Development of Malate Biosensor-Containing Hydrogels and Living Cell-Based Sensors
Abstract
1. Introduction
2. Results
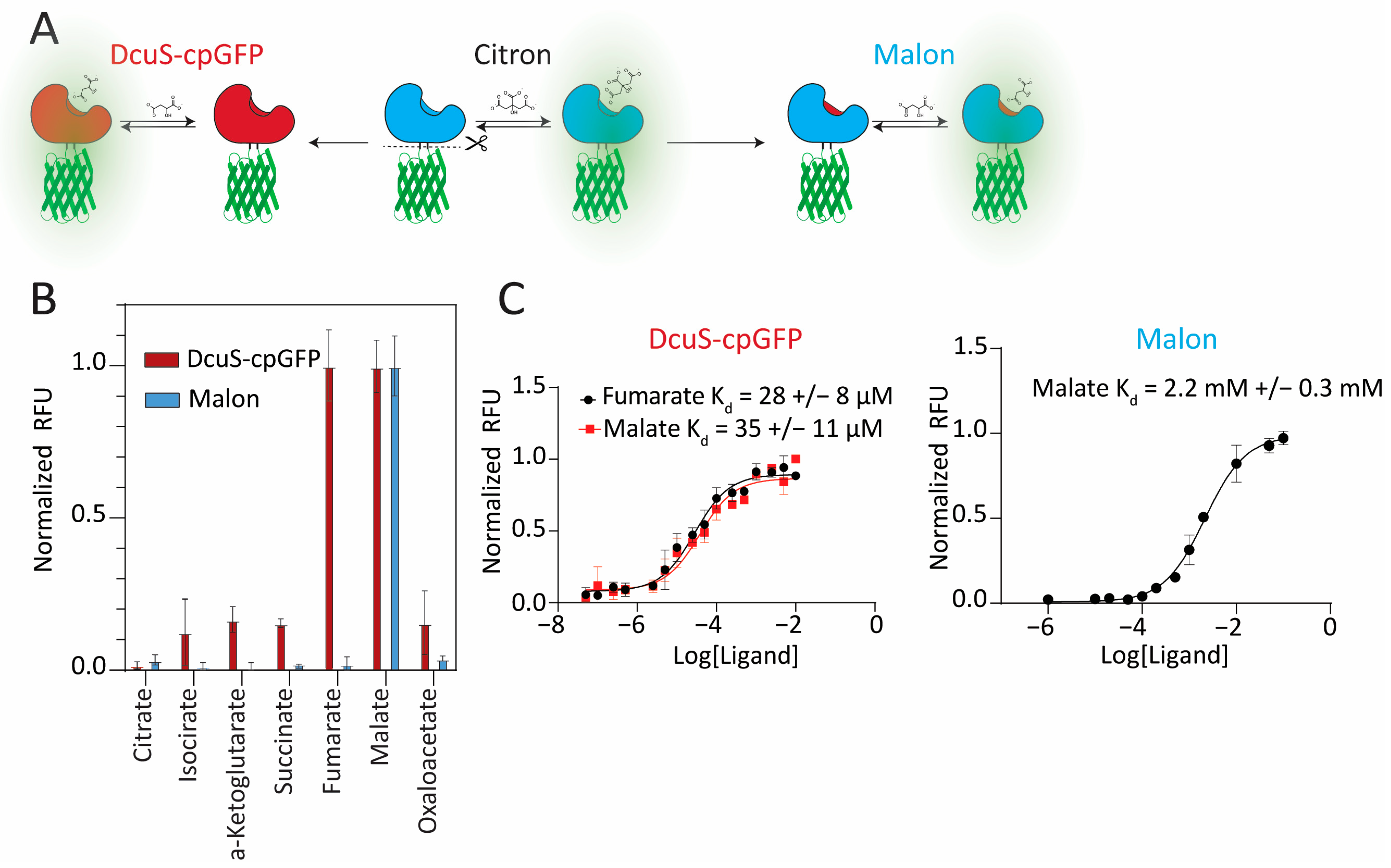
2.1. Design of Malon Based on Domain Replacement and Pocket-Grafting Strategies
2.2. Investigation of Ligand Recognition in the Binding Pocket
2.3. Monitoring and Quantitating Malate in Enzymatic Assays
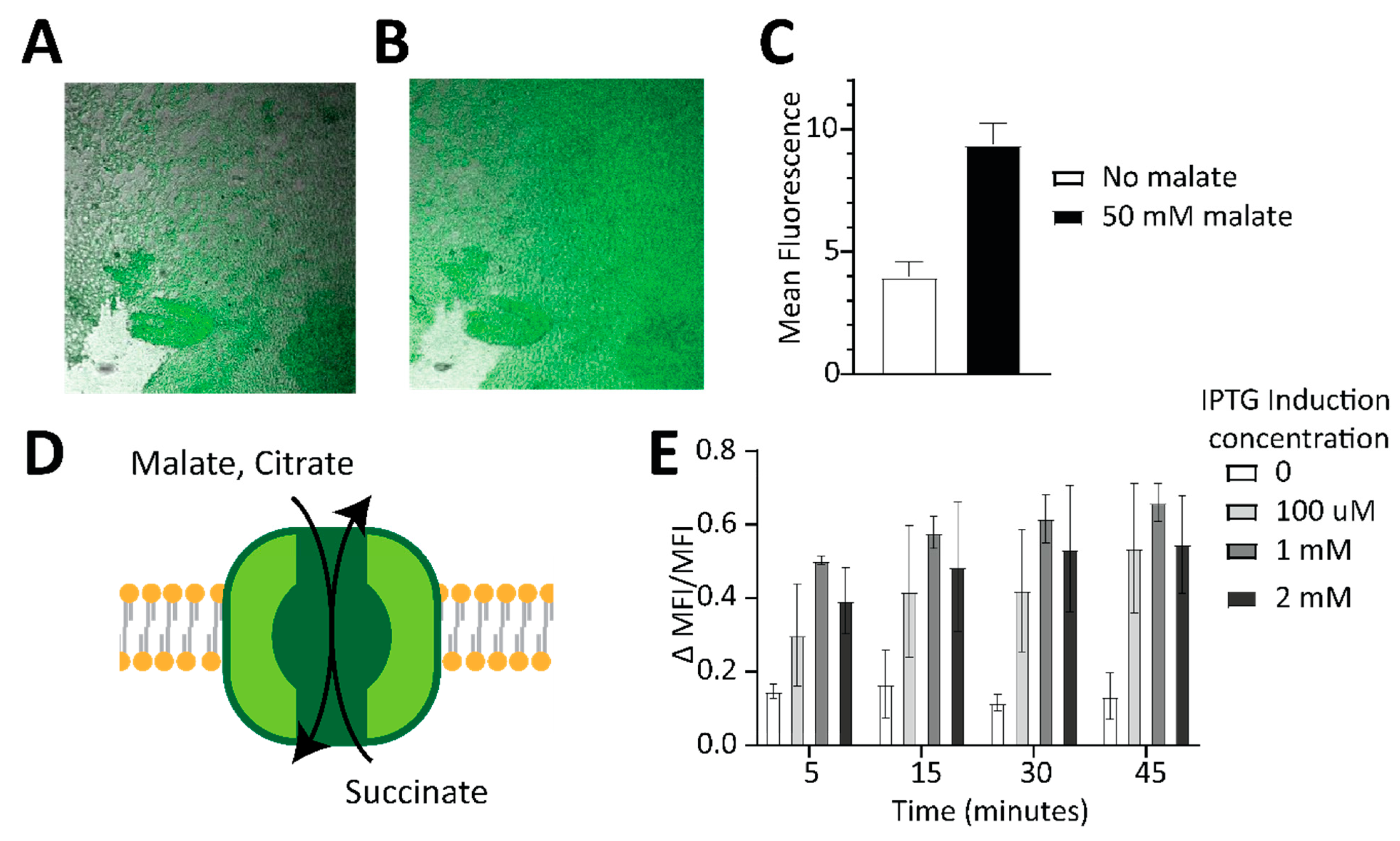
2.4. Incorporating Malon to Make Sensor-Containing Hydrogels and Living Cell-Based Sensors
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krebs, H.A. The Citric Acid Cycle: A Reply to the Criticisms of FL Breusch and of J. Thomas. Biochem. J. 1940, 34, 460. [Google Scholar] [CrossRef] [PubMed]
- Akram, M. Citric Acid Cycle and Role of Its Intermediates in Metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Banta, S.; Chen, W. Functional Assembly of a Multi-Enzyme Methanol Oxidation Cascade on a Surface-Displayed Trifunctional Scaffold for Enhanced NADH Production. Chem. Commun. 2013, 49, 3766–3768. [Google Scholar] [CrossRef] [PubMed]
- Sokic-Lazic, D.; Minteer, S.D. Pyruvate/Air Enzymatic Biofuel Cell Capable of Complete Oxidation. Electrochem. Solid-State Lett. 2009, 12, F26. [Google Scholar] [CrossRef]
- Wheeldon, I.; Minteer, S.D.; Banta, S.; Barton, S.C.; Atanassov, P.; Sigman, M. Substrate Channelling as an Approach to Cascade Reactions. Nat. Chem. 2016, 8, 299–309. [Google Scholar] [CrossRef]
- Xiao, X.; Xia, H.; Wu, R.; Bai, L.; Yan, L.; Magner, E.; Cosnier, S.; Lojou, E.; Zhu, Z.; Liu, A. Tackling the Challenges of Enzymatic (Bio) Fuel Cells. Chem. Rev. 2019, 119, 9509–9558. [Google Scholar] [CrossRef]
- Franco, J.H.; De Andrade, A.R. Bioelectrodes with Enzyme Cascade Reactions. In Advances in Bioelectrochemistry Volume 5: Emerging Techniques and Materials, Biodevice Design and Reactions; Springer: Berlin/Heidelberg, Germany, 2022; pp. 157–179. [Google Scholar]
- Sokic-Lazic, D.; Minteer, S.D. Citric Acid Cycle Biomimic on a Carbon Electrode. Biosens. Bioelectron. 2008, 24, 939–944. [Google Scholar] [CrossRef]
- Fan, S.; Liang, B.; Xiao, X.; Bai, L.; Tang, X.; Lojou, E.; Cosnier, S.; Liu, A. Controllable Display of Sequential Enzymes on Yeast Surface with Enhanced Biocatalytic Activity toward Efficient Enzymatic Biofuel Cells. J. Am. Chem. Soc. 2020, 142, 3222–3230. [Google Scholar] [CrossRef]
- Ganesh, I.; Ravikumar, S.; Yoo, I.-K.; Hong, S.H. Construction of Malate-Sensing Escherichia Coli by Introduction of a Novel Chimeric Two-Component System. Bioprocess Biosyst. Eng. 2015, 38, 797–804. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Xiao, F.; Wang, H.; Zhang, L.; Ding, Z.; Xu, S.; Gu, Z.; Shi, G. Engineering of a Biosensor in Response to Malate in Bacillus Licheniformis. ACS Synth. Biol. 2021, 10, 1775–1784. [Google Scholar] [CrossRef]
- Cormann, K.U.; Baumgart, M.; Bott, M. Structure-Based Design of Versatile Biosensors for Small Molecules Based on the PAS Domain of a Thermophilic Histidine Kinase. ACS Synth. Biol. 2018, 7, 2888–2897. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Shen, Y.; Wen, Y.; Campbell, R.E. High-Performance Intensiometric Direct- and Inverse-Response Genetically Encoded Biosensors for Citrate. ACS Cent. Sci. 2020, 6, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Nasu, Y.; Murphy-Royal, C.; Wen, Y.; Haidey, J.N.; Molina, R.S.; Aggarwal, A.; Zhang, S.; Kamijo, Y.; Paquet, M.E.; Podgorski, K.; et al. A Genetically Encoded Fluorescent Biosensor for Extracellular L-Lactate. Nat. Commun. 2021, 12, 7058. [Google Scholar] [CrossRef] [PubMed]
- Nasu, Y.; Shen, Y.; Kramer, L.; Campbell, R.E. Structure- and Mechanism-Guided Design of Single Fluorescent Protein-Based Biosensors. Nat. Chem. Biol. 2021, 17, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, S.; Liu, X.; Wang, Z.; Jiang, M.; Wang, R.; Xie, L.; Liu, Q.; Xie, X.; Shang, D.; et al. Sensing of Autoinducer-2 by Functionally Distinct Receptors in Prokaryotes. Nat. Commun. 2020, 11, 5371. [Google Scholar] [CrossRef]
- Cheung, J.; Hendrickson, W.A. Crystal Structures of C4-Dicarboxylate Ligand Complexes with Sensor Domains of Histidine Kinases DcuS and DctB. J. Biol. Chem. 2008, 283, 30256–30265. [Google Scholar] [CrossRef]
- Gavira, J.A.; Gumerov, V.M.; Rico-Jiménez, M.; Petukh, M.; Upadhyay, A.A.; Ortega, A.; Matilla, M.A.; Zhulin, I.B.; Krell, T. How Bacterial Chemoreceptors Evolve Novel Ligand Specificities. Mbio 2020, 11, e03066-19. [Google Scholar] [CrossRef]
- Upadhyay, A.A.; Fleetwood, A.D.; Adebali, O.; Finn, R.D.; Zhulin, I.B. Cache Domains That Are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 2016, 12, e10048622. [Google Scholar] [CrossRef]
- Gushchin, I.; Gordeliy, V. Transmembrane Signal Transduction in Two-component Systems: Piston, Scissoring, or Helical Rotation? Bioessays 2018, 40, 1700197. [Google Scholar] [CrossRef]
- Salvi, M.; Schomburg, B.; Giller, K.; Graf, S.; Unden, G.; Becker, S.; Lange, A.; Griesinger, C. Sensory Domain Contraction in Histidine Kinase CitA Triggers Transmembrane Signaling in the Membrane-Bound Sensor. Proc. Natl. Acad. Sci. USA 2017, 114, 3115–3120. [Google Scholar] [CrossRef]
- Monzel, C.; Unden, G. Transmembrane Signaling in the Sensor Kinase DcuS of Escherichia coli: A Long-Range Piston-Type Displacement of Transmembrane Helix 2. Proc. Natl. Acad. Sci. USA 2015, 112, 11042–11047. [Google Scholar] [CrossRef] [PubMed]
- Kramer, J.; Fischer, J.D.; Zientz, E.; Vijayan, V.; Griesinger, C.; Lupas, A.; Unden, G. Citrate Sensing by the C4-Dicarboxylate/Citrate Sensor Kinase DcuS of Escherichia coli: Binding Site and Conversion of DcuS to a C4-Dicarboxylate-or Citrate-Specific Sensor. J. Bacteriol. 2007, 189, 4290–4298. [Google Scholar] [CrossRef] [PubMed]
- Albe, K.R.; Butler, M.H.; Wright, B.E. Cellular Concentrations of Enzymes and Their Substrates. J. Theor. Biol. 1990, 143, 163–195. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A.; van Pelt, S. Enzyme Immobilisation in Biocatalysis: Why, What and How. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Bommarius, A.S.; Paye, M.F. Stabilizing Biocatalysts. Chem. Soc. Rev. 2013, 42, 6534–6565. [Google Scholar] [CrossRef]
- Kummer, M.J.; Lee, Y.S.; Yuan, M.; Alkotaini, B.; Zhao, J.; Blumenthal, E.; Minteer, S.D. Substrate Channeling by a Rationally Designed Fusion Protein in a Biocatalytic Cascade. J. Am. Chem. Soc. 2021, 1, 1187–1197. [Google Scholar] [CrossRef]
- Krebs, H.A. The Equilibrium Constants of the Fumarase and Aconitase Systems. Biochem. J. 1953, 54, 78–82. [Google Scholar] [CrossRef]
- Milton, R.D.; Wang, T.; Knoche, K.L.; Minteer, S.D. Tailoring Biointerfaces for Electrocatalysis. Langmuir 2016, 32, 2291–2301. [Google Scholar] [CrossRef]
- Dippel, A.B.; Anderson, W.A.; Evans, R.S.; Deutsch, S.; Hammond, M.C. Chemiluminescent Biosensors for Detection of Second Messenger Cyclic Di-GMP. ACS Chem. Biol. 2018, 13, 1872–1879. [Google Scholar] [CrossRef]
- Pos, K.M.; Dimroth, P.; Bott, M. The Escherichia Coli Citrate Carrier CitT: A Member of a Novel Eubacterial Transporter Family Related to the 2-Oxoglutarate/Malate Translocator from Spinach Chloroplasts. J. Bacteriol. 1998, 180, 4160–4165. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Abdelfattah, A.S.; Zhou, H.; Ruangkittisakul, A.; Qian, Y.; Ballanyi, K.; Campbell, R.E. Genetically Encoded Glutamate Indicators with Altered Color and Topology. ACS Chem. Biol. 2018, 13, 1832–1837. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Chen, X.; Sheng, H.; Shen, X.; Sun, X.; Yan, Y.; Wang, J.; Yuan, Q. Engineering Probiotics as Living Diagnostics and Therapeutics for Improving Human Health. Microb. Cell Factories 2020, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Isabella, V.M.; Ha, B.N.; Castillo, M.J.; Lubkowicz, D.J.; Rowe, S.E.; Millet, Y.A.; Anderson, C.L.; Li, N.; Fisher, A.B.; West, K.A. Development of a Synthetic Live Bacterial Therapeutic for the Human Metabolic Disease Phenylketonuria. Nat. Biotechnol. 2018, 36, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, C.B.; Millet, Y.A.; Puurunen, M.K.; Perreault, M.; Charbonneau, M.R.; Isabella, V.M.; Kotula, J.W.; Antipov, E.; Dagon, Y.; Denney, W.S. An Engineered E. coli Nissle Improves Hyperammonemia and Survival in Mice and Shows Dose-Dependent Exposure in Healthy Humans. Sci. Transl. Med. 2019, 11, eaau7975. [Google Scholar] [CrossRef]
- Sedlmayer, F.; Hell, D.; Müller, M.; Ausländer, D.; Fussenegger, M. Designer Cells Programming Quorum-Sensing Interference with Microbes. Nat. Commun. 2018, 9, 1822. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Wen, Y.; Serre, N.B.; Laursen, C.C.H.; Dietz, A.G.; Taylor, B.R.; Drobizhev, M.; Molina, R.S.; Aggarwal, A.; Rancic, V. A Sensitive and Specific Genetically-Encoded Potassium Ion Biosensor for in Vivo Applications across the Tree of Life. PLoS Biol. 2022, 20, e3001772. [Google Scholar] [CrossRef]
- Velapoldi, R.A.; Tønnesen, H.H. Corrected Emission Spectra and Quantum Yields for a Series of Fluorescent Compounds in the Visible Spectral Region. J. Fluoresc. 2004, 14, 465–472. [Google Scholar] [CrossRef]
- Abdellaoui, S.; Milton, R.D.; Quah, T.; Minteer, S.D. NAD-dependent dehydrogenase bioelectrocatalysis: The ability of a naphthoquinone redox polymer to regenerate NAD. Chem. Commun. 2015, 52, 1147–1150. [Google Scholar] [CrossRef]
- Tanaka, R.; Ueoka, I.; Takaki, Y.; Kataoka, K.; Saito, S. High molecular weight linear polyethylenimine and poly(N-methylethylenimine). Macromolecules 1983, 16, 849–853. [Google Scholar] [CrossRef]

| Fluorescent Sensor | Ligand | Extinction Coefficient (mM−1 cm−1) | Quantum Yield | Brightness | Excitation and Emission Maximum | Kd | F/Fmin |
|---|---|---|---|---|---|---|---|
| Citron [13] | +citrate | 61.6 | 0.34 | 21 | 499/517 | 1.1 mM | 9 |
| −citrate | 7.2 | 0.29 | 0.21 | 499/516 | |||
| DcuS–cpGFP | +malate | 26 ± 6 | 0.28 ± 0.02 | 7.3 | 493/515 | 35 ± 11 μM | 1.8 ± 0.3 |
| −malate | 13 ± 4 | 0.25 ± 0.01 | 3.3 | 499/516 | |||
| Malon | +malate | 110 ± 10 | 0.32 ± 0.01 | 35 | 499/518 | 2.2 ± 0.3 mM | 10 ± 1.2 |
| −malate | 18 ± 5 | 0.30 ± 0.01 | 5.4 | 496/515 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricks, N.J.; Brachi, M.; McFadden, K.; Jadhav, R.G.; Minteer, S.D.; Hammond, M.C. Development of Malate Biosensor-Containing Hydrogels and Living Cell-Based Sensors. Int. J. Mol. Sci. 2024, 25, 11098. https://doi.org/10.3390/ijms252011098
Ricks NJ, Brachi M, McFadden K, Jadhav RG, Minteer SD, Hammond MC. Development of Malate Biosensor-Containing Hydrogels and Living Cell-Based Sensors. International Journal of Molecular Sciences. 2024; 25(20):11098. https://doi.org/10.3390/ijms252011098
Chicago/Turabian StyleRicks, Nathan J., Monica Brachi, Kevin McFadden, Rohit G. Jadhav, Shelley D. Minteer, and Ming C. Hammond. 2024. "Development of Malate Biosensor-Containing Hydrogels and Living Cell-Based Sensors" International Journal of Molecular Sciences 25, no. 20: 11098. https://doi.org/10.3390/ijms252011098
APA StyleRicks, N. J., Brachi, M., McFadden, K., Jadhav, R. G., Minteer, S. D., & Hammond, M. C. (2024). Development of Malate Biosensor-Containing Hydrogels and Living Cell-Based Sensors. International Journal of Molecular Sciences, 25(20), 11098. https://doi.org/10.3390/ijms252011098







