In Silico Insights Reveal Fibronectin 1 as a Theranostic Marker in Gastric Cancer
Abstract
1. Introduction
2. Results
2.1. Enrichment Analysis Highlights Unfavorable and Favorable Theranostic Genes
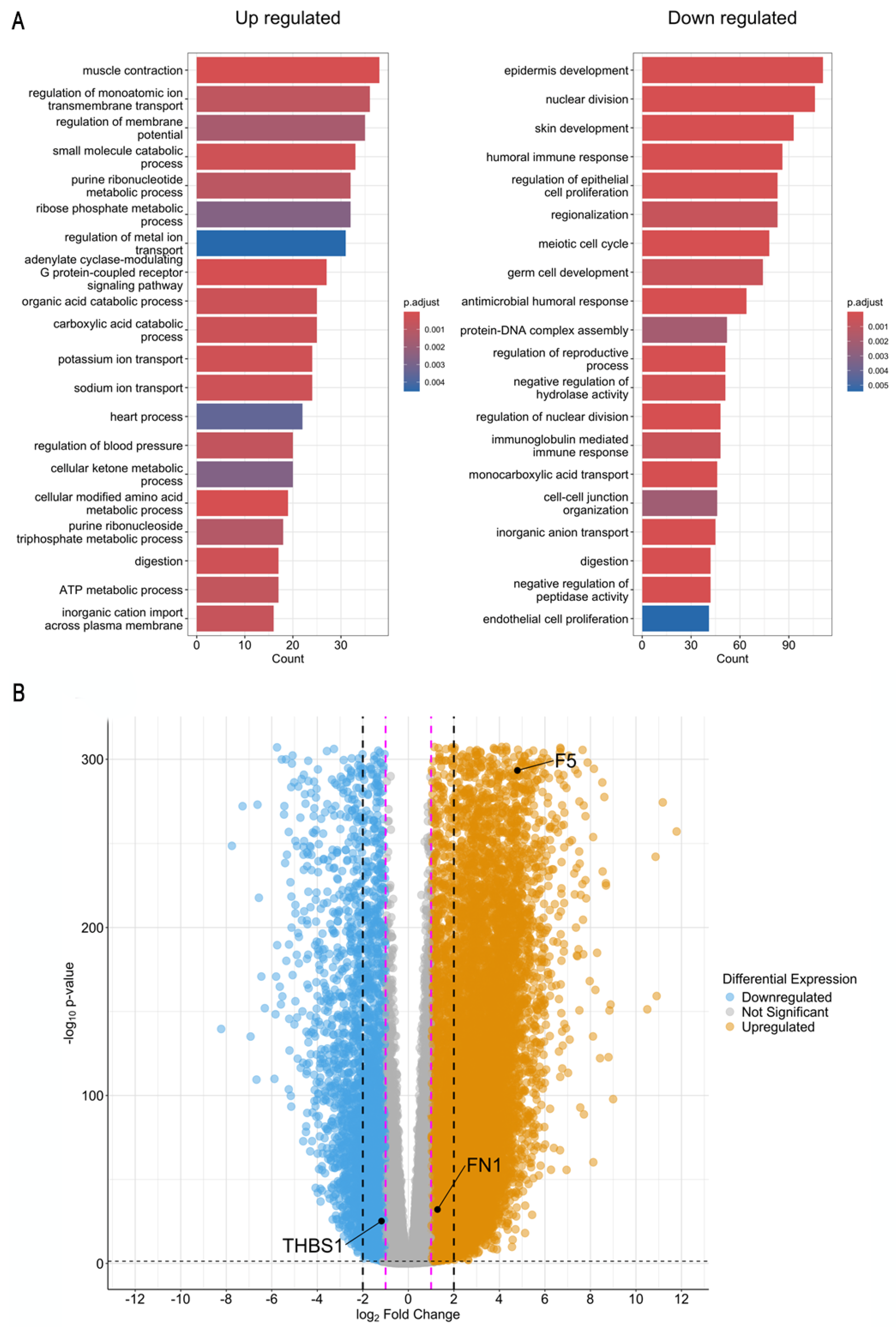
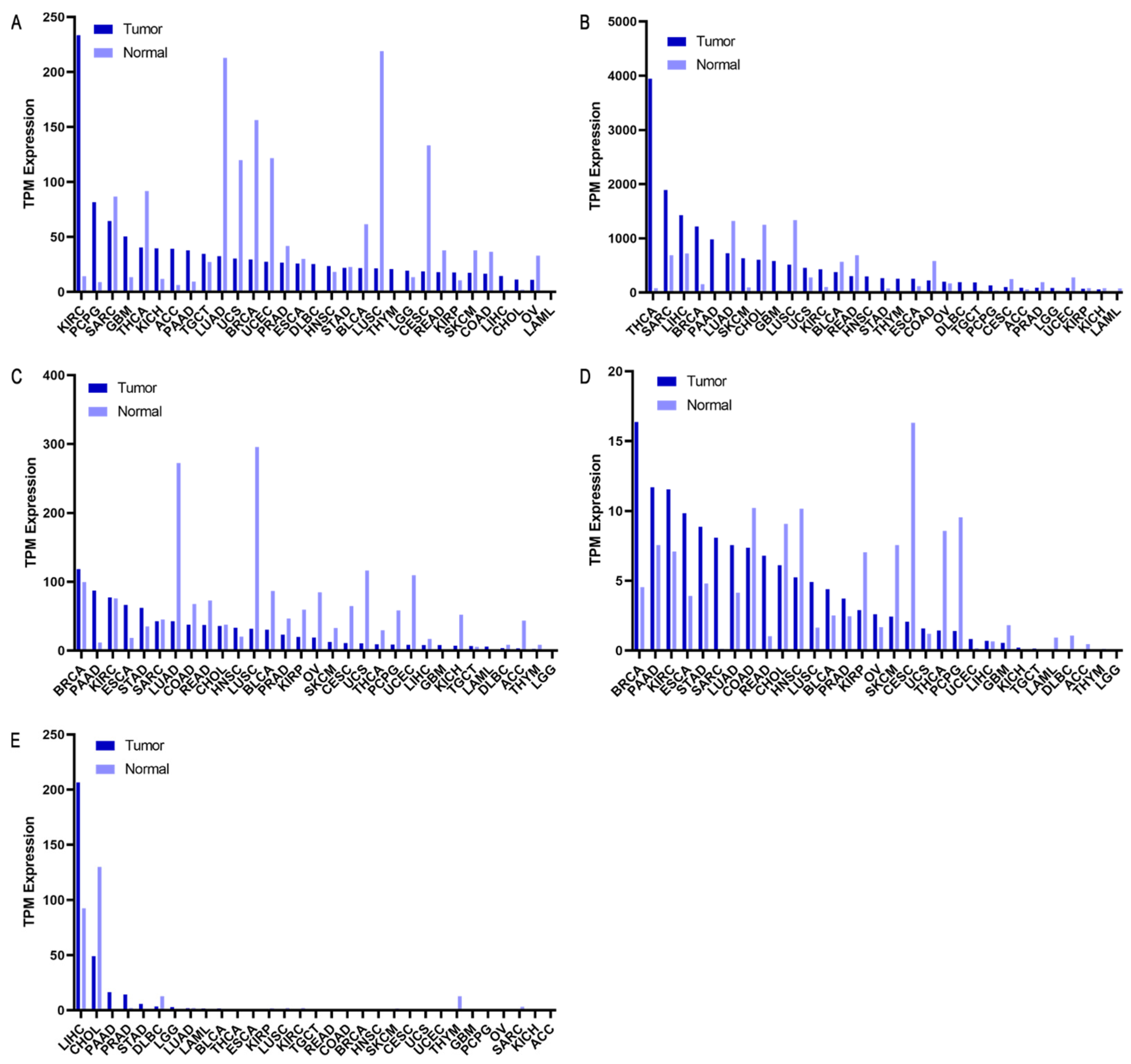
2.2. GC RNA-Seq Analysis from TCGA/GTEx
2.3. Proteomic Expression
2.4. Selected Genes as Biomarkers
3. Discussion
4. Methods and Materials
4.1. Genomic Data
4.2. Enrichment Analysis
4.3. Theranostic Evaluation Criteria
4.4. GC RNA-Seq Analysis from TCGA/GTEx
4.5. Genomic and Proteomic Distribution and Expression of Cancer Cell Lines
4.6. Proteomic Expression in Normal Tissues and Cell Lines
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Ministerio de Salud. Guía de Práctica Clínica Cáncer Gástrico; Ministerio de Salud: Santiago, Chile, 2020; pp. 1–21. Available online: https://diprece.minsal.cl/wp-content/uploads/2021/05/RE_GPC-Ca%CC%81ncer-Ga%CC%81strico_v2.pdf (accessed on 1 September 2024).
- Bornschein, J.; Quante, M.; Fassan, M. Gastric Cancer; Epidemiology and Diagnosis. In Encyclopedia of Gastroenterology, 2nd ed.; Kuipers, E.J., Ed.; Academic Press: Oxford, UK, 2020; pp. 553–564. [Google Scholar] [CrossRef]
- Ilic, M.; Ilic, I. Epidemiology of stomach cancer. World J. Gastroenterol. 2022, 28, 1187. [Google Scholar] [CrossRef]
- Matsuoka, T.; Yashiro, M. Biomarkers of gastric cancer: Current topics and future perspective. World J. Gastroenterol. 2018, 24, 2818–2832. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, I.; Ili, C.; Roa, J.C.; Brebi, P. Long non-coding RNAs in gastric cancer: Mechanisms and potential applications. Oncotarget 2016, 5, 1–14. [Google Scholar] [CrossRef]
- Kojima, R.; Aubel, D.; Fussenegger, M. Novel theranostic agents for next-generation personalized medicine: Small molecules, nanoparticles, and engineered mammalian cells. Curr. Opin. Chem. Biol. 2015, 28, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Kulkarni, H.R.; Singh, A.; Baum, R.P. Theranostics of Neuroendocrine Tumors. Visc. Med. 2017, 33, 358–366. [Google Scholar] [CrossRef]
- Li, X.; Kim, J.; Yoon, J.; Chen, X. Cancer-Associated, Stimuli-Driven, Turn on Theranostics for Multimodality Imaging and Therapy. Adv. Mater. 2017, 29, 1606857. [Google Scholar] [CrossRef]
- Ryu, J.H.; Lee, S.; Son, S.; Kim, S.H.; Leary, J.F.; Choi, K.; Kwon, I.C. Theranostic nanoparticles for future personalized medicine. J. Control. Release 2014, 190, 477–484. [Google Scholar] [CrossRef]
- Yu, G.; Jiang, M.; Huang, F.; Chen, X. Supramolecular coordination complexes as diagnostic and therapeutic agents. Curr. Opin. Chem. Biol. 2021, 61, 19–31. [Google Scholar] [CrossRef]
- Gao, X.; Guo, R.; Li, Y.; Kang, G.; Wu, Y.; Cheng, J.; Jia, J.; Wang, W.; Li, Z.; Wang, A.; et al. Contribution of upregulated aminoacyl-tRNA biosynthesis to metabolic dysregulation in gastric cancer. J. Gastroenterol. Hepatol. 2021, 36, 3113–3126. [Google Scholar] [CrossRef]
- Camacho-Sánchez, M.; Leandro-Vargas, L.A.; Mendoza-Salas, M.; Meza-Gutiérrez, N.; Montero-Zúñiga, F. Biomarcadores en el diagnóstico temprano y tratamiento de cáncer. Rev. Tecnol. En Marcha 2023, 36, 109. [Google Scholar] [CrossRef]
- Mérida de la Torre, F.J.; Moreno Campoy, E.E. Papel diagnóstico de los marcadores tumorales. Med. Clín. (Ed. Impr.) 2019, 152, 185–187. Available online: https://pesquisa.bvsalud.org/portal/resource/pt/ibc-181981 (accessed on 1 September 2024). [CrossRef] [PubMed]
- Socovich, A.M.; Naba, A. The cancer matrisome: From comprehensive characterization to biomarker discovery. Semin. Cell Dev. Biol. 2019, 89, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Goubran, H.A.; Stakiw, J.; Radosevic, M.; Burnouf, T. Platelets Effects on Tumor Growth. Semin. Oncol. 2014, 41, 359–369. [Google Scholar] [CrossRef]
- Li, J.; Han, T. Comprehensive analysis of the oncogenic roles of vascular endothelial growth factors and their receptors in stomach adenocarcinoma. Heliyon 2023, 9, e17687. [Google Scholar] [CrossRef]
- Libby, P.; Kobold, S. Inflammation: A common contributor to cancer, aging, and cardiovascular diseases—Expanding the concept of cardio-oncology. Cardiovasc. Res. 2019, 115, 824–829. [Google Scholar] [CrossRef]
- Chang, J.; Wang, X.; Sun, H.; Li, W.; Zhang, Z.; Zhu, X.; Xu, M. The use of DNA repair genes as prognostic indicators of gastric cancer. J. Cancer 2019, 10, 4866–4875. [Google Scholar] [CrossRef]
- Ebrahimi, V.; Soleimanian, A.; Ebrahimi, T.; Azargun, R.; Yazdani, P.; Eyvazi, S.; Tarhriz, V. Epigenetic modifications in gastric cancer: Focus on DNA methylation. Gene 2020, 742, 144. [Google Scholar] [CrossRef]
- Sonohara, F.; Inokawa, Y.; Hayashi, M.; Kodera, Y.; Nomoto, S. Epigenetic modulation associated with carcinogenesis and prognosis of human gastric cancer (Review). Oncol. Lett. 2017, 13, 3363–3368. [Google Scholar] [CrossRef][Green Version]
- Samadani, A.A.; Noroollahi, S.E.; Mansour-Ghanaei, F.; Rashidy-Pour, A.; Joukar, F.; Bandegi, A.R. Fluctuations of epigenetic regulations in human gastric Adenocarcinoma: How does it affect? Biomed. Pharmacother. 2019, 109, 144–156. [Google Scholar] [CrossRef]
- Tossetta, G.; Avellini, C.; Licini, C.; Giannubilo, S.; Castellucci, M.; Marzioni, D. High temperature requirement A1 and fibronectin: Two possible players in placental tissue remodelling. Eur. J. Histochem. 2016, 60, 2724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, J.; Chen, C.; Chen, B.; Guo, T. High FN1 expression correlates with gastric cancer progression. Pathol. Res. Pract. 2022, 239, 154–179. [Google Scholar] [CrossRef]
- Pan, H.; Luo, Z.; Lin, F.; Zhang, J.; Xiong, T.; Hong, Y.; Sun, B.; Yang, Y. FN1, a reliable prognostic biomarker for thyroid cancer, is associated with tumor immunity and an unfavorable prognosis. Oncol. Lett. 2024, 28, 510. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Yang, B.; Zhang, C.; Zhang, C.; Zang, D.; Gong, L.; Liu, Y.; Li, Z.; Qu, X. The F5 gene predicts poor prognosis of patients with gastric cancer by promoting cell migration identified using a weighted gene co-expression network analysis. Biocell 2021, 45, 911–921. [Google Scholar] [CrossRef]
- Liu, Y.; Liao, X.-W.; Qin, Y.-Z.; Mo, X.-W.; Luo, S.-S. Identification of F5 as a Prognostic Biomarker in Patients with Gastric Cancer. BioMed Res. Int. 2020, 2020, 9280841. [Google Scholar] [CrossRef]
- Yang, X.; Chen, L.; Mao, Y.; Hu, Z.; He, M. Progressive and Prognostic Performance of an Extracellular Matrix-Receptor Interaction Signature in Gastric Cancer. Dis. Markers 2020, 2020, 8816070. [Google Scholar] [CrossRef]
- Hong, B.-B.; Chen, S.-Q.; Qi, Y.-L.; Zhu, J.-W.; Lin, J.-Y. Association of THBS1 rs1478605 T>C in 5′-untranslated regions with the development and progression of gastric cancer. Biomed. Rep. 2015, 3, 207–214. [Google Scholar] [CrossRef]
- López-Cortés, A.; Abarca, E.; Silva, L.; Velastegui, E.; León-Sosa, A.; Karolys, G.; Cabrera, F.; Caicedo, A. Identification of key proteins in the signaling crossroads between wound healing and cancer hallmark phenotypes. Sci. Rep. 2021, 11, 17245. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, J.; Li, H.; Yu, H.; Chen, S.; Liu, S.; Zhang, C.; He, Y. FN1 is a prognostic biomarker and correlated with immune infiltrates in gastric cancers. Front. Oncol. 2022, 12, 918719. [Google Scholar] [CrossRef]
- Eskandarion, M.R.; Eskandarieh, S.; Farahani, A.S.; Mahmoodzadeh, H.; Shahi, F.; Oghabian, M.A.; Shirkoohi, R. Prediction of novel biomarkers for gastric intestinal metaplasia and gastric adenocarcinoma using bioinformatics analysis. Heliyon 2024, 10, e30253. [Google Scholar] [CrossRef]
- Nyakale, N.E.; Aldous, C.; Gutta, A.A.; Khuzwayo, X.; Harry, L.; Sathekge, M.M. Emerging theragnostic radionuclide applications for hepatocellular carcinoma. Front. Nucl. Med. 2023. 3, 1210982. [CrossRef]
- Ye, D.M.; Xu, G.; Ma, W.; Li, Y.; Luo, W.; Xiao, Y.; Liu, Y.; Zhang, Z. Significant function and research progress of biomarkers in gastric cancer (Review). Oncol. Lett. 2020, 19, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; Vettori, R.; Steffan, A.; Cannizzaro, R.; De Re, V. Circulating Proteins as Diagnostic Markers in Gastric Cancer. Int. J. Mol. Sci. 2023, 24, 16931. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Okamoto, K.; Kida, Y.; Mitsui, Y.; Kawano, Y.; Sogabe, M.; Miyamoto, H.; Takayama, T. Overview of Chemotherapy for Gastric Cancer. J. Clin. Med. 2023, 12, 1336. [Google Scholar] [CrossRef] [PubMed]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent advances in gastric cancer early diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef]
- Farhana, A.; Yusuf, N.; Rasheed, Z. Editorial: Cancer genetics and epigenetics: Theranostic targets and mechanisms. Front. Genet. 2024, 15, 1446474. [Google Scholar] [CrossRef]
- Franchini, M.; Frattini, F.; Crestani, S.; Bonfanti, C.; Lippi, G. von Willebrand factor and cancer: A renewed interest. Thromb. Res. 2013, 131, 290–292. [Google Scholar] [CrossRef]
- Yang, A.-J.; Wang, M.; Wang, Y.; Cai, W.; Li, Q.; Zhao, T.-T.; Zhang, L.-H.; Houck, K.; Chen, X.; Jin, Y.-L.; et al. Cancer cell-derived von Willebrand factor enhanced metastasis of gastric adenocarcinoma. Oncogenesis 2018, 7, 12. [Google Scholar] [CrossRef]
- Wang, C.Y.; Wang, M.; Cai, W.; Zhao, C.Y.; Zhou, Q.; Zhang, X.Y.; Wang, F.X.; Zhang, C.L.; Dang, Y.; Yang, A.J.; et al. Von Willebrand Factor Synergizes with Tumor-Derived Extracellular Vesicles to Promote Gastric Cancer Metastasis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Cai, W.; Wang, M.; Wang, C.-Y.; Zhao, C.-Y.; Zhang, X.-Y.; Zhou, Q.; Zhao, W.-J.; Yang, F.; Zhang, C.-L.; Yang, A.-J.; et al. Extracellular vesicles, hyperadhesive von willebrand factor, and outcomes of gastric cancer: A clinical observational study. Med. Oncol. 2023, 40, 140. [Google Scholar] [CrossRef]
- Yang, X.; Sun, H.-J.; Li, Z.-R.; Zhang, H.; Yang, W.-J.; Ni, B.; Wu, Y.-Z. Gastric cancer-associated enhancement of von Willebrand factor is regulated by vascular endothelial growth factor and related to disease severity. BMC Cancer 2015, 15, 80. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhao, C.; Ye, Y.; Wang, Z.; He, Y.; Li, Y.; Mao, H. High expression of fibronectin 1 indicates poor prognosis in gastric cancer. Oncol. Lett. 2020, 19, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Seoane, J.; Gomis, R.R. TGF-β Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb. Perspect. Biol. 2017, 9, a022277. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Zhu, J.; Liu, P.; Wei, Q.; Zhang, S.; An, W.; Tong, Y.; Cheng, Z.; Liu, F. FN1 mRNA 3′-UTR supersedes traditional fibronectin 1 in facilitating the invasion and metastasis of gastric cancer through the FN1 3′-UTR-let-7i-5p-THBS1 axis. Theranostics 2023, 13, 5130–5150. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.-S.; Zhang, T.; Wang, H.-C.; Li, L.-P. Identification of New Therapeutic Targets for Gastric Cancer with Bioinformatics. Front. Genet. 2020, 11, 865. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, T.; Li, Y.; Qiu, H. Upregulation of THBS1 is Related to Immunity and Chemotherapy Resistance in Gastric Cancer. Int. J. Gen. Med. 2021, 14, 4945–4957. [Google Scholar] [CrossRef]
- Khoshdel, F.; Mottaghi-Dastjerdi, N.; Yazdani, F.; Salehi, S.; Ghorbani, A.; Montazeri, H.; Soltany-Rezaee-Rad, M.; Goodarzy, B. CTGF, FN1, IL-6, THBS1, and WISP1 genes and PI3K-Akt signaling pathway as prognostic and therapeutic targets in gastric cancer identified by gene network modeling. Discov. Oncol. 2024, 15, 344. [Google Scholar] [CrossRef]
- Chen, H.-F.; Ma, R.-R.; He, J.-Y.; Zhang, H.; Liu, X.-L.; Guo, X.-Y.; Gao, P. Protocadherin 7 inhibits cell migration and invasion through E-cadherin in gastric cancer. Tumour Biol. 2017, 39, 1010428317697551. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyu, Y.; Chen, L.; Cao, K.; Chen, J.; He, C.; Lyu, X.; Jiang, Y.; Xiang, J.; Liu, B.; et al. Exploring the Prognosis-Related Genetic Variation in Gastric Cancer Based on mGWAS. Int. J. Mol. Sci. 2023, 24, 15259. [Google Scholar] [CrossRef]
- Zheng, Z.; Luan, N.; Tu, K.; Liu, F.; Wang, J.; Sun, J. The roles of protocadherin-7 in colorectal cancer cells on cell proliferation and its chemoresistance. Front. Pharmacol. 2023, 14, 1072033. [Google Scholar] [CrossRef]
- Guan, Y.; Xu, B.; Sui, Y.; Chen, Z.; Luan, Y.; Jiang, Y.; Wei, L.; Long, W.; Zhao, S.; Han, L.; et al. Pan-Cancer Analysis and Validation Reveals that D-Dimer-Related Genes are Prognostic and Downregulate CD8+ T Cells via TGF-Beta Signaling in Gastric Cancer. Front. Mol. Biosci. 2022, 9, 790706. [Google Scholar] [CrossRef] [PubMed]
- Chiurillo, M.A. Role of the Wnt/β-catenin pathway in gastric cancer: An in-depth literature review. World J. Exp. Med. 2015, 5, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Dang, K.; Farooq, H.M.U.; Dong, J.; Yang, H.; Kong, Y.; Wang, H.; Jiang, S.; Gao, Y.; Qian, A. Transcriptomic and proteomic time-course analyses based on Metascape reveal mechanisms against muscle atrophy in hibernating Spermophilus dauricus. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2023, 275, 111336. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Jokerst, J.V.; Gambhir, S.S. Molecular Imaging with Theranostic Nanoparticles. Acc. Chem. Res. 2011, 44, 1050–1060. [Google Scholar] [CrossRef]
- Srinivasarao, M.; Galliford, C.V.; Low, P.S. Low PS. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat. Rev. Drug Discov. 2015, 14, 203–219. [Google Scholar] [CrossRef]
- Espinoza, J.A.; Riquelme, I.; Sagredo, E.A.; Rosa, L.; García, P.; Bizama, C.; Apud-Bell, M.; Leal, P.; Weber, H.; Benavente, F.; et al. Mucin 5B, carbonic anhydrase 9 and claudin 18 are potential theranostic markers of gallbladder carcinoma. Histopathology 2018, 74, 597–607. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 2024, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Chalifa-Caspi, V.; Shmueli, O.; Olender, T.; Lapidot, M.; Rosen, N.; Shmoish, M.; Peter, Y.; Glusman, G.; Feldmesser, E.; et al. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003, 31, 142–146. [Google Scholar] [CrossRef] [PubMed]
- Fishilevich, S.; Zimmerman, S.; Kohn, A.; Stein, T.I.; Olender, T.; Kolker, E.; Safran, M.; Lancet, D. Genic insights from integrated human proteomics in GeneCards. Database 2016, 2016, baw030. [Google Scholar] [CrossRef]
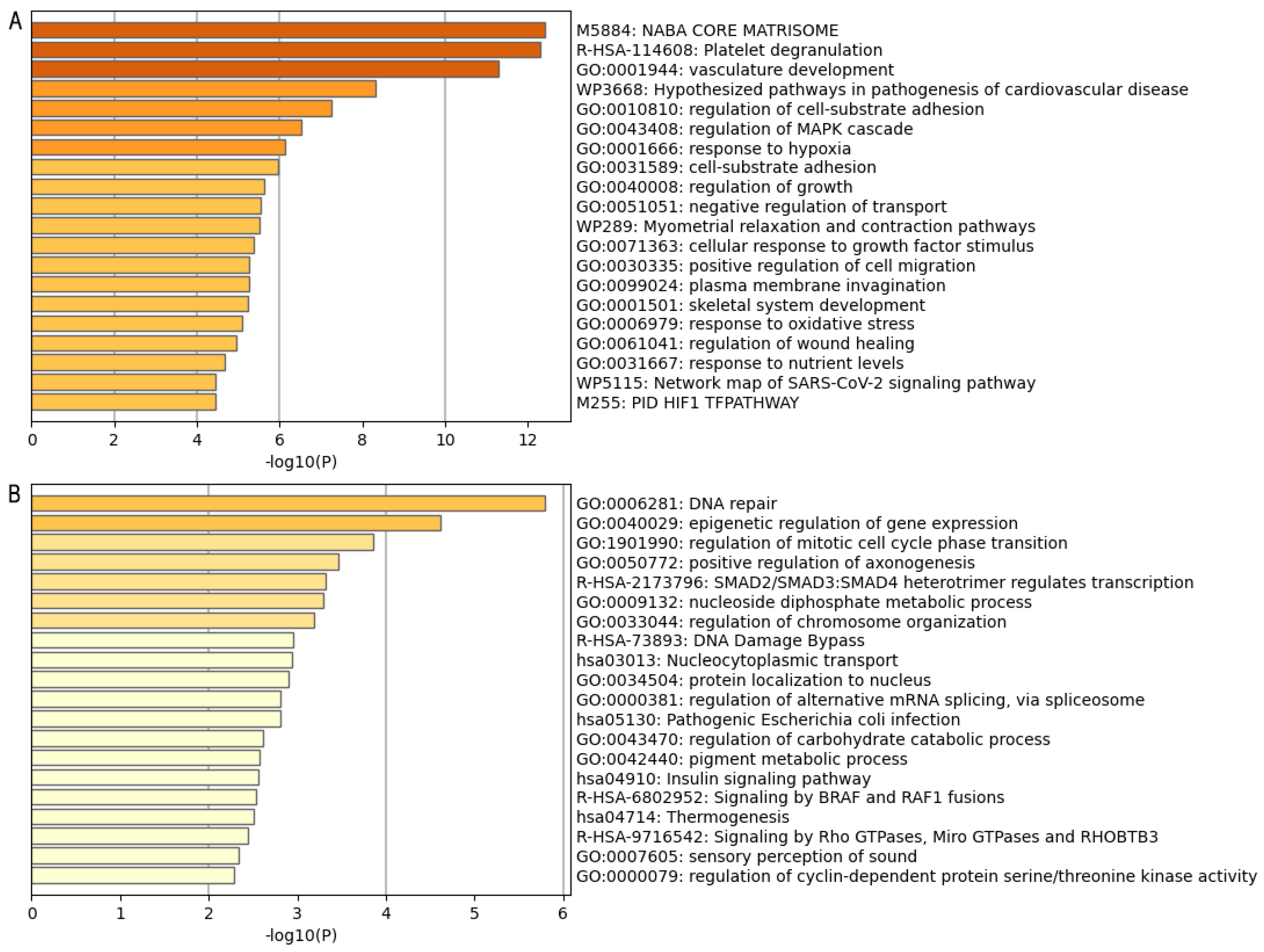
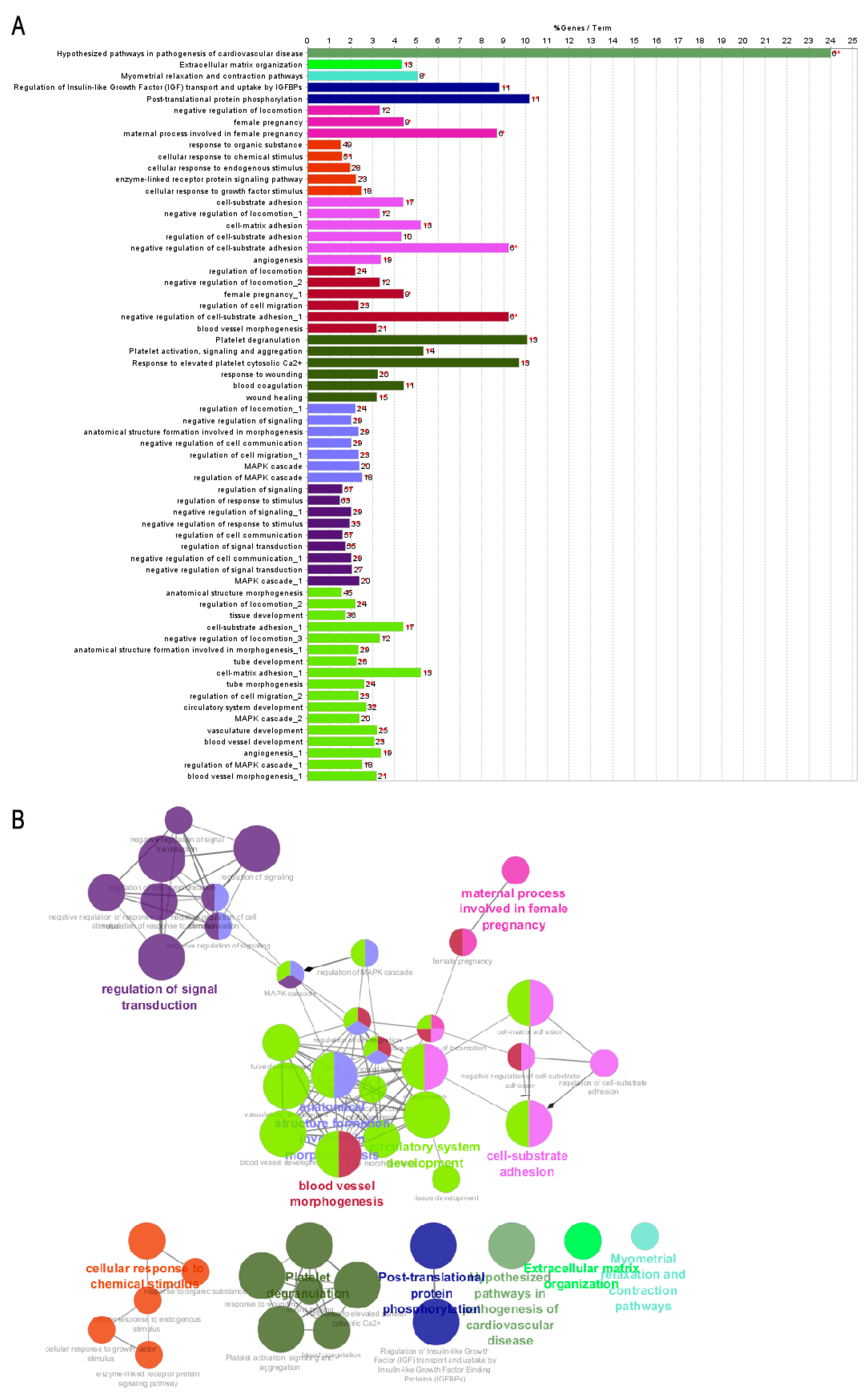
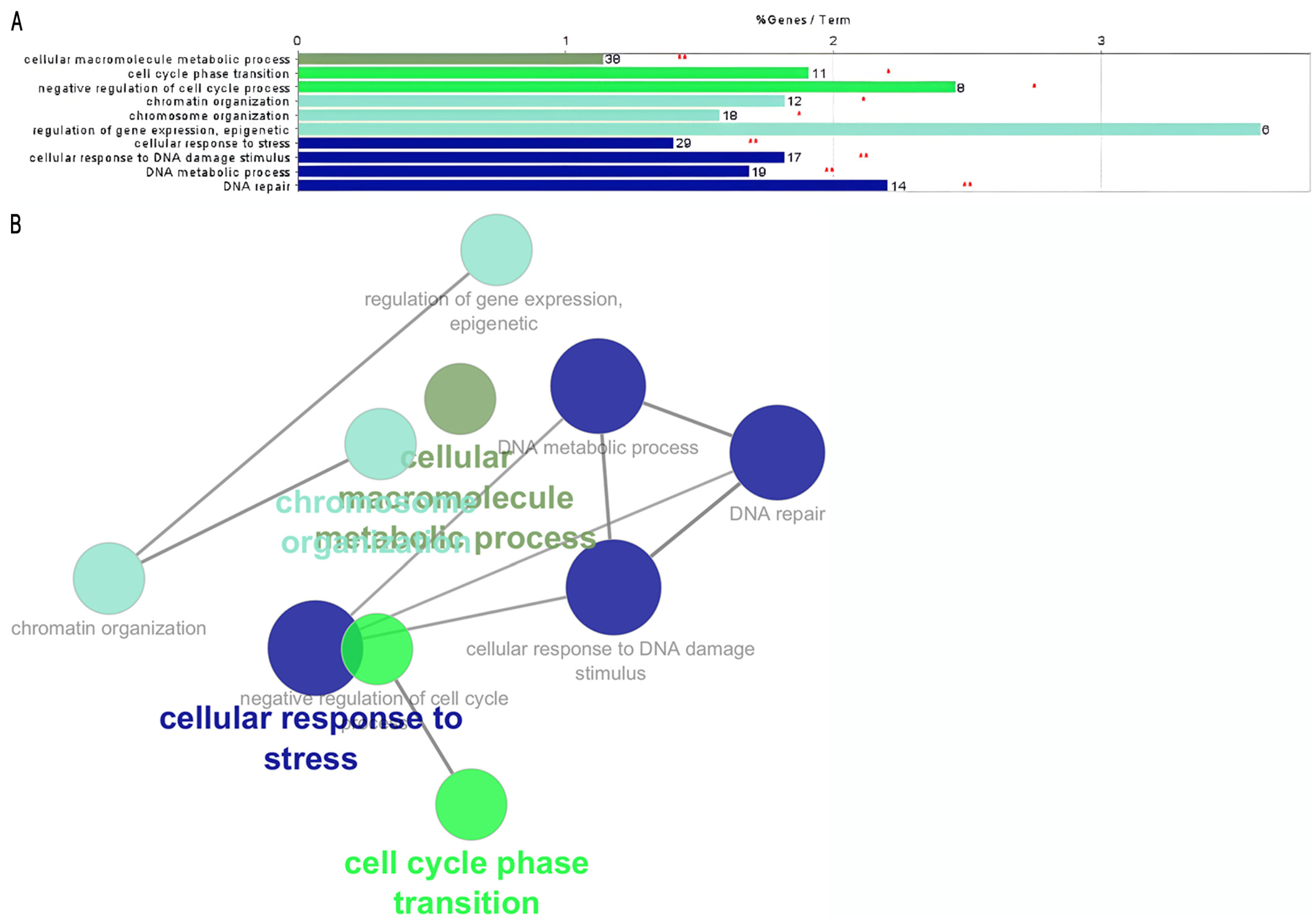
| Group | ID | Enriched Process | Associated Genes |
|---|---|---|---|
| A | WP:3668 | Hypothesized pathways in pathogenesis of cardiovascular disease | ANGPT2, CCN2, FBN1, POSTN, RUNX2, SERPINE1 |
| R-HSA:8957275 | Post-translational protein phosphorylation process | ALB, F5, FBN1, FN1, GPC3, IGFBP1, MATN3, MFGE8, MXRA8, STC2, VCAN | |
| R-HSA:114608 | Platelet degranulation | ALB, ANXA5, CD109, CD36, F5, FN1, MAGED2, MMRN1, PCDH7, PROS1, SERPINE1, THBS1, VWF | |
| B | GO:0040029 | Epigenetic regulation of gene expression | EZH2, KCNQ1, LMNB2, RBM15, SAMD1, SIRT6 |
| GO:0010948 | Negative regulation of cell cycle process | CLSPN, CUL4A, ESPL1, EZH2, FBXO5, INIP, MEN1, NACC2 | |
| GO:0006281 | DNA repair | CHAF1A, CLSPN, CUL4A, FEN1, INIP, INO80B, MEN1, POLQ, SIRT6, SWSAP1, TAF5, TMEM161A, USP43, ZBTB7A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Millapán, T.; Gutiérrez, Á.; Rosas, K.; Buchegger, K.; Ili, C.G.; Brebi, P. In Silico Insights Reveal Fibronectin 1 as a Theranostic Marker in Gastric Cancer. Int. J. Mol. Sci. 2024, 25, 11113. https://doi.org/10.3390/ijms252011113
Millapán T, Gutiérrez Á, Rosas K, Buchegger K, Ili CG, Brebi P. In Silico Insights Reveal Fibronectin 1 as a Theranostic Marker in Gastric Cancer. International Journal of Molecular Sciences. 2024; 25(20):11113. https://doi.org/10.3390/ijms252011113
Chicago/Turabian StyleMillapán, Tatiana, Álvaro Gutiérrez, Krisnna Rosas, Kurt Buchegger, Carmen Gloria Ili, and Priscilla Brebi. 2024. "In Silico Insights Reveal Fibronectin 1 as a Theranostic Marker in Gastric Cancer" International Journal of Molecular Sciences 25, no. 20: 11113. https://doi.org/10.3390/ijms252011113
APA StyleMillapán, T., Gutiérrez, Á., Rosas, K., Buchegger, K., Ili, C. G., & Brebi, P. (2024). In Silico Insights Reveal Fibronectin 1 as a Theranostic Marker in Gastric Cancer. International Journal of Molecular Sciences, 25(20), 11113. https://doi.org/10.3390/ijms252011113







