Prediction of Oral Cancer Biomarkers by Salivary Proteomics Data
Abstract
1. Introduction
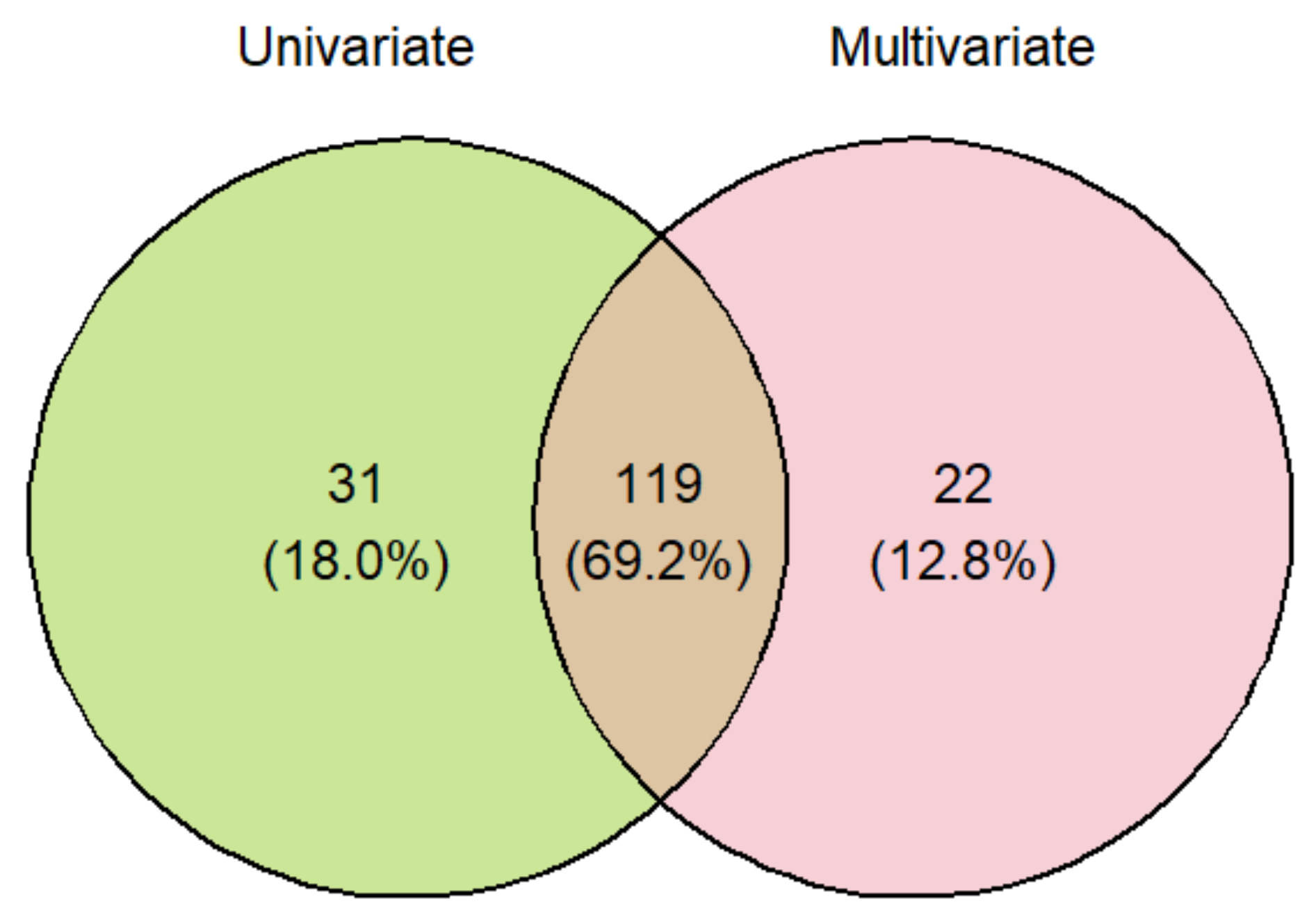
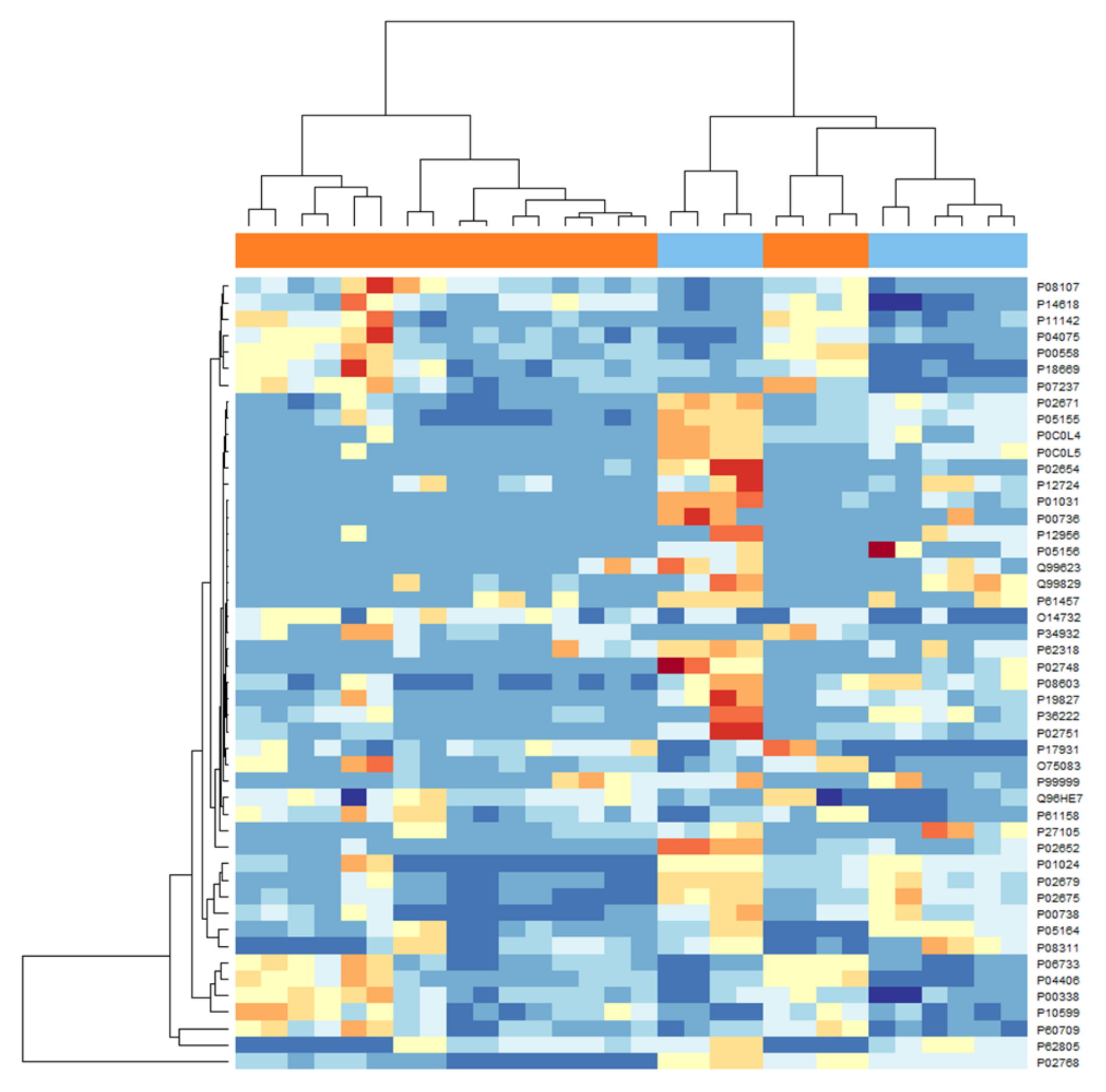
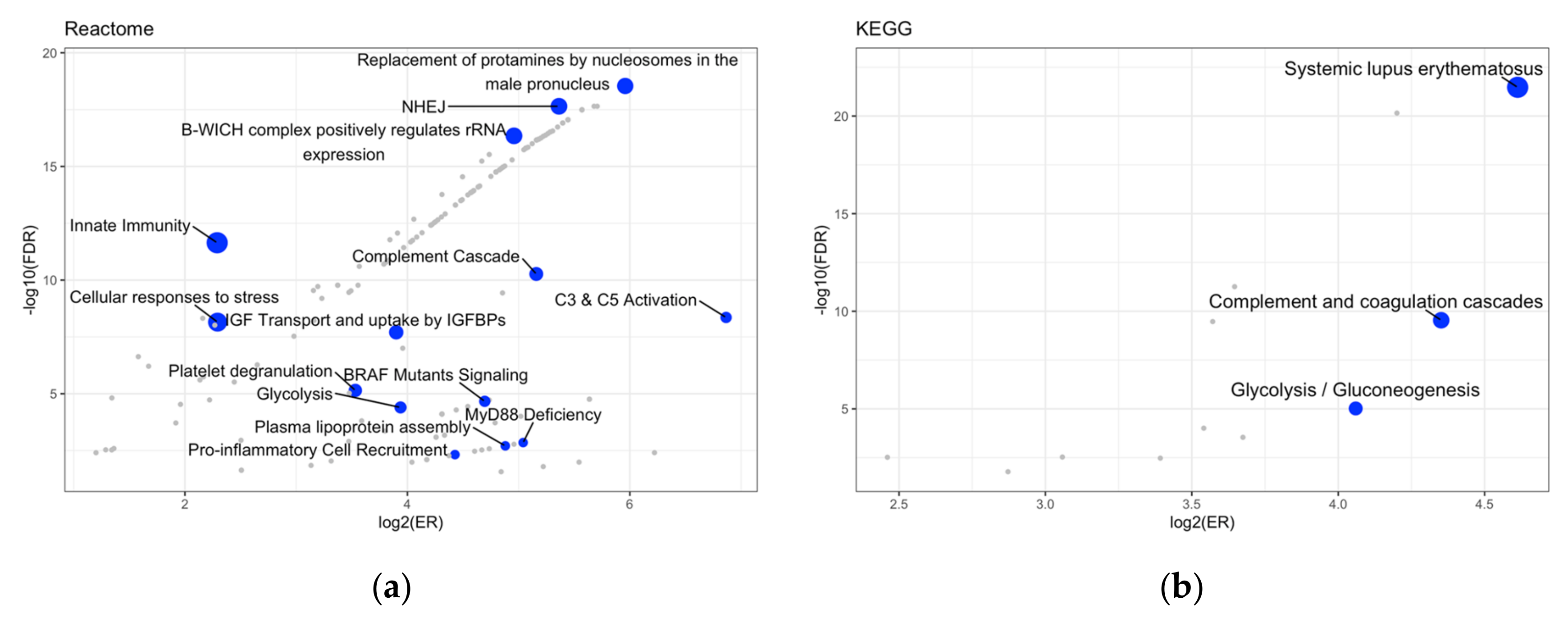
2. Results
3. Discussion
4. Materials and Methods
4.1. Differential Abundance Analysis
4.2. PPI Network Generation Workflow
4.3. Over-Representation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chamoli, A.; Gosavi, A.S.; Shirwadkar, U.P.; Wangdale, K.V.; Behera, S.K.; Kurrey, N.K.; Kalia, K.; Mandoli, A. Overview of Oral Cavity Squamous Cell Carcinoma: Risk Factors, Mechanisms, and Diagnostics. Oral Oncol. 2021, 121, 105451. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer (IARC). Global Cancer Observatory. Available online: https://gco.iarc.fr/ (accessed on 10 August 2024).
- Wyss, A.; Hashibe, M.; Chuang, S.-C.; Lee, Y.-C.A.; Zhang, Z.-F.; Yu, G.-P.; Winn, D.M.; Wei, Q.; Talamini, R.; Szeszenia-Dabrowska, N.; et al. Cigarette, Cigar, and Pipe Smoking and the Risk of Head and Neck Cancers: Pooled Analysis in the International Head and Neck Cancer Epidemiology Consortium. Am. J. Epidemiol. 2013, 178, 679–690. [Google Scholar] [CrossRef]
- Tran, Q.; Maddineni, S.; Arnaud, E.H.; Divi, V.; Megwalu, U.C.; Topf, M.C.; Sunwoo, J.B. Oral Cavity Cancer in Young, Non-Smoking, and Non-Drinking Patients: A Contemporary Review. Crit. Rev. Oncol./Hematol. 2023, 190, 104112. [Google Scholar] [CrossRef] [PubMed]
- Nindra, U.; Hurwitz, J.; Forstner, D.; Chin, V.; Gallagher, R.; Liu, J. A Systematic Review of Neoadjuvant and Definitive Immunotherapy in Locally Advanced Head and Neck Squamous Cell Carcinoma. Cancer Med. 2023, 12, 11234–11247. [Google Scholar] [CrossRef]
- Gissi, D.B.; Rossi, R.; Lenzi, J.; Tarsitano, A.; Gabusi, A.; Balbi, T.; Montebugnoli, L.; Marchetti, C.; Foschini, M.P.; Morandi, L. Thirteen-Gene DNA Methylation Analysis of Oral Brushing Samples: A Potential Surveillance Tool for Periodic Monitoring of Treated Patients with Oral Cancer. Head Neck 2024, 46, 728–739. [Google Scholar] [CrossRef]
- Raslan, S.; Smith, D.H.; Reis, I.M.; Peifer, S.J.; Forman, G.; Ezeh, U.C.; Joshi, P.; Koester, M.; Buitron, I.; Al-Awady, A.; et al. Soluble CD44 in Oral Rinses for the Early Detection of Cancer: A Prospective Cohort Study in High-Risk Individuals. BMC Oral Health 2024, 24, 820. [Google Scholar] [CrossRef]
- Lau, W.W.; Hardt, M.; Zhang, Y.H.; Freire, M.; Ruhl, S. The Human Salivary Proteome Wiki: A Community-Driven Research Platform. J. Dent. Res. 2021, 100, 1510–1519. [Google Scholar] [CrossRef]
- Grishin, N.V. Fold Change in Evolution of Protein Structures. J. Struct. Biol. 2001, 134, 167–185. [Google Scholar] [CrossRef]
- Riccardi, G.; Bellizzi, M.G.; Fatuzzo, I.; Zoccali, F.; Cavalcanti, L.; Greco, A.; de Vincentiis, M.; Ralli, M.; Fiore, M.; Petrella, C.; et al. Salivary Biomarkers in Oral Squamous Cell Carcinoma: A Proteomic Overview. Proteomes 2022, 10, 37. [Google Scholar] [CrossRef]
- Chu, H.-W.; Chang, K.-P.; Hsu, C.-W.; Chang, I.Y.-F.; Liu, H.-P.; Chen, Y.-T.; Wu, C.-C. Identification of Salivary Biomarkers for Oral Cancer Detection with Untargeted and Targeted Quantitative Proteomics Approaches*[S]. Mol. Cell. Proteom. 2019, 18, 1796–1806. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Gulbahce, N.; Loscalzo, J. Network Medicine: A Network-Based Approach to Human Disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, R.; Lu, J.; Jiang, Y.; Huang, T.; Cai, Y.-D. Protein-Protein Interaction Networks as Miners of Biological Discovery. Proteomics 2022, 22, e2100190. [Google Scholar] [CrossRef] [PubMed]
- Monti, C.; Zilocchi, M.; Colugnat, I.; Alberio, T. Proteomics Turns Functional. J. Proteom. 2019, 198, 36–44. [Google Scholar] [CrossRef]
- Curtis, R.K.; Oresic, M.; Vidal-Puig, A. Pathways to the Analysis of Microarray Data. Trends Biotechnol. 2005, 23, 429–435. [Google Scholar] [CrossRef]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten Years of Pathway Analysis: Current Approaches and Outstanding Challenges. PLoS Comput. Biol. 2012, 8, e1002375. [Google Scholar] [CrossRef]
- Yeganeh, P.N.; Mostafavi, M.T. Causal Disturbance Analysis: A Novel Graph Centrality Based Method for Pathway Enrichment Analysis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1613–1624. [Google Scholar] [CrossRef]
- Elhendawy, H.A. Clinical Implications of Heat Shock Protein 70 in Oral Carcinogenesis and Prediction of Progression and Recurrence in Oral Squamous Cell Carcinoma Patients: A Retrospective Clinicopathological Study. Eur. J. Med. Res. 2023, 28, 464. [Google Scholar] [CrossRef]
- Kasioumi, P.; Vrazeli, P.; Vezyraki, P.; Zerikiotis, S.; Katsouras, C.; Damalas, A.; Angelidis, C. Hsp70 (HSP70A1A) Downregulation Enhances the Metastatic Ability of Cancer Cells. Int. J. Oncol. 2019, 54, 821–832. [Google Scholar] [CrossRef]
- Kurihara-Shimomura, M.; Sasahira, T.; Nakashima, C.; Kuniyasu, H.; Shimomura, H.; Kirita, T. The Multifarious Functions of Pyruvate Kinase M2 in Oral Cancer Cells. Int. J. Mol. Sci. 2018, 19, 2907. [Google Scholar] [CrossRef]
- Chen, Y.; Cen, L.; Guo, R.; Huang, S.; Chen, D. Roles and Mechanisms of Phosphoglycerate Kinase 1 in Cancer. Bull. Du Cancer 2022, 109, 1298–1307. [Google Scholar] [CrossRef]
- Ajona, D.; Pajares, M.; Chiara, M.; Rodrigo, J.; Jantus-Lewintre, E.; Camps, C.; Suarez, C.; Bagán, J.; Montuenga, L.; Pio, R. Complement Activation Product C4d in Oral and Oropharyngeal Squamous Cell Carcinoma. Oral Dis. 2015, 21, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Kalló, G.; Bertalan, P.M.; Márton, I.; Kiss, C.; Csősz, É. Salivary Chemical Barrier Proteins in Oral Squamous Cell Carcinoma—Alterations in the Defense Mechanism of the Oral Cavity. Int. J. Mol. Sci. 2023, 24, 13657. [Google Scholar] [CrossRef] [PubMed]
- Caponio, V.C.A.; Zhurakivska, K.; Muzio, L.L.; Troiano, G.; Cirillo, N. The Immune Cells in the Development of Oral Squamous Cell Carcinoma. Cancers 2023, 15, 3779. [Google Scholar] [CrossRef] [PubMed]
- Afshar-Kharghan, V. The Role of the Complement System in Cancer. J. Clin. Investig. 2017, 127, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Gallenkamp, J.; Spanier, G.; Wörle, E.; Englbrecht, M.; Kirschfink, M.; Greslechner, R.; Braun, R.; Schäfer, N.; Bauer, R.J.; Pauly, D. A Novel Multiplex Detection Array Revealed Systemic Complement Activation in Oral Squamous Cell Carcinoma. Oncotarget 2017, 9, 3001–3013. [Google Scholar] [CrossRef]
- Falanga, A.; Marchetti, M.; Vignoli, A. Coagulation and Cancer: Biological and Clinical Aspects. J. Thromb. Haemost. 2013, 11, 223–233. [Google Scholar] [CrossRef]
- Racine, F.; Soudet, S.; Sevestre, M.-A.; Galmiche, A.; Saidak, Z. The Coagulome of Oral Squamous Cell Carcinoma: Examining the Role and Regulation of Coagulation in Oral Cancers Using a Systems Approach. Curr. Opin. Otolaryngol. Head Neck Surg. 2023, 31, 73. [Google Scholar] [CrossRef]
- Ren, J.G.; Zhang, W.; Liu, B.; Man, Q.W.; Xiong, X.P.; Li, C.; Zhu, J.Y.; Wang, W.M.; Jia, J.; Sun, Z.J.; et al. Clinical Significance and Roles in Angiogenesis of Circulating Microparticles in Oral Cancer. J. Dent. Res. 2016, 95, 860–867. [Google Scholar] [CrossRef]
- Ogata, S.; Masuda, T.; Ito, S.; Ohtsuki, S. Targeted Proteomics for Cancer Biomarker Verification and Validation. Cancer Biomark. 2022, 33, 427–436. [Google Scholar] [CrossRef]
- Lualdi, M.; Fasano, M. Statistical Analysis of Proteomics Data: A Review on Feature Selection. J. Proteom. 2019, 198, 18–26. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Eguez, R.V.; Torralba, M.G.; Singh, H.; Golusinski, W.; Masternak, M.; Nelson, K.E.; Freire, M.; Yu, Y. Self-Assembled STrap for Global Proteomics and Salivary Biomarker Discovery. J. Proteome Res. 2019, 18, 1907–1915. [Google Scholar] [CrossRef] [PubMed]
- Wold, S.; Ruhe, A.; Wold, H.; Dunn, W.J., III. The Collinearity Problem in Linear Regression. The Partial Least Squares (PLS) Approach to Generalized Inverses. SIAM J. Sci. Stat. Comput. 1984, 5, 735–743. [Google Scholar] [CrossRef]
- Su, G.; Morris, J.H.; Demchak, B.; Bader, G.D. Biological Network Exploration with Cytoscape 3. Curr. Protoc. Bioinform. 2014, 47, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Aranda, B.; Blankenburg, H.; Kerrien, S.; Brinkman, F.S.L.; Ceol, A.; Chautard, E.; Dana, J.M.; De Las Rivas, J.; Dumousseau, M.; Galeota, E.; et al. PSICQUIC and PSISCORE: Accessing and Scoring Molecular Interactions. Nat. Methods 2011, 8, 528–529. [Google Scholar] [CrossRef]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. WebGestalt 2024: Faster Gene Set Analysis and New Support for Metabolomics and Multi-Omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology Knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]




| Uniprot Accession | Protein Name | log2 FC | FDR |
|---|---|---|---|
| P08107 | Heat shock 70 kDa protein 1A/1B | −0.62 | 0.000075 |
| P14618 | Pyruvate kinase PKM | −1.12 | 0.000187 |
| P00558 | Phosphoglycerate kinase 1 | −2.28 | 0.000396 |
| P0C0L5 | Complement C4-B | 2.48 | 0.001869 |
| P07237 | Protein disulfide-isomerase | −0.68 | 0.002520 |
| P02671 | Fibrinogen alpha chain | 1.73 | 0.003736 |
| P02748 | Complement component C9 | 1.39 | 0.003736 |
| P05156 | Complement factor I | 0.47 | 0.003736 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | −1.50 | 0.003736 |
| P02675 | Fibrinogen beta chain | 1.48 | 0.003894 |
| P02751 | Fibronectin | 1.63 | 0.005073 |
| P01031 | Complement C5 | 0.59 | 0.007604 |
| P02679 | Fibrinogen gamma chain | 1.60 | 0.007648 |
| P05155 | Plasma protease C1 inhibitor | 1.49 | 0.007648 |
| P01024 | Complement C3 | 1.08 | 0.008591 |
| P08603 | Complement factor H | 0.97 | 0.010488 |
| P10599 | Thioredoxin | −1.02 | 0.012139 |
| P05164 | Myeloperoxidase | 1.23 | 0.012872 |
| P34932 | Heat shock 70 kDa protein 4 | −0.73 | 0.019327 |
| P02768 | Serum albumin | 1.05 | 0.019750 |
| P62805 | Histone H4 | 1.38 | 0.020198 |
| P18669 | Phosphoglycerate mutase 1 | −1.07 | 0.021136 |
| P04075 | Fructose-bisphosphate aldolase A | −0.89 | 0.021136 |
| P02652 | Apolipoprotein A-II | 3.34 | 0.023220 |
| P60709 | Actin, cytoplasmic 1 | −0.60 | 0.023220 |
| Q96HE7 | ERO1-like protein alpha | −0.90 | 0.025640 |
| P06733 | Alpha-enolase | −1.04 | 0.029925 |
| P12956 | X-ray repair cross-complementing protein 6 | 0.40 | 0.030974 |
| P11142 | Heat shock cognate 71 kDa protein | −0.56 | 0.032450 |
| P00338 | L-lactate dehydrogenase A chain | −0.83 | 0.032450 |
| Q99829 | Copine-1 | 0.85 | 0.032993 |
| P17931 | Galectin-3 | −1.32 | 0.036387 |
| P61158 | Actin-related protein 3 | −1.28 | 0.036525 |
| Q99623 | Prohibitin-2 | 0.66 | 0.040969 |
| P99999 | Cytochrome c | 1.19 | 0.040969 |
| O75083 | WD repeat-containing protein 1 | −1.08 | 0.040969 |
| P12724 | Eosinophil cationic protein | 1.69 | 0.042247 |
| P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | 0.69 | 0.043209 |
| P00738 | Haptoglobin | 1.18 | 0.044182 |
| P62318 | Small nuclear ribonucleoprotein Sm D3 | 1.09 | 0.045532 |
| P61457 | Pterin-4-alpha-carbinolamine dehydratase | 0.87 | 0.050922 |
| P02654 | Apolipoprotein C-I | 2.52 | 0.051262 |
| P00736 | Complement C1r subcomponent | 0.33 | 0.051262 |
| P08311 | Cathepsin G | 1.38 | 0.053989 |
| O14732 | Inositol monophosphatase 2 | −0.43 | 0.101858 |
| P0C0L4 | Complement C4-A | 1.60 | 0.115126 |
| P36222 | Chitinase-3-like protein 1 | 1.09 | 0.125089 |
| P27105 | Erythrocyte band 7 integral membrane protein | 1.50 | 0.150589 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remori, V.; Airoldi, M.; Alberio, T.; Fasano, M.; Azzi, L. Prediction of Oral Cancer Biomarkers by Salivary Proteomics Data. Int. J. Mol. Sci. 2024, 25, 11120. https://doi.org/10.3390/ijms252011120
Remori V, Airoldi M, Alberio T, Fasano M, Azzi L. Prediction of Oral Cancer Biomarkers by Salivary Proteomics Data. International Journal of Molecular Sciences. 2024; 25(20):11120. https://doi.org/10.3390/ijms252011120
Chicago/Turabian StyleRemori, Veronica, Manuel Airoldi, Tiziana Alberio, Mauro Fasano, and Lorenzo Azzi. 2024. "Prediction of Oral Cancer Biomarkers by Salivary Proteomics Data" International Journal of Molecular Sciences 25, no. 20: 11120. https://doi.org/10.3390/ijms252011120
APA StyleRemori, V., Airoldi, M., Alberio, T., Fasano, M., & Azzi, L. (2024). Prediction of Oral Cancer Biomarkers by Salivary Proteomics Data. International Journal of Molecular Sciences, 25(20), 11120. https://doi.org/10.3390/ijms252011120







