Early-Stage Infection-Specific Heterobasidion annosum (Fr.) Bref. Transcripts in H. annosum–Pinus sylvestris L. Pathosystem
Abstract
1. Introduction
2. Results
2.1. Sequencing Statistics
2.2. Most Transcribed Genes at Each Time Point
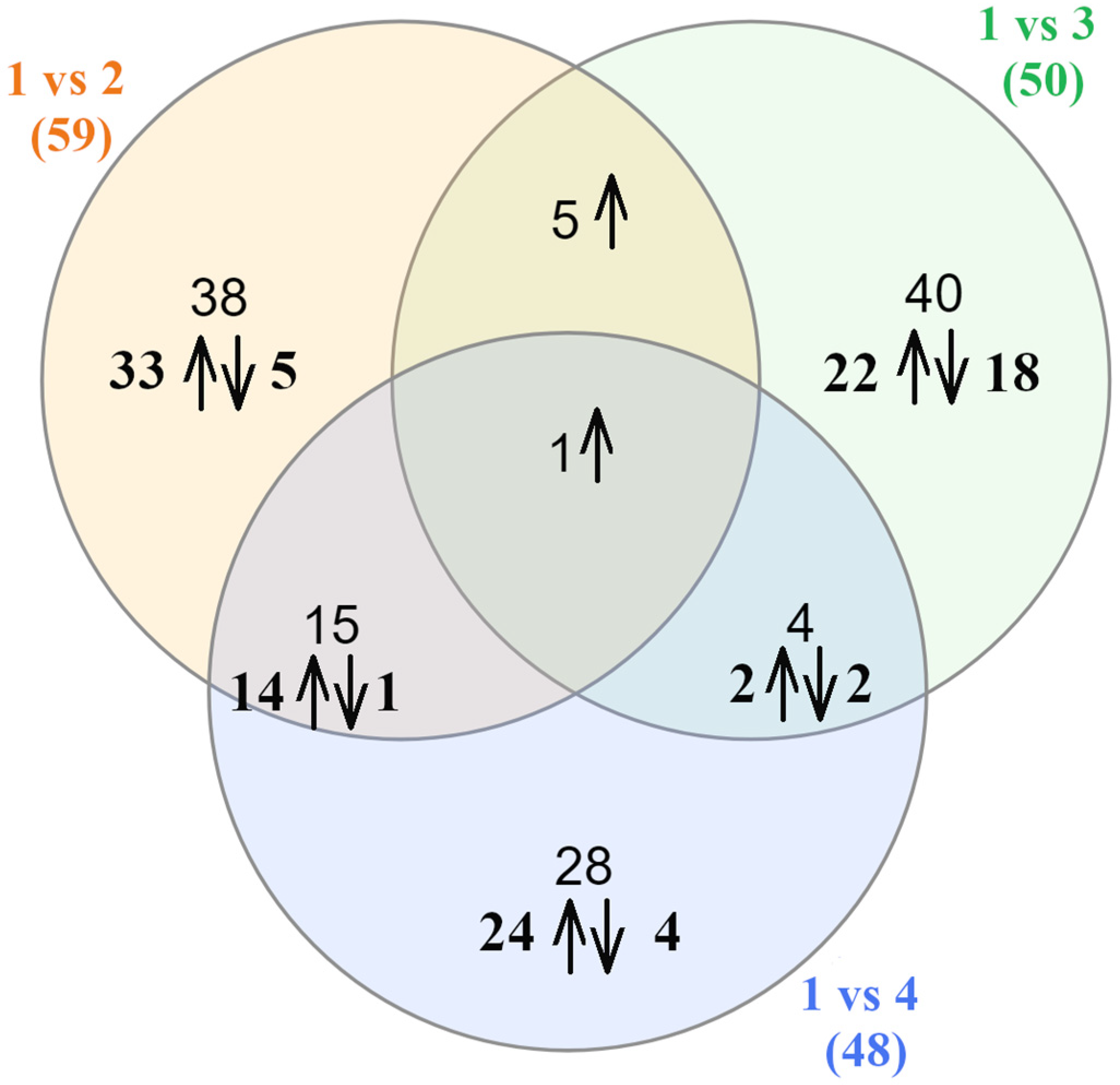
2.3. Differential Gene Expression
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- San-Miguel-Ayanz, J.; de Rigo, D.; Caudullo, G.; Durrant, T.H.; Mauri, A. European Atlas of Forest Tree Species; Publication Office of the European Union: Luxembourg, 2016; p. 202. ISSN 9279528335. [Google Scholar]
- Farjon, A. A Handbook of the World’s Conifers; Brill: Leiden, The Netherlands, 2010. [Google Scholar] [CrossRef]
- González Díaz, P. Development and Maintenance of Genetic Diversity in Scots Pine, Pinus sylvestris (L.). Ph.D. Thesis, University of Stirling, Stirling, UK, 2018. [Google Scholar]
- Tyrmi, J.S.; Vuosku, J.; Acosta, J.J.; Li, Z.; Sterck, L.; Cervera, M.T.; Savolainen, O.; Pyhäjärvi, T. Genomics of Clinal Local Adaptation in Pinus sylvestris under Continuous Environmental and Spatial Genetic Setting. G3 Genes Genomes Genet. 2020, 10, 2683–2696. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.; Uchman, A. Spatially Associated or Composite Life Traces from Holocene Paleosols and Dune Sands Provide Evidence for Past Biotic Interactions. Sci. Nat. 2023, 110, 9. [Google Scholar] [CrossRef]
- Siitonen, J. Ips Acuminatus Kills Pines in Southern Finland. Silva Fenn. 2014, 48, 1145. [Google Scholar] [CrossRef]
- Hlávková, D.; Doležal, P. Cambioxylophagous Pests of Scots Pine: Ecological Physiology of European Populations—A Review. Front. For. Glob. Chang. 2022, 5, 864651. [Google Scholar] [CrossRef]
- Garbelotto, M.; Gonthier, P. Biology, Epidemiology, and Control of Heterobasidion Species Worldwide. Annu. Rev. Phytopathol. 2013, 51, 39–59. [Google Scholar] [CrossRef]
- Asiegbu, F.O.; Adomas, A.; Stenlid, J. Conifer Root and Butt Rot Caused by Heterobasidion annosum (Fr.) Bref. s.l. Mol. Plant Pathol. 2005, 6, 395–409. [Google Scholar] [CrossRef]
- Samils, N. Monitoring the Control Methods of Heterobasidion annosum s.l. Root Rot. Ph.D. Thesis, Swedish University of Agricultural Sciences, Uppsala, Sweden, 2008. [Google Scholar]
- Piri, T. Silvicultural Control of Heterobasidion Root Rot in Norway Spruce Forests in Southern Finland: Regeneration and Vitality Fertilization of Infected Stands. Ph.D. Thesis, University of Helsinki, Finnish Forest Research Institute, Vantaa Research Centre, Juupajoki, Finland, 2003. [Google Scholar]
- Piri, T. Response of Compensatory-Fertilized Pinus sylvestris to Infection by Heterobasidion annosum. Scand. J. For. Res. 2010, 15, 218–224. [Google Scholar] [CrossRef]
- Rönnberg, J.; Berglund, M.; Johansson, U.; Cleary, M. Incidence of Heterobasidion spp. Following Different Thinning Regimes in Norway Spruce in Southern Sweden. For. Ecol. Manag. 2013, 289, 409–415. [Google Scholar] [CrossRef]
- Blomquist, M.; Cleary, M.; Sherwood, P.; Pinto, W.; Herrera, S.L.; Marčiulynienė, D.; Elsafy, M.; Bakal, I.; Nilsson, A.; Rönnberg, J. The Potential of Biological Control against Heterobasidion Root Rot Is Not Realized in Practical Forestry. For. Ecol. Manag. 2023, 531, 120778. [Google Scholar] [CrossRef]
- Kenigsvalde, K.; Brauners, I.; Zaļuma, A.; Jansons, J.; Gaitnieks, T. Biological Protection of Conifers against Heterobasidion Infection—Interaction between Root-Rot Fungus and Phlebiopsis Gigantea. Res. Rural. Dev. 2017, 1, 69–75. [Google Scholar] [CrossRef]
- Piri, T.; Saarinen, M.; Hamberg, L.; Hantula, J.; Gaitnieks, T. Efficacy of Biological and Chemical Control Agents against Heterobasidion Spore Infections of Norway Spruce and Scots Pine Stumps on Drained Peatland. J. Fungi 2023, 9, 346. [Google Scholar] [CrossRef] [PubMed]
- Korshikov, I.I.; Demkovich, A.E. Genetic Features of Root Fungus-Resistant Scotch Pine Trees in Artificial Stands in the Steppe Zone of Ukraine. Cytol. Genet. 2008, 42, 323–328. [Google Scholar] [CrossRef]
- Vasiliauskas, A. Šakninės Pinties (Heterobasidion annosum (Fr.) Bref.) Paplitimas Pušynuose, Įveistuose Žemės Ūkio Naudmenuose, Kovos Su Ja Priemonės Ir Kovos Rezultatai [Root Rot Caused by Heterobasidion annosum in Pinus sylvestris Plantations on Former Agricultural Land]. Miskininkyste 2001, 2, 42–59. [Google Scholar]
- Rieksts-Riekstiņš, R.; Zeltiņš, P.; Baliuckas, V.; Brūna, L.; Zaļuma, A.; Kāpostiņš, R. Pinus sylvestris Breeding for Resistance against Natural Infection of the Fungus Heterobasidion annosum. Forests 2020, 11, 23. [Google Scholar] [CrossRef]
- Adomas, A.; Heller, G.; Li, G.; Olson, Å.; Chu, T.M.; Osborne, J.; Craig, D.; Van Zyl, L.; Wolfinger, R.; Sederoff, R.; et al. Transcript Profiling of a Conifer Pathosystem: Response of Pinus sylvestris Root Tissues to Pathogen (Heterobasidion annosum) Invasion. Tree Physiol. 2007, 27, 1441–1458. [Google Scholar] [CrossRef]
- Mukrimin, M.; Kovalchuk, A.; Ghimire, R.P.; Kivimäenpää, M.; Sun, H.; Holopainen, J.K.; Asiegbu, F.O. Evaluation of Potential Genetic and Chemical Markers for Scots Pine Tolerance against Heterobasidion annosum Infection. Planta 2019, 250, 1881–1895. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Escribano, L.; Visser, E.A.; Iturritxa, E.; Raposo, R.; Naidoo, S. The Transcriptome of Pinus pinaster under Fusarium Circinatum Challenge. BMC Genom. 2020, 21, 28. [Google Scholar] [CrossRef]
- Šķipars, V.; Ruņģis, D. Transcript Dynamics in Wounded and Inoculated Scots Pine. Int. J. Mol. Sci. 2021, 22, 1505. [Google Scholar] [CrossRef]
- Liu, M.; Wang, K.; Haapanen, M.; Ghimire, R.P.; Kivimäenpää, M.; Asiegbu, F.O. Analysis of Transcriptome and Terpene Constituents of Scots Pine Genotypes Inherently Resistant or Susceptible to Heterobasidion annosum. Front. Plant Sci. 2022, 13, 947734. [Google Scholar] [CrossRef]
- Dalman, K.; Himmelstrand, K.; Olson, Å.; Lind, M.; Brandström-Durling, M.; Stenlid, J. A Genome-Wide Association Study Identifies Genomic Regions for Virulence in the Non-Model Organism Heterobasidion annosum s.s. PLoS ONE 2013, 8, e53525. [Google Scholar] [CrossRef]
- Adomas, A.; Eklund, M.; Johansson, M.; Asiegbu, F.O. Identification and Analysis of Differentially Expressed CDNAs during Nonself-Competitive Interaction between Phlebiopsis Gigantea and Heterobasidion parviporum. FEMS Microbiol. Ecol. 2006, 57, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Sun, H.; Vainio, E.J.; Raffaello, T.; Kovalchuk, A.; Morin, E.; Duplessis, S.; Asiegbu, F.O. Intraspecific Comparative Genomics of Isolates of the Norway Spruce Pathogen (Heterobasidion parviporum) and Identification of Its Potential Virulence Factors. BMC Genom. 2018, 19, 220. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-Base: The Pathogen–Host Interactions Database. Nucleic Acids Res. 2020, 48, D613–D620. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Kim, J.E.; Lee, Y.W.; Son, H. Fungal Cytochrome P450s and the P450 Complement (CYPome) of Fusarium Graminearum. Toxins 2018, 10, 112. [Google Scholar] [CrossRef]
- Kovalchuk, A.; Zeng, Z.; Ghimire, R.P.; Kivimäenpää, M.; Raffaello, T.; Liu, M.; Mukrimin, M.; Kasanen, R.; Sun, H.; Julkunen-Tiitto, R.; et al. Dual RNA-Seq Analysis Provides New Insights into Interactions between Norway Spruce and Necrotrophic Pathogen Heterobasidion annosum s.l. BMC Plant Biol. 2019, 19, 2. [Google Scholar] [CrossRef]
- Wen, Z.; Zeng, Z.; Ren, F.; Asiegbu, F.O. The Conifer Root and Stem Rot Pathogen (Heterobasidion parviporum): Effectome Analysis and Roles in Interspecific Fungal Interactions. Microorganisms 2019, 7, 658. [Google Scholar] [CrossRef]
- Zamora-Ballesteros, C.; Pinto, G.; Amaral, J.; Valledor, L.; Alves, A.; Diez, J.J.; Martín-García, J. Dual Rna-Sequencing Analysis of Resistant (Pinus pinea) and Susceptible (Pinus radiata) Hosts during Fusarium Circinatum Challenge. Int. J. Mol. Sci. 2021, 22, 5231. [Google Scholar] [CrossRef]
- Lundén, K.; Danielsson, M.; Durling, M.B.; Ihrmark, K.; Nemesio Gorriz, M.; Stenlid, J.; Asiegbu, F.O.; Elfstrand, M. Transcriptional Responses Associated with Virulence and Defence in the Interaction between Heterobasidion annosum s.s. and Norway Spruce. PLoS ONE 2015, 10, e0131182. [Google Scholar] [CrossRef]
- Asiegbu, F.O.; Johansson, M.; Woodward, S.; Hüttermann, A. Biochemistry of the Host—Parasite Interaction. In Heterobasidion annosum: Biology, Ecology, Impact and Control; Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A., Eds.; CABI Publishing: Wallingford, UK, 1998; pp. 176–180. [Google Scholar]
- Coenye, T. Do results obtained with RNA-sequencing require independent verification? Biofilm 2021, 3, 100043. [Google Scholar] [CrossRef]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef]
- Brutyn, M.; D’Herde, K.; Dhaenens, M.; Van Rooij, P.; Verbrugghe, E.; Hyatt, A.D.; Croubels, S.; Deforce, D.; Ducatelle, R.; Haesebrouck, F.; et al. Batrachochytrium Dendrobatidis Zoospore Secretions Rapidly Disturb Intercellular Junctions in Frog Skin. Fungal Genet. Biol. 2012, 49, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Panevska, A.; Hodnik, V.; Skočaj, M.; Novak, M.; Modic, Š.; Pavlic, I.; Podržaj, S.; Zarić, M.; Resnik, N.; Maček, P.; et al. Pore-Forming Protein Complexes from Pleurotus Mushrooms Kill Western Corn Rootworm and Colorado Potato Beetle through Targeting Membrane Ceramide Phosphoethanolamine. Sci. Rep. 2019, 9, 5073. [Google Scholar] [CrossRef] [PubMed]
- Wynn, J.P.; Hamid, A.B.A.; Ratledge, C. The Role of Malic Enzyme in the Regulation of Lipid Accumulation in Filamentous Fungi. Microbiology 1999, 145, 1911–1917. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Griest, T.A.; Harter, T.M.; Mark Petrash, J. Functional Studies of Aldo-Keto Reductases in Saccharomyces Cerevisiae. Biochim. Biophys. Acta 2007, 1773, 321–329. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, H.; Zhao, G.; Yang, J.; Luo, Y.; Sun, S.; Wang, Z.; Li, S.; Jin, C. Genetical and O-Glycoproteomic Analyses Reveal the Roles of Three Protein O-Mannosyltransferases in Phytopathogen Fusarium Oxysporum f.sp. Cucumerinum. Fungal Genet. Biol. 2020, 134, 103285. [Google Scholar] [CrossRef]
- Gerke, J.; Bayram, Ö.; Braus, G.H. Fungal S-Adenosylmethionine Synthetase and the Control of Development and Secondary Metabolism in Aspergillus Nidulans. Fungal Genet. Biol. 2012, 49, 443–454. [Google Scholar] [CrossRef]
- Watanabe, T.; Tsuda, S.; Nishimura, H.; Honda, Y.; Watanabe, T. Characterization of a Delta12-Fatty Acid Desaturase Gene from Ceriporiopsis Subvermispora, a Selective Lignin-Degrading Fungus. Appl. Microbiol. Biotechnol. 2010, 87, 215–224. [Google Scholar] [CrossRef]
- Van Den Brink, J.; De Vries, R.P. Fungal Enzyme Sets for Plant Polysaccharide Degradation. Appl. Microbiol. Biotechnol. 2011, 91, 1477. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.Y.; Ye, R.Y.; Yu, H.L.; Li, A.T.; Xu, J.H. Mining Methods and Typical Structural Mechanisms of Terpene Cyclases. Bioresour. Bioprocess. 2021, 8, 66. [Google Scholar] [CrossRef]
- González-Hernández, R.A.; Valdez-Cruz, N.A.; Macías-Rubalcava, M.L.; Trujillo-Roldán, M.A. Overview of Fungal Terpene Synthases and Their Regulation. World J. Microbiol. Biotechnol. 2023, 39, 194. [Google Scholar] [CrossRef]
- Palanimurugan, R.; Scheel, H.; Hofmann, K.; Dohmen, R.J. Polyamines Regulate Their Synthesis by Inducing Expression and Blocking Degradation of ODC Antizyme. EMBO J. 2004, 23, 4857–4867. [Google Scholar] [CrossRef] [PubMed]
- Beccaccioli, M.; Reverberi, M.; Scala, V. Fungal Lipids: Biosynthesis and Signalling during Plant-Pathogen Interaction. Front. Biosci. Landmark 2019, 24, 172–185. [Google Scholar] [CrossRef]
- Cardoza, R.E.; McCormick, S.P.; Lindo, L.; Mayo-Prieto, S.; González-Cazón, D.; Martínez-Reyes, N.; Carro-Huerga, G.; Rodríguez-González, Á.; Proctor, R.H.; Casquero, P.A.; et al. Effect of Farnesol in Trichoderma Physiology and in Fungal-Plant Interaction. J. Fungi 2022, 8, 1266. [Google Scholar] [CrossRef] [PubMed]
- Vorapreeda, T.; Thammarongtham, C.; Cheevadhanarak, S.; Laoteng, K. Repertoire of Malic Enzymes in Yeast and Fungi: Insight into Their Evolutionary Functional and Structural Significance. Microbiology 2013, 159, 2548–2557. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://genome.jgi.doe.gov/portal/Hetan2/Hetan2.download.html (accessed on 28 June 2023).
- Hao, G.; Chen, H.; Wang, L.; Gu, Z.; Song, Y.; Zhang, H.; Chen, W.; Chen, Y.Q. Role of Malic Enzyme during Fatty Acid Synthesis in the Oleaginous Fungus Mortierella Alpina. Appl. Environ. Microbiol. 2014, 80, 2672. [Google Scholar] [CrossRef]
- Raffaello, T.; Chen, H.; Kohler, A.; Asiegbu, F.O. Transcriptomic Profiles of Heterobasidion annosum under Abiotic Stresses and during Saprotrophic Growth in Bark, Sapwood and Heartwood. Environ. Microbiol. 2014, 16, 1654–1667. [Google Scholar] [CrossRef]
- Looi, H.K.; Toh, Y.F.; Yew, S.M.; Na, S.L.; Tan, Y.C.; Chong, P.S.; Khoo, J.S.; Yee, W.Y.; Ng, K.P.; Kuan, C.S. Genomic Insight into Pathogenicity of Dematiaceous Fungus Corynespora cassiicola. PeerJ 2017, 5, e2841. [Google Scholar] [CrossRef][Green Version]
- Gong, L.; Liu, Y.; Xiong, Y.; Li, T.; Yin, C.; Zhao, J.; Yu, J.; Yin, Q.; Gupta, V.K.; Jiang, Y.; et al. New Insights into the Evolution of Host Specificity of Three Penicillium Species and the Pathogenicity of P. Italicum Involving the Infection of Valencia Orange (Citrus Sinensis). Virulence 2020, 11, 748–768. [Google Scholar] [CrossRef]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the Large Glycoside Hydrolase Family 43 into Subfamilies: A Motivation for Detailed Enzyme Characterization. Appl. Environ. Microbiol. 2016, 82, 1686. [Google Scholar] [CrossRef]
- Sahu, P.K.; Jayalakshmi, K.; Tilgam, J.; Gupta, A.; Nagaraju, Y.; Kumar, A.; Hamid, S.; Singh, H.V.; Minkina, T.; Rajput, V.D.; et al. ROS Generated from Biotic Stress: Effects on Plants and Alleviation by Endophytic Microbes. Front. Plant Sci. 2022, 13, 1042936. [Google Scholar] [CrossRef]
- Heller, J.; Tudzynski, P. Reactive Oxygen Species in Phytopathogenic Fungi: Signaling, Development, and Disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.; Jung, J.; Jang, I.A.; Madsen, E.L.; Park, W. Role of Glyoxylate Shunt in Oxidative Stress Response. J. Biol. Chem. 2016, 291, 11928. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L. Isolation of Plant DNA from Fresh Tissue. Focus 1987, 12, 13–15. [Google Scholar]
- Asiegbu, F.; Daniel, G.; Johansson, M. Studies on the Infection of Norway Spruce Roots by Heterobasidion annosum. Canad. J. Bot. 1993, 71, 1552–1561. [Google Scholar] [CrossRef]
- Karlsson, M.; Hietala, A.M.; Kvaalen, H.; Solheim, H.; Olson, Å.; Stenlid, J.; Fossdal, C.G. Quantification of Host and Pathogen DNA and RNA Transcripts in the Interaction of Norway Spruce with Heterobasidion parviporum. Physiol. Mol. Plant Pathol. 2007, 70, 99–109. [Google Scholar] [CrossRef]
- Olson, Å.; Aerts, A.; Asiegbu, F.; Belbahri, L.; Bouzid, O.; Broberg, A.; Canbäck, B.; Coutinho, P.M.; Cullen, D.; Dalman, K.; et al. Insight into Trade-off between Wood Decay and Parasitism from the Genome of a Fungal Forest Pathogen. New Phytol. 2012, 194, 1001–1013. [Google Scholar] [CrossRef]
- Wachowiak, W.; Trivedi, U.; Perry, A.; Cavers, S. Comparative Transcriptomics of a Complex of Four European Pine Species. BMC Genom. 2015, 16, 234. [Google Scholar] [CrossRef]
- CLC Bio. CLC Genomics Workbench User Manual for CLC Genomics Workbench 23.0.5; CLC: Aarhus, Denmark, 2023; p. 1165. [Google Scholar]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
| Sample Name 1 | Total Reads | Percentage of Reads Mapping onto H. annosum Transcriptome | Percentage of Reads Mapping onto P. sylvestris Transcriptome |
|---|---|---|---|
| 1_5 | 330,514,752 | 0.32 | 26.69 |
| 1_9 | 148,181,900 | 0.47 | 69.69 |
| 1_10 | 127,532,184 | 0.73 | 44.92 |
| 1_14 | 265,534,720 | 0.44 | 45.81 |
| 2_4 | 302,911,430 | 0.17 | 26.15 |
| 2_10 | 200,927,014 | 0.36 | 54.40 |
| 2_14 | 138,048,364 | 0.12 | 23.87 |
| 2_15 | 297,177,502 | 0.38 | 42.04 |
| 3_3 | 183,571,350 | 0.65 | 55.76 |
| 3_5 | 175,591,306 | 0.38 | 57.62 |
| 3_7 | 214,847,606 | 0.28 | 41.78 |
| 3_12 | 176,541,044 | 0.32 | 29.99 |
| 4_1 | 163,295,084 | 0.27 | 56.09 |
| 4_3 | 123,870,986 | 0.52 | 49.52 |
| 4_10 | 400,583,236 | 0.11 | 23.70 |
| 4_13 | 115,595,890 | 0.24 | 32.19 |
| Biological Process | Unique for Time Point |
|---|---|
| Histidine biosynthetic process | 1 WPI |
| Arginine biosynthetic process | 1 WPI |
| Coenzyme A metabolic process | 1 WPI |
| Response to heat | 1 WPI |
| Response to oxygen-containing compound | 1 WPI |
| Isoprenoid biosynthetic process | 1 WPI |
| Response to osmotic stress | 1 WPI |
| Response to oxidative stress | 1 WPI |
| Glyoxylate cycle | 1 WPI |
| Carboxylic acid metabolic process | 1 WPI |
| Protein-containing complex assembly | 1 WPI |
| S-adenosylmethionine biosynthetic process | 1 WPI |
| Proton transmembrane transport | 2 WPI |
| Negative regulation of protein modification process | 2 WPI |
| Negative regulation of phosphate metabolic process | 2 WPI |
| DNA-templated transcription | 2 WPI |
| Macroautophagy | 2 WPI |
| Regulation of translation | 2 WPI |
| Cellular response to amino acid starvation | 2 WPI |
| Regulation of protein dephosphorylation | 2 WPI |
| Positive regulation of transcription by RNA polymerase II | 2 WPI |
| Acetyl-coa biosynthetic process | 3 WPI |
| Citrate metabolic process | 3 WPI |
| Cellular biosynthetic process | 4 WPI |
| Protein import into mitochondrial matrix | 4 WPI |
| Signal transduction | 4 WPI |
| Ergosterol biosynthetic process | 4 WPI |
| Mapping Reference ID | Annotation | Fold Change | p-Value |
|---|---|---|---|
| Upregulated | |||
| CCPA1999.b1 | Hypothetical protein HETIRDRAFT_426980 | 181.85 | 4.72 × 10−5 |
| CCPB2345.b1 | Carotenoid ester lipase precursor | 145.16 | 2.55 × 10−4 |
| CCOZ2064.b1 | ATP-utilizing phosphoenolpyruvate carboxykinase | 64.23 | 9.74 × 10−5 |
| CCPA2867.g1 | Aldo/keto reductase | 45.65 | 1.76 × 10−4 |
| CCPB993.g1 | Terpenoid cyclases/protein prenyltransferase alpha-alpha toroid | 43.52 | 5.23 × 10−3 |
| CCPC5739.g1 | NAD-P-binding protein | 34.14 | 1.76 × 10−3 |
| CCPC2435.b1 | Na * | 33.93 | 2.87 × 10−4 |
| CCPA4098.g1 | GPI mannosyltransferase 3 | 29.59 | 2.71 × 10−3 |
| CCPB1601.b1 | Na | 29.53 | 8.77 × 10−4 |
| CCOZ1600.b1 | Methionine adenosyltransferase | 28.24 | 4.23 × 10−3 |
| CCPC3360.b1 | Isocitrate lyase | 27.32 | 9.33 × 10−5 |
| CCPA4929.b1 | Alpha/beta hydrolase | 25.73 | 5.18 × 10−4 |
| Downregulated | |||
| CCPC8078.b1 | Transcription regulator | −23.45 | 7.99 × 10−3 |
| CCOZ5192.g1 | Dnaj domain-containing protein | −20.35 | 3.12 × 10−3 |
| CCPB3914.b1 | Hypothetical protein HETIRDRAFT_426907 | −18.18 | 4.34 × 10−3 |
| CCOZ3764.b1 | Negative regulator of differentiation 1 | −12.93 | 9.40 × 10−3 |
| CCPC2832.b1 | Ornithine decarboxylase antizyme domain-containing protein | −12.24 | 8.22 × 10−3 |
| CCPA3492.b1 | Hypothetical protein HETIRDRAFT_477666 | −8.36 | 3.06 × 10−3 |
| Mapping Reference ID | Annotation | Fold Change | p-Value |
|---|---|---|---|
| Upregulated | |||
| CCPC2187.b1 | Malic enzyme | 61.69 | 2.06 × 10−4 |
| CCOZ3444.b1 | Heat shock protein 70 | 55.59 | 3.80 × 10−3 |
| CCPB993.g1 | Terpenoid cyclases/protein prenyltransferase alpha-alpha toroid | 44.46 | 1.40 × 10−3 |
| CCPC2829.b1 | Pali domain-containing protein | 32.82 | 2.45 × 10−4 |
| CCPA4010.b1 | Fatty acid desaturase domain-containing protein | 27.64 | 4.38 × 10−3 |
| CCPC4213.b1 | Hypothetical protein HETIRDRAFT_325943 | 25.72 | 5.16 × 10−3 |
| 11E44-04-08 | Predicted protein | 24.54 | 8.11 × 10−4 |
| CCPC4213.g1 | Hypothetical protein HETIRDRAFT_325943 | 24.37 | 6.08 × 10−3 |
| CCPA4569.b1 | Hypothetical protein HETIRDRAFT_441917 | 23.75 | 3.45 × 10−4 |
| CCPC6772.g1 | Putative BAG domain-containing protein | 23.30 | 7.21 × 10−3 |
| Downregulated | |||
| CCPC3479.g1 | Groes-like protein | −41.47 | 1.64 × 10−3 |
| CCPA5017.g1 | Protein arginine N-methyltransferase | −39.95 | 3.01 × 10−4 |
| CCOZ3601.b1 | Secy protein | −27.45 | 6.26 × 10−3 |
| 16D10 | HSP20-like chaperone | −25.79 | 2.78 × 10−3 |
| CCOZ4082.b1 | Glucoamylase | −23.26 | 2.19 × 10−3 |
| CCPA3011.b1 | Cell division control/GTP-binding protein | −20.94 | 8.14 × 10−3 |
| CCPA3961.g1 | Glutamate decarboxylase | −18.18 | 7.47 × 10−3 |
| CCPC7984.b1 | Na * | −17.48 | 5.70 × 10−3 |
| D69E9 | 40S ribosomal protein S26 | −16.73 | 3.76 × 10−3 |
| CCPB4097.b1 | Hypothetical protein HETIRDRAFT_439855 | −14.76 | 4.80 × 10−3 |
| CCPC6286.b1 | Leucine aminopeptidase | −13.02 | 7.79 × 10−3 |
| Mapping Reference ID | Annotation | Fold Change | p-Value |
|---|---|---|---|
| Upregulated | |||
| CCPA575.b1 | Delta-12 fatty acid desaturase | 48.12 | 7.94 × 10−5 |
| CCPA1686.b1 | Polysaccharide lyase family 1 protein | 47.55 | 4.24 × 10−3 |
| CCPA1999.b1 | Hypothetical protein HETIRDRAFT_426980 | 46.41 | 1.60 × 10−4 |
| 10F24-03-16 | Elongase of fatty acids ELO | 45.14 | 9.47 × 10−3 |
| CCPC993.b1 | Erylysin B | 38.75 | 5.07 × 10−3 |
| CCPC1268.b1 | Delta-12 fatty acid desaturase protein | 38.60 | 1.89 × 10−3 |
| CCPA3999.b1 | Fatty acid desaturase domain-containing protein | 35.32 | 4.51 × 10−3 |
| CCPC5436.b1 | Hypothetical protein EW146_g3762 | 33.84 | 4.25 × 10−3 |
| CCPA5235.g1 | Hypothetical protein HETIRDRAFT_468348 | 33.44 | 3.52 × 10−3 |
| CCPC8046.b1 | Delta-12 fatty acid desaturase | 33.02 | 1.59 × 10−4 |
| Downregulated | |||
| D128H9 | RS27A protein | −128.69 | 2.57 × 10−4 |
| CCPA2234.b1 | Hypothetical protein HETIRDRAFT_409605 | −50.89 | 7.84 × 10−4 |
| CCOZ3606.b1 | Glycoside hydrolase superfamily | −21.73 | 5.62 × 10−3 |
| CCPB3914.b1 | Hypothetical protein HETIRDRAFT_426907 | −20.09 | 3.36 × 10−3 |
| CCPA5017.g1 | Protein arginine N-methyltransferase | −18.89 | 5.61 × 10−3 |
| CCPA2400.b1 | General substrate transporter | −13.93 | 8.68 × 10−3 |
| CCPB4930.b1 | Family 43 glycosylhydrolase | −11.52 | 3.40 × 10−3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramanenka, M.; Ruņģis, D.E.; Šķipars, V. Early-Stage Infection-Specific Heterobasidion annosum (Fr.) Bref. Transcripts in H. annosum–Pinus sylvestris L. Pathosystem. Int. J. Mol. Sci. 2024, 25, 11375. https://doi.org/10.3390/ijms252111375
Ramanenka M, Ruņģis DE, Šķipars V. Early-Stage Infection-Specific Heterobasidion annosum (Fr.) Bref. Transcripts in H. annosum–Pinus sylvestris L. Pathosystem. International Journal of Molecular Sciences. 2024; 25(21):11375. https://doi.org/10.3390/ijms252111375
Chicago/Turabian StyleRamanenka, Maryna, Dainis Edgars Ruņģis, and Vilnis Šķipars. 2024. "Early-Stage Infection-Specific Heterobasidion annosum (Fr.) Bref. Transcripts in H. annosum–Pinus sylvestris L. Pathosystem" International Journal of Molecular Sciences 25, no. 21: 11375. https://doi.org/10.3390/ijms252111375
APA StyleRamanenka, M., Ruņģis, D. E., & Šķipars, V. (2024). Early-Stage Infection-Specific Heterobasidion annosum (Fr.) Bref. Transcripts in H. annosum–Pinus sylvestris L. Pathosystem. International Journal of Molecular Sciences, 25(21), 11375. https://doi.org/10.3390/ijms252111375







