Role of IL3RA in a Family with Lumbar Spinal Stenosis
Abstract
1. Introduction
2. Results
2.1. Clinical Features of the Family with Lumbar Stenosis
2.2. Filtering of the Candidate Mutations in the Family
2.3. Sequencing Validation of the Candidate Mutation
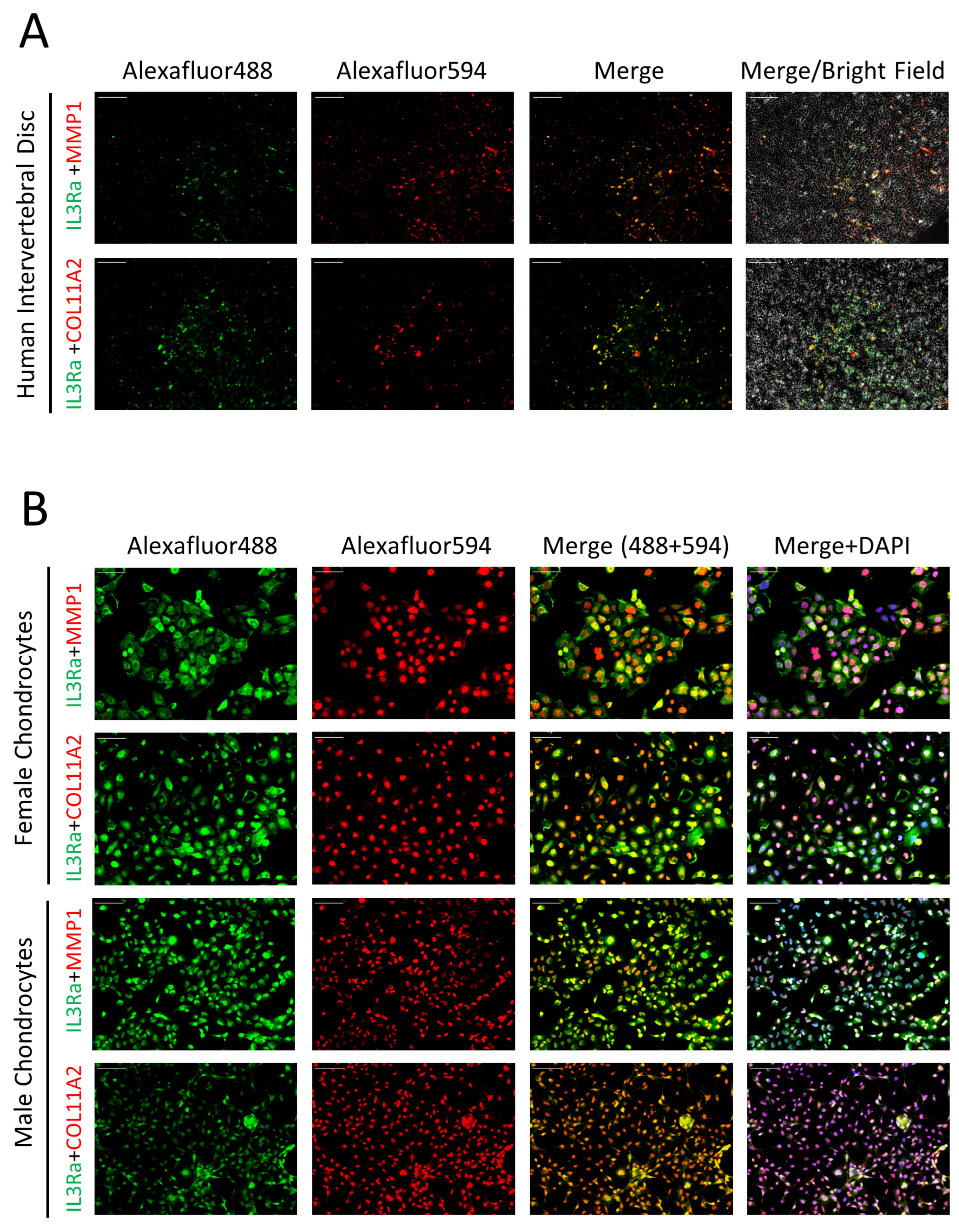
2.4. Expression of IL3RA in Human IVD and Chondrocytes
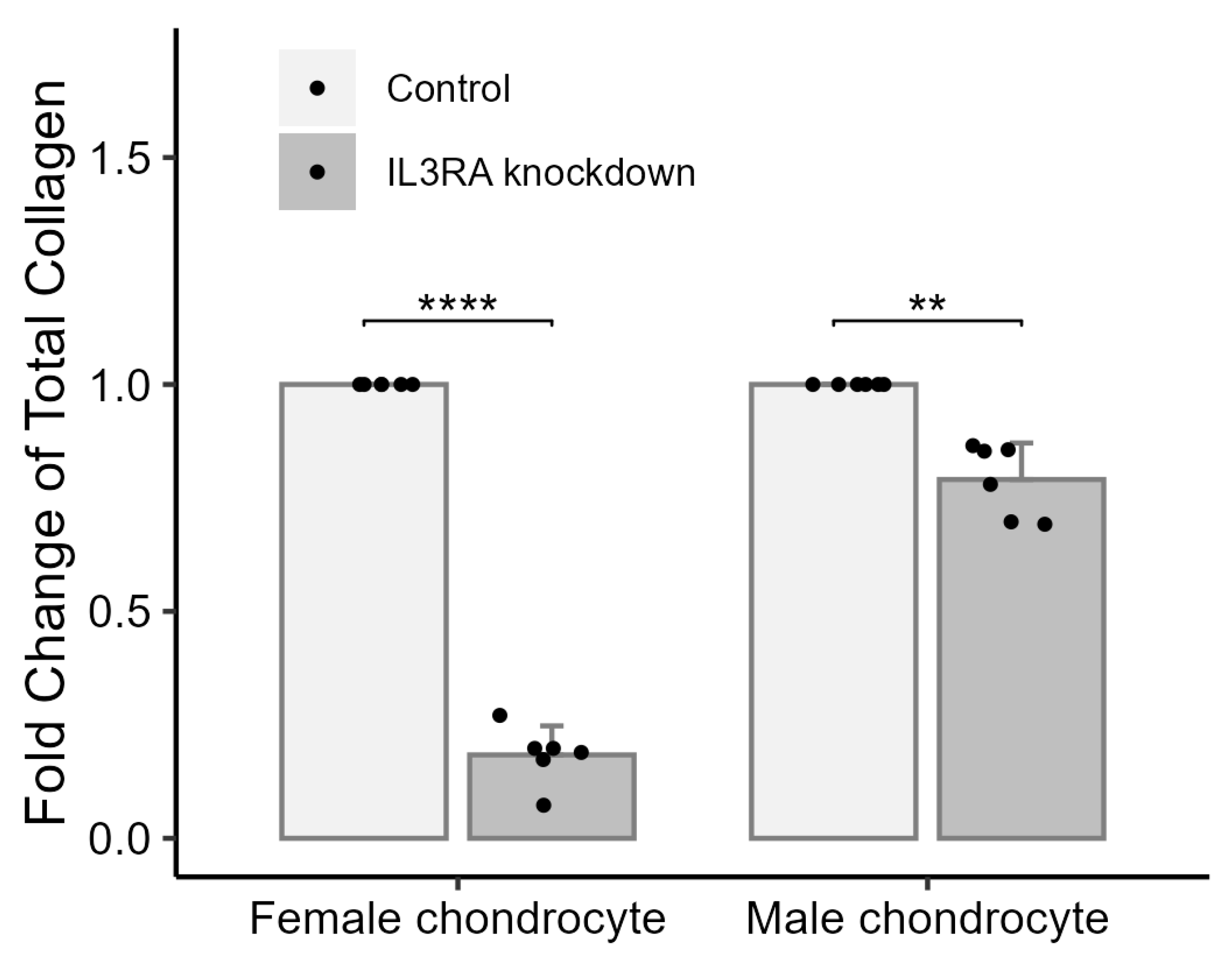
2.5. Knockdown of IL3RA Affects the Total Amount of Collagen in Chondrocytes from Human Females and Males
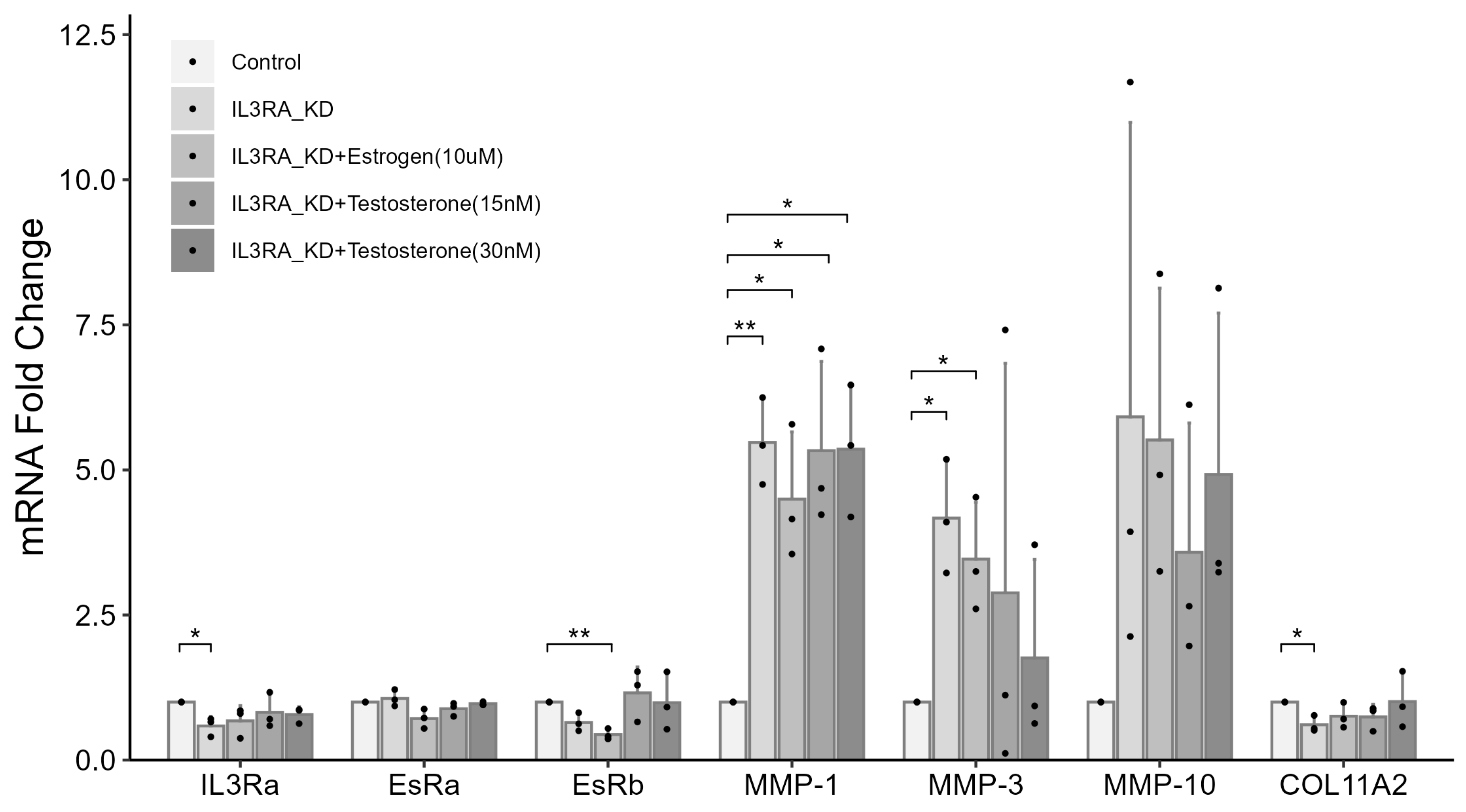
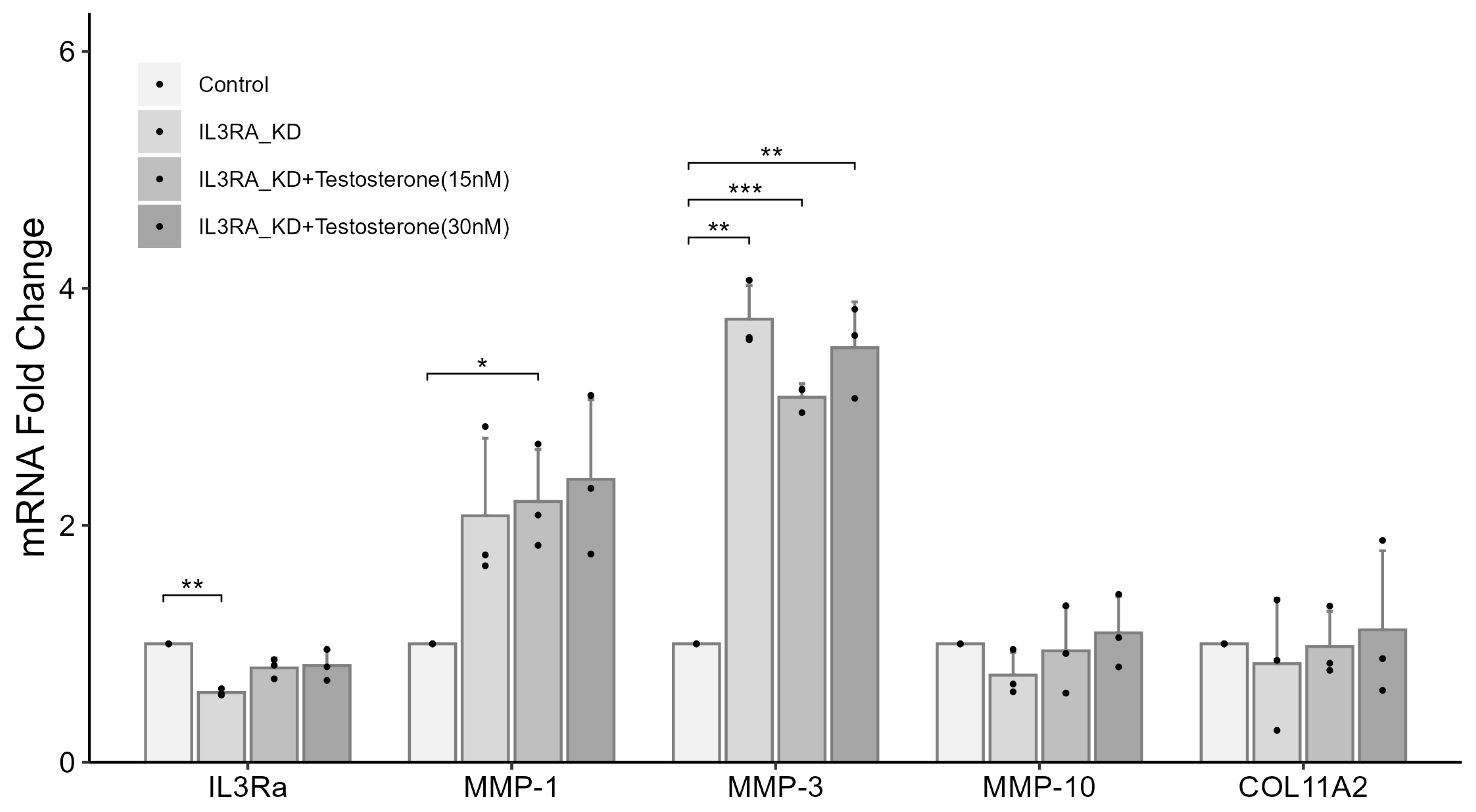
2.6. Knockdown of IL3RA Affects the mRNA Expression of MMP1, MMP3, MMP10, and COL11A2 in Human Chondrocytes
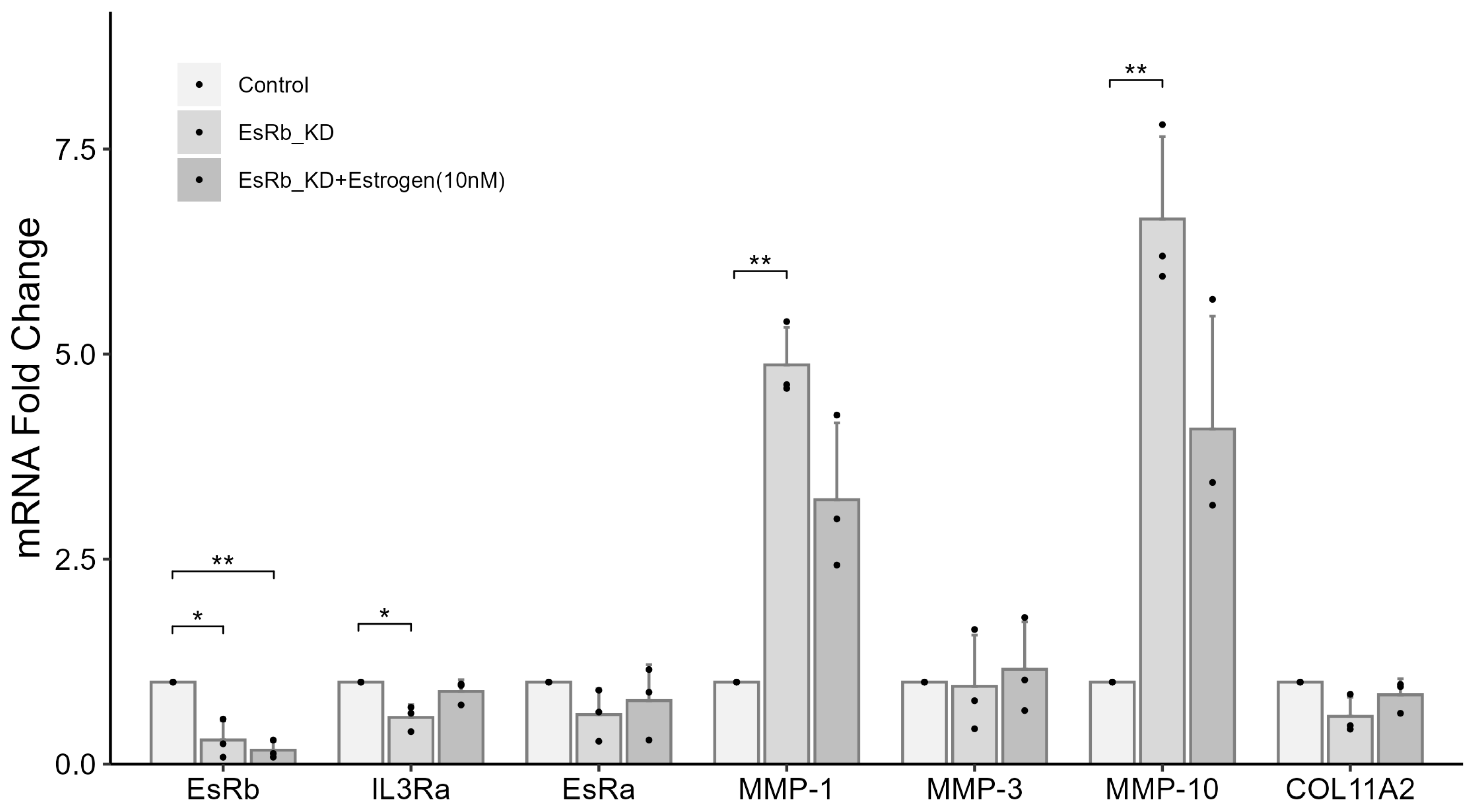
2.7. IL3RA Knockdown Reduces the Expression of EsRb and Affects the Expression of MMP-1, MMP-10, and COL11A2 in Female Chondrocytes
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Exome Sequencing
4.3. Polymerase Chain Reaction and Sanger Sequencing
4.4. Cell Culture
4.5. Immunofluorescence and Imaging
4.6. Determination of Collagen Content
4.7. siRNA and Transfection
4.8. Real-Time PCR
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwon, J.W.; Moon, S.H.; Park, S.Y.; Park, S.J.; Park, S.R.; Suk, K.S.; Kim, H.S.; Lee, B.H. Lumbar Spinal Stenosis: Review Update 2022. Asian Spine J. 2022, 16, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Siebert, E.; Pruss, H.; Klingebiel, R.; Failli, V.; Einhaupl, K.M.; Schwab, J.M. Lumbar spinal stenosis: Syndrome, diagnostics and treatment. Nat. Rev. Neurol. 2009, 5, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Konno, S.; Kikuchi, S.; Tanaka, Y.; Yamazaki, K.; Shimada, Y.; Takei, H.; Yokoyama, T.; Okada, M.; Kokubun, S. A diagnostic support tool for lumbar spinal stenosis: A self-administered, self-reported history questionnaire. BMC Musculoskelet. Disord. 2007, 8, 102. [Google Scholar] [CrossRef]
- Jensen, R.K.; Jensen, T.S.; Koes, B.; Hartvigsen, J. Prevalence of lumbar spinal stenosis in general and clinical populations: A systematic review and meta-analysis. Eur. Spine J. 2020, 29, 2143–2163. [Google Scholar] [CrossRef]
- Gagne, A.R.; Hasson, S.M. Lumbar extension exercises in conjunction with mechanical traction for the management of a patient with a lumbar herniated disc. Physiother. Theory Pract. 2010, 26, 256–266. [Google Scholar] [CrossRef]
- Hennemann, S.; de Abreu, M.R. Degenerative Lumbar Spinal Stenosis. Rev. Bras. Ortop. 2021, 56, 9–17. [Google Scholar]
- Browner, M.F.; Smith, W.W.; Castelhano, A.L. Matrilysin-inhibitor complexes: Common themes among metalloproteases. Biochemistry 1995, 34, 6602–6610. [Google Scholar] [CrossRef]
- Takahashi, M.; Haro, H.; Wakabayashi, Y.; Kawa-uchi, T.; Komori, H.; Shinomiya, K. The association of degeneration of the intervertebral disc with 5a/6a polymorphism in the promoter of the human matrix metalloproteinase-3 gene. J. Bone Jt. Surg. Br. 2001, 83, 491–495. [Google Scholar] [CrossRef]
- Eser, B.; Eser, O.; Yuksel, Y.; Aksit, H.; Karavelioglu, E.; Tosun, M.; Sekerci, Z. Effects of MMP-1 and MMP-3 gene polymorphisms on gene expression and protein level in lumbar disc herniation. Genet. Mol. Res. 2016, 15, gmr.15038669. [Google Scholar] [CrossRef]
- Goupille, P.; Jayson, M.I.; Valat, J.P.; Freemont, A.J. Matrix metalloproteinases: The clue to intervertebral disc degeneration? Spine 1998, 23, 1612–1626. [Google Scholar] [CrossRef]
- Roberts, S.; Caterson, B.; Menage, J.; Evans, E.H.; Jaffray, D.C.; Eisenstein, S.M. Matrix metalloproteinases and aggrecanase: Their role in disorders of the human intervertebral disc. Spine 2000, 25, 3005–3013. [Google Scholar] [CrossRef] [PubMed]
- Vo, N.V.; Hartman, R.A.; Yurube, T.; Jacobs, L.J.; Sowa, G.A.; Kang, J.D. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 2013, 13, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Sahlman, J.; Inkinen, R.; Hirvonen, T.; Lammi, M.J.; Lammi, P.E.; Nieminen, J.; Lapvetelainen, T.; Prockop, D.J.; Arita, M.; Li, S.W.; et al. Premature vertebral endplate ossification and mild disc degeneration in mice after inactivation of one allele belonging to the Col2a1 gene for Type II collagen. Spine 2001, 26, 2558–2565. [Google Scholar] [CrossRef] [PubMed]
- Sarver, J.J.; Elliott, D.M. Altered disc mechanics in mice genetically engineered for reduced type I collagen. Spine 2004, 29, 1094–1098. [Google Scholar] [CrossRef]
- Boyd, L.M.; Richardson, W.J.; Allen, K.D.; Flahiff, C.; Jing, L.; Li, Y.; Chen, J.; Setton, L.A. Early-onset degeneration of the intervertebral disc and vertebral end plate in mice deficient in type IX collagen. Arthritis Rheumatol. 2008, 58, 164–171. [Google Scholar] [CrossRef]
- Kibble, M.J.; Domingos, M.; Hoyland, J.A.; Richardson, S.M. Importance of Matrix Cues on Intervertebral Disc Development, Degeneration, and Regeneration. Int. J. Mol. Sci. 2022, 23, 6915. [Google Scholar] [CrossRef]
- Cui, Y.; Yu, J.; Urban, J.P.; Young, D.A. Differential gene expression profiling of metalloproteinases and their inhibitors: A comparison between bovine intervertebral disc nucleus pulposus cells and articular chondrocytes. Spine 2010, 35, 1101–1108. [Google Scholar] [CrossRef]
- Wang, G.; Huang, K.; Dong, Y.; Chen, S.; Zhang, J.; Wang, J.; Xie, Z.; Lin, X.; Fang, X.; Fan, S. Lycorine Suppresses Endplate-Chondrocyte Degeneration and Prevents Intervertebral Disc Degeneration by Inhibiting NF-κB Signalling Pathway. Cell. Physiol. Biochem. 2018, 45, 1252–1269. [Google Scholar] [CrossRef]
- Sakai, D.; Andersson, G.B. Stem cell therapy for intervertebral disc regeneration: Obstacles and solutions. Nat. Rev. Rheumatol. 2015, 11, 243–256. [Google Scholar] [CrossRef]
- Sambrook, P.N.; MacGregor, A.J.; Spector, T.D. Genetic influences on cervical and lumbar disc degeneration: A magnetic resonance imaging study in twins. Arthritis Rheumatol. 1999, 42, 366–372. [Google Scholar] [CrossRef]
- Battie, M.C.; Videman, T.; Gibbons, L.E.; Fisher, L.D.; Manninen, H.; Gill, K. 1995 Volvo Award in clinical sciences. Determinants of lumbar disc degeneration. A study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine 1995, 20, 2601–2612. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y. Genetic background of degenerative disc disease in the lumbar spine. Spine Surg. Relat. Res. 2018, 2, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Yang, Q.; Jiang, J.; Zhan, X.; Xiao, Z. Association between COL11A1 (rs1337185) and ADAMTS5 (rs162509) gene polymorphisms and lumbar spine pathologies in Chinese Han population: An observational study. BMJ Open 2017, 7, e015644. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Ishidou, Y.; Koga, H.; Taketomi, E.; Ikari, K.; Komiya, S.; Takeda, J.; Sakou, T.; Inoue, I. Functional impact of human collagen alpha2(XI) gene polymorphism in pathogenesis of ossification of the posterior longitudinal ligament of the spine. J. Bone Miner. Res. 2001, 16, 948–957. [Google Scholar] [CrossRef] [PubMed]
- Annunen, S.; Paassilta, P.; Lohiniva, J.; Perälä, M.; Pihlajamaa, T.; Karppinen, J.; Tervonen, O.; Kröger, H.; Lähde, S.; Vanharanta, H.; et al. An allele of COL9A2 associated with intervertebral disc disease. Science 1999, 285, 409–412. [Google Scholar] [CrossRef]
- Paassilta, P.; Lohiniva, J.; Göring, H.H.; Perälä, M.; Räinä, S.S.; Karppinen, J.; Hakala, M.; Palm, T.; Kröger, H.; Kaitila, I.; et al. Identification of a novel common genetic risk factor for lumbar disk disease. JAMA 2001, 285, 1843–1849. [Google Scholar] [CrossRef] [PubMed]
- Postacchini, F.; Massobrio, M.; Ferro, L. Familial lumbar stenosis. Case report of three siblings. J. Bone Jt. Surg. Am. 1985, 67, 321–323. [Google Scholar] [CrossRef]
- Helena Mangs, A.; Morris, B.J. The Human Pseudoautosomal Region (PAR): Origin, Function and Future. Curr. Genom. 2007, 8, 129–136. [Google Scholar] [CrossRef]
- Kour, S.; Garimella, M.G.; Shiroor, D.A.; Mhaske, S.T.; Joshi, S.R.; Singh, K.; Pal, S.; Mittal, M.; Krishnan, H.B.; Chattopadhyay, N.; et al. IL-3 Decreases Cartilage Degeneration by Downregulating Matrix Metalloproteinases and Reduces Joint Destruction in Osteoarthritic Mice. J. Immunol. 2016, 196, 5024–5035. [Google Scholar] [CrossRef]
- Dowdell, J.; Erwin, M.; Choma, T.; Vaccaro, A.; Iatridis, J.; Cho, S.K. Intervertebral Disk Degeneration and Repair. Neurosurgery 2017, 80 (Suppl. S3), S46–S54. [Google Scholar] [CrossRef]
- Jehan, F.; Zarka, M.; de la Houssaye, G.; Veziers, J.; Ostertag, A.; Cohen-Solal, M.; Geoffroy, V. New insights into the role of matrix metalloproteinase 3 (MMP3) in bone. FASEB Bioadv. 2022, 4, 524–538. [Google Scholar] [CrossRef] [PubMed]
- Roman-Blas, J.A.; Castañeda, S.; Largo, R.; Herrero-Beaumont, G. Osteoarthritis associated with estrogen deficiency. Arthritis Res. Ther. 2009, 11, 241. [Google Scholar] [CrossRef] [PubMed]
- Martín-Millán, M.; Castañeda, S. Estrogens, osteoarthritis and inflammation. Jt. Bone Spine 2013, 80, 368–373. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, E.B.; Kwon, Y.E.; Lee, J.J.; Cho, W.S.; Kim, H.A.; Song, Y.W. Effect of estrogen on the expression of matrix metalloproteinase (MMP)-1, MMP-3, and MMP-13 and tissue inhibitor of metalloproternase-1 in osteoarthritis chondrocytes. Rheumatol. Int. 2003, 23, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Burrage, P.S.; Mix, K.S.; Brinckerhoff, C.E. Matrix metalloproteinases: Role in arthritis. Front. Biosci. 2006, 11, 529–543. [Google Scholar] [CrossRef]
- Mei, Y.; Williams, J.S.; Webb, E.K.; Shea, A.K.; MacDonald, M.J.; Al-Khazraji, B.K. Roles of Hormone Replacement Therapy and Menopause on Osteoarthritis and Cardiovascular Disease Outcomes: A Narrative Review. Front. Rehabil. Sci. 2022, 3, 825147. [Google Scholar] [CrossRef]
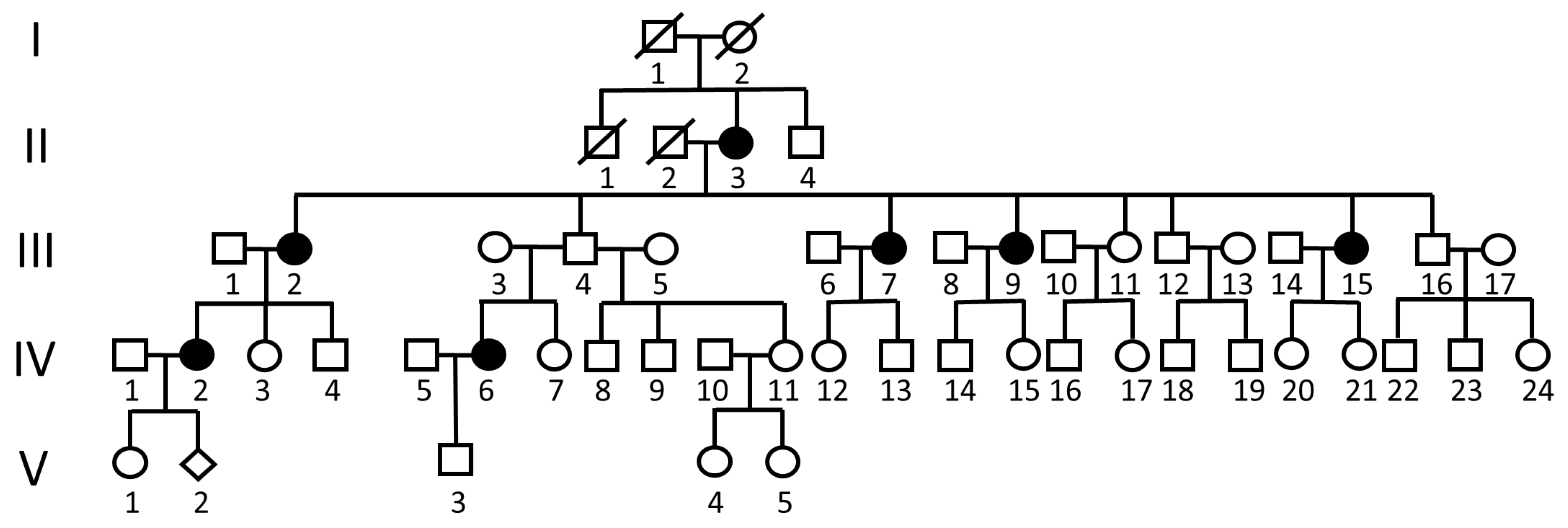
| II-3 | III-2 | III-7 | III-9 | III-15 | IV-2 | IV-6 | |
|---|---|---|---|---|---|---|---|
| Sex | Female | Female | Female | Female | Female | Female | Female |
| Birth year | 1926 | 1946 | 1951 | 1954 | 1964 | 1977 | 1972 |
| Clinical symptoms and management | Low back pain after prolonged standing or walking | Low back pain since her 20s; numbness in the right leg; limp while walking; prone to falls; spine surgery at 56 and 63 years of age | Low back pain with muscle weakness since her 20s; persistent pain even after surgery; sciatica | Occasional back pain and soreness began at 37 years of age; soreness and numbness in lower limb; surgery for herniated intervertebral disk at 47 and 48 years of age | Occasional low back pain since her 20s; symptoms worsened significantly after 53 years of age | Low back pain, herniated disk, and bone spurs at 28 years of age | Low back pain and leg numbness after 45 years of age |
| Diagnosis | LSS | LSS | LSS | LSS | - | - | - |
| Age at diagnosis (years) | 76 | 56 | 50 | 47 | - | - | - |
| Radiograph | Confirmed using X-ray and MRI | Confirmed using X-ray and MRI | Confirmed using X-ray and MRI | Confirmed using X-ray and MRI | MRI failed to indicate a spinal anomaly | MRI failed to indicate a spinal anomaly | MRI failed to indicate a spinal anomaly |
| IL3RA variant | c.417C > G (p.Y139X) | c.417C > G (p.Y139X) | c.417C > G (p.Y139X) | c.417C > G (p.Y139X) | c.417C > G (p.Y139X) | c.417C > G (p.Y139X) | c.417C > G (p.Y139X) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, K.-M.; Yang, C.-F.; H’ng, W.-S.; Chuang, H.-P.; Khor, E.H.X.; Tsai, P.-C.; Khosasih, V.; Lu, L.-S.; Yeh, E.-C.; Lin, W.-J.; et al. Role of IL3RA in a Family with Lumbar Spinal Stenosis. Int. J. Mol. Sci. 2024, 25, 10915. https://doi.org/10.3390/ijms252010915
Liu K-M, Yang C-F, H’ng W-S, Chuang H-P, Khor EHX, Tsai P-C, Khosasih V, Lu L-S, Yeh E-C, Lin W-J, et al. Role of IL3RA in a Family with Lumbar Spinal Stenosis. International Journal of Molecular Sciences. 2024; 25(20):10915. https://doi.org/10.3390/ijms252010915
Chicago/Turabian StyleLiu, Kai-Ming, Chi-Fan Yang, Weng-Siong H’ng, Hui-Ping Chuang, Eunice Han Xian Khor, Pei-Chun Tsai, Vivia Khosasih, Liang-Suei Lu, Erh-Chan Yeh, Wan-Jia Lin, and et al. 2024. "Role of IL3RA in a Family with Lumbar Spinal Stenosis" International Journal of Molecular Sciences 25, no. 20: 10915. https://doi.org/10.3390/ijms252010915
APA StyleLiu, K.-M., Yang, C.-F., H’ng, W.-S., Chuang, H.-P., Khor, E. H. X., Tsai, P.-C., Khosasih, V., Lu, L.-S., Yeh, E.-C., Lin, W.-J., Hsieh, F.-J., Chen, C.-H., Hwang, S.-L., & Wu, J.-Y. (2024). Role of IL3RA in a Family with Lumbar Spinal Stenosis. International Journal of Molecular Sciences, 25(20), 10915. https://doi.org/10.3390/ijms252010915






