman Spectroscopic Criteria for Assessing Candidiasis Severity
3.11. Limitations of This Study and Directions for Future Work
4. Materials and Methods
4.1. Clinical Samples and Assessment of Their Clinical Characteristics
4.2. Reference Samples
4.3. Raman Spectroscopy
4.4. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Talapko, J.; Juzbasic, M.; Matijevic, T.; Pustijanac, E.; Bekic, S.; Kotris, I.; Skrlec, I. Candida albicans—The virulence factors and clinical manifestations of infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.-Z.; Yan, L.; Jiang, Y.-Y. The synthesis, regulation, and functions of sterols in Candida albicans: Well-known but still lots to learn. Virulence 2016, 7, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Fresneda, R.; Guirao-Abad, J.P.; Arguelles, A.; Gonzales-Parraga, P.; Valentin, E.; Arguelles, J.-C. Specific stress-induced storage of trehalose, glycerol and D-arabitol in response to oxidative and osmotic stress in Candida albicans. Biochem. Biophys. Res. Commun. 2013, 430, 1334–1339. [Google Scholar] [CrossRef]
- Noble, S.M.; Gianetti, B.A.; Witchley, J.N. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat. Rev. Microbiol. 2017, 15, 96–108. [Google Scholar] [CrossRef]
- Bonhomme, J.; d’Enfert, C. Candida albicans biofilms: Building a heterogeneous, drug-tolerant environment. Curr. Opin. Microbiol. 2013, 16, 398–403. [Google Scholar] [CrossRef]
- Sav, H.; Altinbas, R.; Dursun, Z.B. Fungal profile and antifungal susceptibility pattern in patients with oral candidiasis. Infez. Med. 2020, 28, 392–396. [Google Scholar]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman spectroscopy of oral Candida species: Molecular-scale analyses, chemometrics, and barcode identification. Int. J. Mol. Sci. 2023, 23, 5359. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Fujii, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; et al. Raman metabolomics of Candida auris clades: Profiling and barcode identification. Int. J. Mol. Sci. 2022, 23, 11736. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Asai, T.; Nakaya, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman imaging of pathogenic Candida auris: Visualization of structural characteristics and machine-learning identification. Front. Microbiol. 2021, 12, 769597. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Asai, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; et al. Raman study of pathogenic Candida auris: Imaging metabolic machineries in reaction to antifungal drugs. Front. Microbiol. 2022, 13, 896359. [Google Scholar] [CrossRef]
- Varki, A. Biological roles of glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Yadav, V.; Prasad, R. Comparative lipidomics in clinical isolates of Candida albicans reveal crosstalk between mitochondria, cell wall integrity and azole resistance. PLoS ONE 2012, 7, e39812. [Google Scholar]
- McAvan, B.S.; Bowsher, L.A.; Powell, T.; O’Hara, J.F.; Spitali, R.; Goodacre, M.; Doig, A.J. Raman spectroscopy to monitor post-translational modifications and degradation in monoclonal antibody therapeutics. Anal. Chem. 2020, 92, 10381–10389. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.; Muhamadali, H.; Goodacre, R. The role of Raman spectroscopy within quantitative metabolomics. Annu. Rev. Anal. Chem. 2020, 14, 323–345. [Google Scholar] [CrossRef]
- Pezzotti, G. Raman spectroscopy in cell biology and microbiology. J. Raman Spectrosc. 2021, 52, 2348–2443. [Google Scholar] [CrossRef]
- Yao, X.; Höppener, C.; Schneidewind, H.; Hoeppener, S.; Tang, Z.; Buchholz, A.; König, A.; Mogavero, S.; Diegel, M.; Dellith, J.; et al. Targeted suppression of peptide degradation in Ag-based surface-enhanced Raman spectra by depletion of hot carriers. Small 2022, 18, 2205080. [Google Scholar] [CrossRef]
- Trentin, G.; Bitencourt, T.A.; Guedes, A.; Pessoni, A.M.; Brauer, V.S.; Pereira, A.K.; Costa, J.H.; Fill, T.P.; Almeida, F. Mass spectrometry analysis reveals lipids induced by oxidative stress in Candida albicans. Microorganisms 2023, 11, 1669. [Google Scholar] [CrossRef]
- Himmelreich, U.; Somorjai, R.L.; Dolenko, B.; Lee, O.C.; Daniel, H.-M.; Murray, R.; Mountford, C.E.; Sorrell, T.C. Rapid identification of Candida species by using nuclear magnetic resonance spectroscopy and a statistical classification strategy. Appl. Environ. Microbiol. 2003, 69, 4566–4574. [Google Scholar] [CrossRef]
- Lipke, P.N. Glycomics for microbes and microbiologists. mBio 2016, 7, e01224-16. [Google Scholar] [CrossRef]
- Garcia, B.; Bermejo, B.; Owens, R.A.; Briones, A.; Arevalo-Villena, M. Proteomic profiling and glycomic analysis of the yeast cell wall in strains with Aflatoxin B1 elimination ability. Environ. Microbiol. 2021, 23, 5305–5319. [Google Scholar] [CrossRef]
- Opathy, C.; Gabaldon, T. Recent trends in molecular diagnostics of yeast infections: From PCR to NGS. FEMS Microbiol. Rev. 2019, 43, 517–547. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, M.; Kumar, N.; Kaur, R. Global secretome characterization of the pathogenic yeast Candida glabrata. J. Proteome Res. 2020, 19, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Costa-Orlandi, C.B.; Sardi, J.C.O.; Pitangui, N.S.; de Oliveira, H.C.; Scorzoni, L.; Galeane, M.C.; Medina-Alarcon, K.P.; Melo, W.C.M.A.; Marcelino, M.Y.; Braz, J.D.; et al. Fungal biofilms and polymicrobial Diseases. J. Fungi 2017, 3, 22. [Google Scholar] [CrossRef]
- Pezzotti, G.; Ofuji, S.; Imamura, H.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; Zhu, W.; Mazda, O.; et al. In situ Raman analysis of biofilm exopolysaccharides formed in Streptococcus mutans and Streptococcus sanguinis commensal cultures. Int. J. Mol. Sci. 2023, 24, 6694. [Google Scholar] [CrossRef]
- Alnuaimi, A.D.; O’Brien-Simpson, N.M.; Reynolds, E.C.; McCullough, M.J. Clinical isolates and laboratory reference Candida species and strains have varying abilities to form biofilms. FEMS Yeast Res. 2013, 13, 689–699. [Google Scholar] [CrossRef]
- Nejad, E.E.; Nejad Almani, P.G.; Mohammadi, M.A.; Salari, S. Molecular identification of Candida isolates by real-time PCR-high-resolution melting analysis and investigation of the genetic diversity of Candida species. J. Clin. Lab. Anal. 2020, 34, e23444. [Google Scholar] [CrossRef]
- Odds, F.C.; Brown, A.J.P.; Gow, N.A.R. Candida albicans genome sequence: A platform for genomics in the absence of genetics. Genome Biol. 2004, 5, 230. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, J.; Tian, L.; Zhang, Q.; Guan, Y.; Chen, L.; Liu, G.; Yu, H.-q.; Tian, Y.; Huang, Q. Raman micro-spectroscopy monitoring of cytochrome c redox state in Candida utilis during cell death under low-temperature plasma-induced oxidative stress. Analyst 2020, 145, 3922–3930. [Google Scholar] [CrossRef]
- Bin, P.; Huang, R.; Zhou, X. Oxidation resistance of the sulfur amino acids: Methionine and cysteine. BioMed Res. Int. 2017, 2017, 9584932. [Google Scholar] [CrossRef]
- Poole, L.B. The basics of thiols and cysteines in redox biology and chemistry. Free Radic. Biol. Med. 2015, 80, 148–157. [Google Scholar] [CrossRef]
- Liang, X.; Kaya, A.; Zhang, Y.; Le, D.T.; Hua, D.; Gladyshev, V.N. Characterization of methionine oxidation and methionine sulfoxide reduction using methionine-rich cysteine-free proteins. BMC Biochem. 2012, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, R.O.; Prado, F.M.; Massafera, M.P.; Mascio, P.D.; Ronsein, G.E. Dehydromethionine is a common product of methionine oxidation by singlet molecular oxygen and hypohalous acid. Free Radic. Biol. Med. 2022, 187, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.A.; de la Lastra, J.M.P.; Plou, F.J.; Perez-Lebena, E. The chemistry of reactive oxygen species (ROS) revisited: Outlining their role in biological macromolecules (DNA, Lipids and proteins) and induced pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Adachi, T.; Imamura, H.; Bristol, D.R.; Adachi, K.; Yamamoto, T.; Kanamura, N.; Marin, E.; Zhu, W.; Kawai, T.; et al. In situ Raman study of neurodegenerated human neuroblastoma cells exposed to outer-membrane vesicles isolated from Porphyromonas gingivalis. Int. J. Mol. Sci. 2023, 24, 13351. [Google Scholar] [CrossRef]
- Pezzotti, G.; Ohgitani, E.; Imamura, H.; Ikegami, S.; Shin-Ya, M.; Adachi, T.; Adachi, K.; Yamamoto, T.; Kanamura, N.; Marin, E.; et al. Raman multi-omic snapshot and statistical validation of structural differences between Herpes simplex Type I and Epstein-Barr viruses. Int. J. Mol. Sci. 2023, 24, 15567. [Google Scholar] [CrossRef]
- Kurouski, D.; Van Duyne, R.P.; Lednev, I.K. Exploring structure and formation mechanism of amyloid fibrils by Raman spectrocopy. A review. Analyst 2015, 140, 4967–4980. [Google Scholar] [CrossRef]
- Xie, Y.; Jiang, Y.; Ben-Amotz, D. Detection of amino acid and peptide phosphate protonation using Raman spectroscopy. Anal. Biochem. 2005, 343, 223–230. [Google Scholar] [CrossRef]
- Shiota, M.; Naya, M.; Yamamoto, T.; Hishiki, T.; Tani, T.; Takahashi, H.; Kubo, A.; Koike, D.; Itoh, M.; Ohmura, M.; et al. Gold-nanofeve surface-enhanced Raman spectroscopy visualizes hypotaurine as a robust anti-oxidant consumed in cancer survival. Nat. Commun. 2018, 9, 1561. [Google Scholar] [CrossRef]
- Hernandez, B.; Pflueger, F.; Kruglik, S.G.; Ghomi, M. Characteristic Raman lines of phenylalanine analyzed by a multiconformal approach. J. Raman Spectrosc. 2013, 44, 827–833. [Google Scholar] [CrossRef]
- Paulsen, C.E.; Carroll, K.S. Cysteine-mediated redox signaling: Chemistry, biology, and tools for discovery. Chem. Rev. 2013, 113, 4633–4679. [Google Scholar] [CrossRef]
- Sato, S.; Higuchi, S.; Tanaka, S. Identification and determination of oxygen-containing inorganosulfur compounds by laser Raman spectrometry. Appl. Spectrosc. 1985, 39, 822–827. [Google Scholar] [CrossRef]
- Yucesoy, M.; Marol, S. Performance of CHROMAGAR candida and BIGGY agar for identification of yeast species. Ann. Clin. Microbiol. Antimicrobiol. 2003, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Huycke, M.M.; Abrams, V.; Moore, D.R. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis 2002, 23, 529–536. [Google Scholar] [CrossRef]
- Fujishima, K.; Kawada, M.; Oogai, Y.; Tokuda, M.; Torii, M.; Komatsuzawa, H. Dpr and sod in Streptococcus mutans are involved in coexistence with Streptococcus sanguinis, and PerR is associated with resistance to H2O2. Appl. Environ. Microbiol. 2013, 79, 1436–1443. [Google Scholar] [CrossRef]
- Fitzsimmons, N.; Berry, D.R. Inhibition of Candida albicans by Lactobacillus acidophilus: Evidence for the involvement of a peroxidase system. Microbios 1994, 80, 125–133. [Google Scholar]
- Cruz, M.R.; Graham, C.E.; Gagliano, B.C.; Lorenz, M.C.; Garsin, D.A. Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 2013, 81, 189–200. [Google Scholar] [CrossRef]
- Phillips, A.J.; Sudbery, I.; Ramsdale, M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 2003, 100, 14327–14332. [Google Scholar] [CrossRef]
- Halliwell, B. Oxidative stress and cancer: Have we moved forward? Biochem. J. 2007, 401, 1–11. [Google Scholar] [CrossRef]
- Brown, G.C.; Borutaite, V. Regulation of apoptosis by the redox state of cytochrome c. Biochim. Biophys. Acta 2008, 1777, 877–881. [Google Scholar] [CrossRef]
- Guerra-Castellano, A.; Marquez, I.; Perez-Mejias, G.; Diaz-Quintana, A.; De la Rosa, M.A.; Diaz-Moreno, I. Post-translational modifications of cytochrome c in cell life and disease. Int. J. Mol. Sci. 2020, 21, 8483. [Google Scholar] [CrossRef]
- Trewhella, J.; Carlson, V.A.; Curtis, E.H.; Heidorn, D.B. Differences in the solution structures of oxidized and reduced cytochrome c measured by small-angle X-ray scattering. Biochemistry 1988, 27, 1121–1125. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Ogura, T. Resonance Raman spectra of cytochrome c oxidase in whole mitochondria. Bull. Chem. Soc. Jpn. 2002, 75, 1001–1004. [Google Scholar] [CrossRef]
- Piccoli, C.; Perna, G.; Scrima, R.; Cela, O.; Rinaldi, R.; Boffoli, D.; Capozzi, V.; Capitanio, N. A novel redox state Heme a marker in cytochrome c oxidase revealed by Raman spectroscopy. Phys. Scr. 2005, 2005, 199–204. [Google Scholar] [CrossRef]
- Hu, S.; Morris, I.K.; Singh, J.P.; Smith, K.M.; Spiro, T.G. Complete assignment of cytochrome c resonance Raman spectra via enzymatic reconstitution with isotopically labeled hemes. J. Am. Chem. Soc. 1993, 115, 12446–12458. [Google Scholar] [CrossRef]
- Kitagawa, T.; Orii, Y. Resonance Raman studies of cytochrome oxidase. J. Biochem. 1978, 84, 1245–1252. [Google Scholar] [CrossRef]
- Kakita, M.; Kaliaperumal, V.; Hamaguchi, H. Resonance Raman quantification of the redox state of cytochromes b and c in-vivo and in-vitro. J. Biophotonics 2012, 5, 20–24. [Google Scholar] [CrossRef]
- Okada, M.; Smith, N.I.; Palonpon, A.F.; Endo, H.; Kawata, S.; Sodeoka, M.; Fujita, K. Label-free Raman observation of cytochrome c dynamics during apoptosis. Proc. Natl. Acad. Sci. USA 2012, 109, 28–32. [Google Scholar] [CrossRef]
- Morimoto, T.; Kanda, H.; Kawagoe, H.; Ozawa, T.; Nakamura, M.; Nishida, K.; Fujita, K.; Fujikado, T. Using redox-sensitive mitochondrial cytochrome Raman bands for label-free detection of mitochondrial dysfunction. Analyst 2019, 144, 2531–2540. [Google Scholar] [CrossRef]
- Tournu, H.; Van Dijck, P. Candida biofilm and the host: Models and new concepts for eradication. Int. J. Microbiol. 2012, 2012, 845352. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Al-Fattani, M.A.; Douglas, L.J. Biofilm matrix of Candida albicans and Candida tropicalis: Chemical composition and role in drug resistance. J. Med. Microbiol. 2006, 55, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Mickova, K.; Synytsya, A.; Jablonsky, I.; Spevacek, J.; Erban, V.; Kovarikova, E.; Copikova, J. Glucans from fruit bodies of cultivated mushrooms Pleurotus ostreatus and Pleurotus eryngii: Structure and potential prebiotic activity. Carbohydr. Polym. 2009, 76, 548–556. [Google Scholar] [CrossRef]
- Mikkelsen, M.S.; Jespersen, B.M.; Moller, B.L.; Laerke, H.N.; Larsen, F.H.; Engelsen, S.B. Comparative spectroscopic and rheological studies on crude and purified soluble barley and oat β-glucan preparations. Food Res. Int. 2010, 43, 2417–2424. [Google Scholar] [CrossRef]
- Hanada, S.B.; Kuramitsu, H.K. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect. Immun. 1988, 56, 1999–2005. [Google Scholar] [CrossRef]
- Hanada, N.; Kuramitsu, H.K. Isolation and characterization of the Streptococcus mutans gtfD gene, coding for primer-dependent soluble glucan synthesis. Infect. Immun. 1989, 57, 2079–2085. [Google Scholar] [CrossRef]
- Wangpaiboon, K.; Padungros, P.; Nakapong, S.; Charoenwongpaiboon, T.; Rejzek, H.; Field, R.A.; Pichyangkura, R. An α-1,6-and α-1,3-linked glucan produced by Leuconostoc citreum ABK-1 alternansucrase with nanoparticle and film-forming properties. Sci. Rep. 2018, 8, 8340. [Google Scholar] [CrossRef]
- Gantner, B.N.; Simmons, R.M.; Underhill, D.M. Dectin-1 mediates macrophage recognition of Candida albicans yeast but not filaments. Embo J. 2005, 24, 1277–1286. [Google Scholar] [CrossRef]
- Kang, X.; Kirui, A.; Muszynski, A.; Dickwella-Widanage, M.C.; Chen, A.; Azadi, P.; Wang, P.; Mentink-Vigier, F.; Wang, T. Molecular architecture of fungal cell walls revealed by solid-state NMR. Nat. Commun. 2018, 9, 2747. [Google Scholar] [CrossRef]
- De Gussem, K.; Vandenabeele, P.; Verbeken, A.; Moens, L. Raman spectroscopic study of Lactarius spores (Russulales, Fungi). Spectrochim. Acta Part A 2005, 61, 2896–2908. [Google Scholar] [CrossRef]
- Focher, B.; Naggi, A.; Torri, G.; Cosani, A.; Terbojevich, M. Structural differences between chitin polymorphs and their precipitates from solutions—Evidence from CP-MAS13C-NMR, FT-IR and FT-Raman spectroscopy. Carbohydr. Polym. 1992, 17, 97–102. [Google Scholar] [CrossRef]
- Zhang, K.; Geissler, A.; Fischer, S.; Brendler, E.; Baucker, E. Solid-state spectroscopic characterization of α-chitins deacetylated in homogeneous solutions. J. Phys. Chem. B 2012, 116, 4584–4592. [Google Scholar] [CrossRef] [PubMed]
- Minke, R.; Blackwell, J. The structure of α–chitin. J. Mol. Biol. 1978, 120, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sasaki, T.; Irie, S.; Sakurai, K. A novel biomass-ionic liquid platform for the utilization of native chitin. Polymer 2008, 49, 2321–2327. [Google Scholar] [CrossRef]
- Okuyama, K.; Noguchi, K.; Miyazawa, T.; Yui, T.; Ogawa, K. Molecular and crystal structure of hydrated chitosan. Macromolecules 1997, 30, 5849–5855. [Google Scholar] [CrossRef]
- Okuyama, K.; Noguchi, K.; Kanenari, M.; Egawa, T.; Osawa, K. Structural diversity of chitosan and its complexes. Carbohydr. Polym. 2000, 41, 237–247. [Google Scholar] [CrossRef]
- Van de Velde, K.; Kiekens, P. Structure analysis and degree of substitution of chitin, chitosan, and dibutyrylchitin by FT-IR spectroscopy and solid state 13C NMR. Carbohydr. Polym. 2004, 58, 409–416. [Google Scholar] [CrossRef]
- Kameda, T.; Miyazawa, M.; Ono, H.; Yoshida, M. Hydrogen bonding structure and stability of -chitin studied by 13C Solid-State NMR. Macromol. Biosci. 2005, 5, 103–106. [Google Scholar] [CrossRef]
- Deringer, V.L.; Englert, U.; Dronskowski, R. Nature, strength, and cooperativity of the hydrogen-bonding network in α-chitin. Biomacromolecules 2016, 17, 996–1003. [Google Scholar] [CrossRef]
- Sawada, D.; Nishiyama, Y.; Langan, P.; Forsyth, V.T.; Kimura, S.; Wada, M. Water in crystalline fibers of dehydrate β-chitin results in unexpected absence of intramolecular hydrogen bonding. PLoS ONE 2012, 7, e39376. [Google Scholar] [CrossRef]
- Sawada, D.; Nishiyama, Y.; Langan, P.; Forsyth, V.T.; Kimura, S.; Wada, M. Direct Determination of the Hydrogen Bonding Arrangement in Anhydrous β-Chitin by Neutron Fiber Diffraction. Biomacromolecules 2012, 13, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto-Nakano, M. Role of Streptococcus mutans surface proteins for biofilm formation. Jpn. Dent. Sci. Rev. 2018, 54, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Li, M.; Gregory, R.L. Bacterial interactions in dental biofilm. Virulence 2011, 2, 435–444. [Google Scholar] [CrossRef]
- Mack, D.; Fischer, W.; Krokotsch, A.; Leopold, K.; Hartmann, R.; Egge, H.; Laufs, R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta–1,6–linked glucosaminoglycan: Purification and structural analysis. J. Bacteriol. 1996, 178, 175–183. [Google Scholar] [CrossRef]
- Maira-Litrán, T.; Kropec, A.; Abeygunawardana, C.; Joyce, J.; Mark III, G.; Goldmann, D.A.; Pier, G.B. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 2002, 70, 4433–4440. [Google Scholar] [CrossRef]
- Schilcher, K.; Horswill, A.R. Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiol. Mol. Biol. Rev. 2020, 84, e00026-19. [Google Scholar] [CrossRef]
- Gieroba, B.; Stroka-Bartnicka, A.; Kazimierczak, P.; Kalisz, G.; Pieta, I.S.; Nowakowski, R.; Pisarek, M.; Przekora, A. Effect of gelation temperature on the molecular structure and physicochemical properties of the curdlan matrix: Spectroscopic and microscopic analyses. Int. J. Mol. Sci. 2020, 21, 6154. [Google Scholar] [CrossRef]
- Vuong, C.; Kocianova, S.; Voyich, J.M.; Yao, Y.; Fischer, E.R.; DeLeo, F.R.; Otto, M. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 2004, 279, 54881–54886. [Google Scholar] [CrossRef]
- Tsui, C.; Kong, E.F.; Jabra-Rizk, M.A. Pathogenesis of Candida albicans biofilm. Pathog. Dis. 2016, 74, ftw018. [Google Scholar] [CrossRef]
- Maidan, M.M.; De Rop, L.; Serneels, J.; Exler, S.; Rupp, S.; Tournu, H.; Thevelein, J.M.; Van Dijck, P. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 2005, 16, 1971–1986. [Google Scholar] [CrossRef]
- Chen, J.; Lane, S.; Liu, H. A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol. Microbiol. 2002, 46, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Eisman, B.; Alonso-Monge, R.; Roman, E.; Arana, D.; Nombela, C.; Pla, J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot. Cell 2006, 5, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Baek, Y.U.; Martin, S.J.; Davis, D.A. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall beta-glycosidase Phr2. Eukaryot. Cell 2006, 5, 1550–1559. [Google Scholar] [CrossRef] [PubMed]
- Lotz, H.; Sohn, K.; Brunner, H.; Muhlschlegel, F.A.; Rupp, S. RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryot. Cell 2004, 3, 776–784. [Google Scholar] [CrossRef]
- Stichternoth, C.; Fraund, A.; Setiadi, E.; Giasson, L.; Vecchiarelli, A.; Ernst, J.F. Sch9 kinase integrates hypoxia and CO2 sensing to suppress hyphal morphogenesis in Candida albicans. Eukaryot. Cell 2011, 10, 502–511. [Google Scholar] [CrossRef]
- Stichternoth, C.; Ernst, J.F. Hypoxic adaptation by Efg1 regulates biofilm formation by Candida albicans. Appl. Environ. Microbiol. 2009, 75, 3663–3672. [Google Scholar] [CrossRef]
- Shi, Q.M.; Wang, Y.M.; Zheng, X.D.; Lee, R.T.; Wang, Y. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 2007, 18, 815–826. [Google Scholar] [CrossRef]
- Bachewich, C.; Thomas, D.Y.; Whiteway, M. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 2003, 14, 2163–2180. [Google Scholar] [CrossRef]
- da Silva Dantas, A.; Patterson, M.J.; Smith, D.A.; MacCallum, D.M.; Erwig, L.P.; Morgan, B.A.; Quinn, J. Thioredoxin regulates multiple hydrogen peroxide-induced signaling pathways in Candida albicans. Mol. Cell. Biol. 2010, 30, 4550–4563. [Google Scholar] [CrossRef]
- Chattaway, F.W.; Holmes, M.R.; Barlow, A.J. Cell wall composition of the mycelial and blastospore forms of Candida albicans. J. Gen. Microbiol. 1968, 51, 367–376. [Google Scholar] [CrossRef]
- Braun, P.C.; Calderone, R.A. Chitin synthesis in Candida albicans: Comparison of yeast and hyphal forms. J. Bacteriol. 1978, 133, 1472–1477. [Google Scholar] [CrossRef]
- Nather, K.; Munro, C.A. Generating cell surface diversity in Candida albicans and other fungal pathogens. FEMS Microbiol. Lett. 2008, 285, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Machova, E.; Fiacanova, L.; Cizova, A.; Korkova, J. Mannoproteins from yeast and hyphal form of Candida albicans considerably differ in mannan and protein content. Carbohydr. Res. 2015, 408, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Dudek, M.; Zajac, G.; Szafraniec, E.; Wiercigroch, E.; Tott, S.; Malek, K.; Kaczor, A.; Baranska, M. Raman optical activity and Raman spectroscopy of carbohydrates in solution. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2019, 206, 597–612. [Google Scholar] [CrossRef] [PubMed]
- Czamara, K.; Majzner, K.; Pacia, M.Z.; Kochan, K.; Kaczor, A.; Baranska, M. Raman spectroscopy of lipids: A review. J. Raman Spectrosc. 2015, 46, 4–20. [Google Scholar] [CrossRef]
- Moyes, D.L.; Wilson, D.; Richardson, J.P.; Mogavero, S.; Tang, S.X.; Wernecke, J.; Hoefs, S.; Gratacap, R.L.; Robbins, J.; Runglall, M.; et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 2016, 532, 64–68. [Google Scholar] [CrossRef]
- Nikou, S.A.; Zhou, C.; Griffiths, J.S.; Kotowicz, N.K.; Coleman, B.M.; Green, M.J.; Moyes, D.L.; Gaffen, S.L.; Naglik, J.R.; Parker, P.J. The Candida albicans toxin candidalysin mediates distinct epithelial inflammatory responses through p38 and EGFR-ERK pathways. Sci. Signal. 2022, 15, eabj6915. [Google Scholar] [CrossRef]
- Li, X.V.; Leonardi, I.; Putzel, G.G.; Semon, A.; Fiers, W.D.; Kusakabe, T.; Lin, W.-Y.; Gao, I.H.; Doron, I.; Gutierrez-Guerrero, A.; et al. Immune regulation by fungal strain diversity in inflammatory bowel disease. Nature 2022, 603, 672–678, Erratum in Nature 2022, 608, E21. [Google Scholar] [CrossRef]
- Blagojevic, M.; Camilli, G.; Maxson, M.; Hube, B.; Moyes, D.L.; Richardson, J.P.; Naglik, J.R. Candidalysin triggers epithelial cellular stresses that induce necrotic death. Cell. Microbiol. 2021, 23, e13371. [Google Scholar] [CrossRef]
- Russell, C.M.; Schaefer, K.G.; Dixson, A.; Gray, A.L.H.; Pyron, R.J.; Alves, D.S.; Moore, N.; Conley, E.A.; Schuck, R.J.; White, T.A.; et al. The Candida albicans virulence factor candidalysin polymerizes in solution to form membrane pores and damage epithelial cells. eLife 2022, 11, e75490. [Google Scholar] [CrossRef]
- Wilson, D.; Naglik, J.R.; Hube, B. The missing link between Candida albicans hyphal morphogenesis and host cell damage. PLoS Pathog. 2016, 12, e1005867. [Google Scholar] [CrossRef] [PubMed]
- Ibelings, M.S.; Maquelin, K.; Endtz, H.P.; Bruining, H.A.; Puppels, G.J. Rapid identification of Cansisa spp. in peritonitis patients by Raman spectroscopy. Clin. Microbiol. Infect. 2005, 11, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.P.; Brown, R.; Kichik, N.; Lee, S.; Priest, E.; Mogavero, S.; Maufreis, C.; Wickramasinghe, D.N.; Tsavou, A.; Kotowicz, N.K.; et al. Candidalysins are a new family of cytolytic fungal peptide toxins. mBio 2022, 13, e03510-21. [Google Scholar] [CrossRef] [PubMed]
- Gieroba, B.; Krysa, M.; Wojtowicz, K.; Wiater, A.; Pleszczyńska, M.; Tomczyk, M.; Sroka-Bartnicka, A. The FT-IR and Raman spectroscopies as tools for biofilm characterization created by cariogenic Streptococci. Int. J. Mol. Sci. 2020, 21, 3811. [Google Scholar] [CrossRef]
- Verma, S.P.; Wallach, D.F.H. Raman spectra of some saturated, unsaturated and deuterated C18 fatty acids in the hch-deformation and ch-stretching regions. Biochim. Biophys. Acta 1977, 486, 217–227. [Google Scholar] [CrossRef]
- Hrubanova, K.; Krzyzanek, V.; Nebesarova, J.; Ruzicka, F.; Pilat, Z.; Samek, O. Monitoring Candida parapsilosis and Staphylococcus epidermidis biofilms by a combination of scanning electron microscopy and Raman spectroscopy. Sensors 2018, 18, 4089. [Google Scholar] [CrossRef]
- Ramirez-Mora, T.; Dávila-Pérez, C.; Torres-Méndez, F.; Valle-Bourrouet, G. Raman spectroscopic characterization of endodontic biofilm matrices. J. Spectrosc. 2019, 2019, 1307397. [Google Scholar] [CrossRef]
- Wagner, M.; Ivleva, N.P.; Haisch, C.; Niessner, R.; Horn, H. Combined use of confocal laser scanning microscopy (CLSM) and Raman microscopy (RM): Investigations on EPS–matrix. Water Res. 2009, 43, 63–76. [Google Scholar] [CrossRef]
- Zeng, G.; Xu, X.; Kok, Y.J.; Deng, F.-S.; Chow, E.W.L.; Gao, J.; Bi, X.; Wang, Y. Cytochrome c regulates hyphal morphogenesis by interfering with cAMP-PKA signaling in Candida albicans. Cell Rep. 2023, 42, 113473. [Google Scholar] [CrossRef]
- Ren, Z.; Jeckel, H.; Simon-Soro, A.; Koo, H. Interkingdom assemblages in human saliva display group-level surface mobility and disease-promoting emergent functions. Proc. Natl. Acad. Sci. USA 2022, 119, e2209699119. [Google Scholar] [CrossRef]
- Maruyama, K.; Takayama, Y.; Sugisawa, E.; Yamanoi, Y.; Yokawa, T.; Kondo, T.; Ishibashi, K.-I.; Sahoo, B.R.; Takemura, N.; Mori, Y.; et al. The ATP transporter VNUT mediates induction of Dectin-1-triggered Candida nociception. iScience 2018, 6, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Cheng, L.; Lei, Y.L.; Ren, B.; Zhou, X. The Interactions Between Candida albicans and Mucosal Immunity. Front. Microbiol. 2021, 12, 652725. [Google Scholar] [CrossRef] [PubMed]
- Nikou, S.-A.; Kichik, N.; Brown, R.; Ponde, N.O.; Ho, J.; Naglik, J.R.; Richardson, J.P. Candida albicans interactions with mucosal surfaces during health and disease. Pathogens 2019, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Boschetto, F.; Ohgitani, E.; Fujita, Y.; Shin-Ya, M.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Marin, E.; Zhu, W.; et al. Raman molecular fingerprints of SARS-CoV-2 British variant and the concept of Raman barcode. Adv. Sci. 2022, 9, 2103287. [Google Scholar] [CrossRef]





| Candida Species * | Candida CFU/mL | Age (y) | Sex | Number of Teeth | Additional Bacteria Species | |
|---|---|---|---|---|---|---|
| Sample 1 | C. albicans | 1 × 104~5 | 79 | Male | 20 | α-Streptococcus (3+) Neisseria sp. (1+) |
| Sample 2 | C. albicans C. parapsilosis # | 1 × 105 | 79 | Female | 8 | α-Streptococcus (3+) |
| Sample 3 | C. glabrata C. albicans # | 1 × 108 | 80 | Male | 3 | α-Streptococcus (3+) Corynebacterium sp. (2+) |
| Sample 4 | C. albicans Filamentous fungi | 1 × 104~5 | 39 | Female | 12 | α-Streptococcus (3+) Neisseria sp. (1+) |
| Sample 5 | C. parapsilosis | 1 × 107 | 92 | Female | 16 | S. aureus MSSA (3+) α-Streptococcus (3+) |
| Sample 6 | C. glabrata | 1 × 105 | 79 | Female | 0 | α-Streptococcus (2+) |
| Sample 7 | C. albicans C. glabrata # Filamentous fungi | 1 × 108 | 81 | Female | 6 | α-Streptococcus (3+) |
| Sample 8 | C. tropicalis C. albicans # Filamentous fungi | 1 × 107 | 49 | Male | 26 | γ-Streptococcus (3+) α-Streptococcus (2+) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pezzotti, G.; Adachi, T.; Imamura, H.; Ikegami, S.; Kitahara, R.; Yamamoto, T.; Kanamura, N.; Zhu, W.; Ishibashi, K.-i.; Okuma, K.; et al. Raman Spectroscopic Algorithms for Assessing Virulence in Oral Candidiasis: The Fight-or-Flight Response. Int. J. Mol. Sci. 2024, 25, 11410. https://doi.org/10.3390/ijms252111410
Pezzotti G, Adachi T, Imamura H, Ikegami S, Kitahara R, Yamamoto T, Kanamura N, Zhu W, Ishibashi K-i, Okuma K, et al. Raman Spectroscopic Algorithms for Assessing Virulence in Oral Candidiasis: The Fight-or-Flight Response. International Journal of Molecular Sciences. 2024; 25(21):11410. https://doi.org/10.3390/ijms252111410
Chicago/Turabian StylePezzotti, Giuseppe, Tetsuya Adachi, Hayata Imamura, Saki Ikegami, Ryo Kitahara, Toshiro Yamamoto, Narisato Kanamura, Wenliang Zhu, Ken-ichi Ishibashi, Kazu Okuma, and et al. 2024. "Raman Spectroscopic Algorithms for Assessing Virulence in Oral Candidiasis: The Fight-or-Flight Response" International Journal of Molecular Sciences 25, no. 21: 11410. https://doi.org/10.3390/ijms252111410
APA StylePezzotti, G., Adachi, T., Imamura, H., Ikegami, S., Kitahara, R., Yamamoto, T., Kanamura, N., Zhu, W., Ishibashi, K.-i., Okuma, K., Mazda, O., Komori, A., Komatsuzawa, H., & Makimura, K. (2024). Raman Spectroscopic Algorithms for Assessing Virulence in Oral Candidiasis: The Fight-or-Flight Response. International Journal of Molecular Sciences, 25(21), 11410. https://doi.org/10.3390/ijms252111410






