Effects of Aminomethylphosphonic Acid on the Transcriptome and Metabolome of Red Swamp Crayfish, Procambarus clarkii
Abstract
1. Introduction
2. Results
2.1. Identification of AMPA Dosage-Related DEGs in P. clarkii
2.2. Functional Enrichment Analysis of AMPA Doses Related DEGs
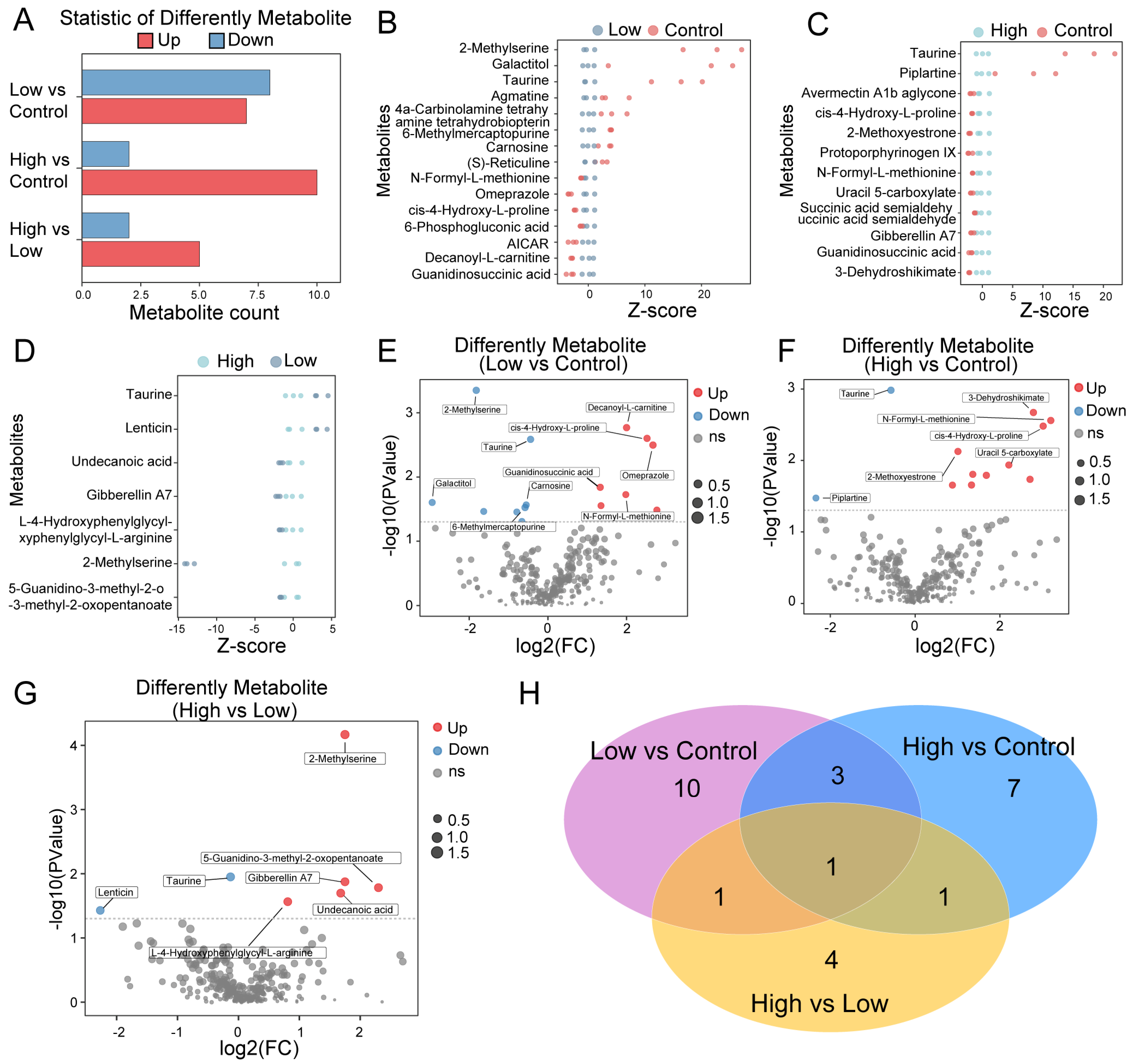
2.3. Identifying AMPA Dosage-Related Metabolites in P. clarkii
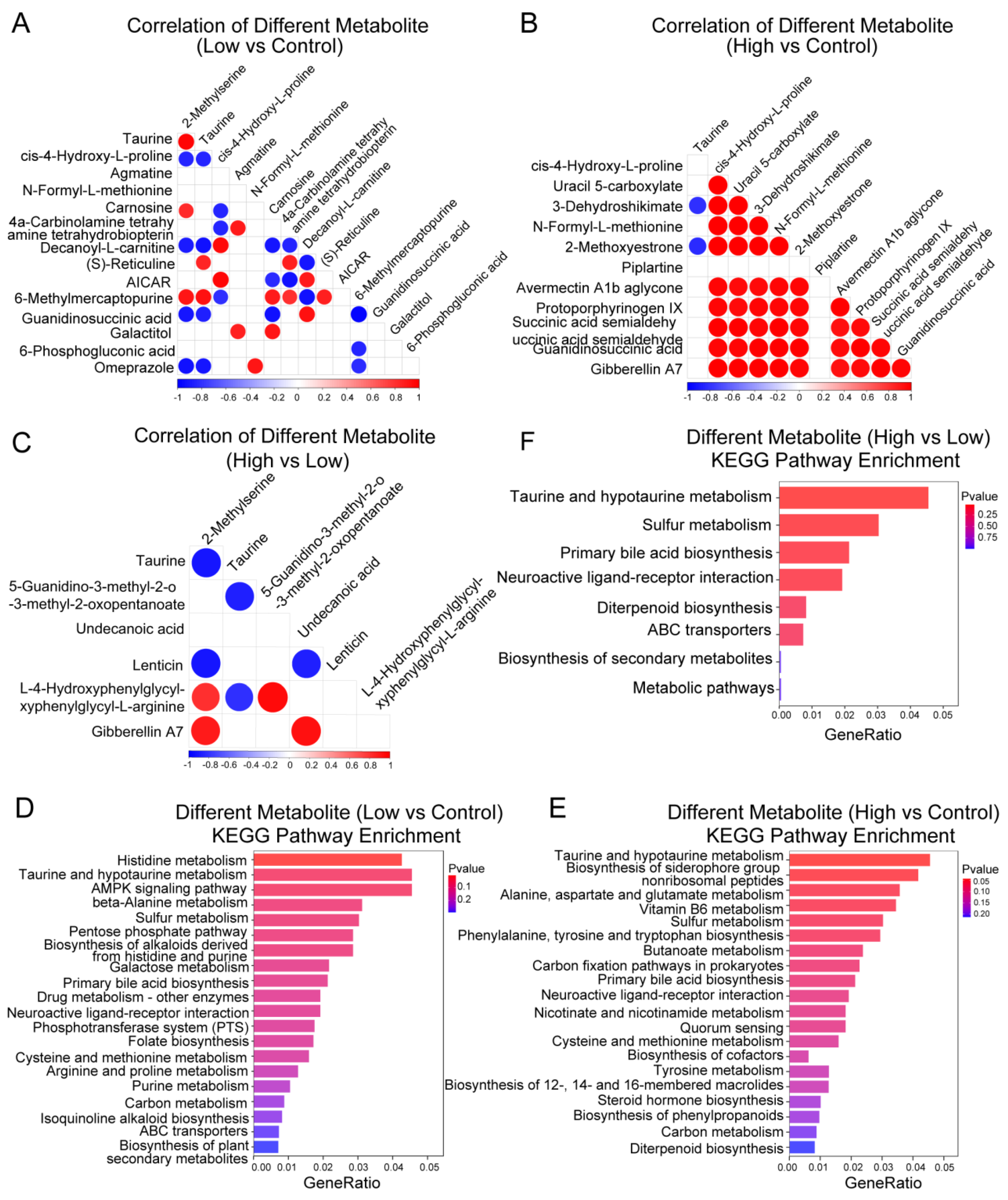
2.4. Functional Enrichment Analysis of AMPA Dosage-Related Differential Metabolites
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. P. clarkii Exposure to AMPA
4.3. RNA Extraction, Transcriptome Library Construction and Sequencing
4.4. Chemicals and Reagents
4.5. Sample Preparation and Standards
4.6. Differential Expression Genes and Differential Metabolite Abundance in Three Dose Groups of AMPA
4.7. Real-Time Quantitative PCR Quantify Differential Expression Genes
4.8. GO Term Annotation and KEGG Pathway Enrichment
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lopes, A.R.; Moraes, J.S.; Martins, C.d.M.G. Effects of the herbicide glyphosate on fish from embryos to adults: A review addressing behavior patterns and mechanisms behind them. Aquat. Toxicol. 2022, 251, 106281. [Google Scholar] [CrossRef] [PubMed]
- Lares, B.A.; Vignatti, A.M.; Echaniz, S.A.; Gutiérrez, M.F. Effects of glyphosate on cladocera: A synthetic review. Aquat. Toxicol. 2022, 249, 106232. [Google Scholar] [CrossRef] [PubMed]
- Rawat, D.; Bains, A.; Chawla, P.; Kaushik, R.; Yadav, R.; Kumar, A.; Sridhar, K.; Sharma, M. Hazardous impacts of glyphosate on human and environment health: Occurrence and detection in food. Chemosphere 2023, 329, 138676. [Google Scholar] [CrossRef] [PubMed]
- Masotti, F.; Garavaglia, B.S.; Gottig, N.; Ottado, J. Bioremediation of the herbicide glyphosate in polluted soils by plant-associated microbes. Curr. Opin. Microbiol. 2023, 73, 102290. [Google Scholar] [CrossRef] [PubMed]
- Grandcoin, A.; Piel, S.; Baurès, E. AminoMethylPhosphonic acid (AMPA) in natural waters: Its sources, behavior and environmental fate. Water Res. 2017, 117, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Poiger, T.; Buerge, I.J.; Bächli, A.; Müller, M.D.; Balmer, M.E. Occurrence of the herbicide glyphosate and its metabolite AMPA in surface waters in Switzerland determined with on-line solid phase extraction LC-MS/MS. Environ. Sci. Pollut. Res. Int. 2017, 24, 1588–1596. [Google Scholar] [CrossRef] [PubMed]
- Desmet, N.; Touchant, K.; Seuntjens, P.; Tang, T.; Bronders, J. A hybrid monitoring and modelling approach to assess the contribution of sources of glyphosate and AMPA in large river catchments. Sci. Total Environ. 2016, 573, 1580–1588. [Google Scholar] [CrossRef]
- Coupe, R.H.; Kalkhoff, S.J.; Capel, P.D.; Gregoire, C. Fate and transport of glyphosate and aminomethylphosphonic acid in surface waters of agricultural basins. Pest Manag. Sci. 2012, 68, 16–30. [Google Scholar] [CrossRef]
- Mendoza-Vargas, L.; Guarneros-Bañuelos, E.; Báez-Saldaña, A.; Galicia-Mendoza, F.; Flores-Soto, E.; Fuentes-Pardo, B.; Alvarado, R.; Valdés-Tovar, M.; Sommer, B.; Benítez-King, G.; et al. Involvement of Melatonin in the Regulation of the Circadian System in Crayfish. Int. J. Mol. Sci. 2018, 19, 2147. [Google Scholar] [CrossRef]
- Hou, J.; Styles, D.; Cao, Y.; Ye, X. The sustainability of rice-crayfish coculture systems: A mini review of evidence from Jianghan plain in China. J. Sci. Food Agric. 2021, 101, 3843–3853. [Google Scholar] [CrossRef]
- Marçal, R.; Pacheco, M.; Guilherme, S. DNA of crayfish spermatozoa as a target of waterborne pesticides—An ex vivo approach as a tool to short-term spermiotoxicity screening. J. Hazard. Mater. 2020, 400, 123300. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Lei, L.; Chen, X.; Men, J.; Sun, Y.; Guo, Y.; Yang, L.; Wang, Q.; Han, J.; Zhou, B. Glyphosate and glufosinate-ammonium in aquaculture ponds and aquatic products: Occurrence and health risk assessment. Environ. Pollut. 2022, 296, 118742. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L.; Zhou, F.; Li, J.; Wu, X.; Zhong, X.; Lv, H.; Yi, S.; Gao, Q.; Yang, Z.; et al. Integrated comparative transcriptome and weighted gene co-expression network analysis provide valuable insights into the response mechanisms of crayfish (P. clarkii) to copper stress. J. Hazard. Mater. 2023, 448, 130820. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Gao, T.; Li, X.; Shi, L.; Chen, S.; Ding, A.; Li, X.; Qiao, Y.; Liao, L.; Xiong, G.; et al. Integrating transcriptomic and metabolomic analysis to understand muscle qualities of red swamp crayfish (P. clarkii) under transport stress. Food Res. Int. 2023, 164, 112361. [Google Scholar] [CrossRef] [PubMed]
- Ou-Yang, K.; Feng, T.; Han, Y.; Li, J.; Ma, H. Cyhalofop-butyl and pyribenzoxim-induced oxidative stress and transcriptome changes in the muscle of crayfish (P. clarkii). Sci. Total Environ. 2023, 864, 161170. [Google Scholar] [CrossRef]
- Nian, Y.-Y.; Chen, B.-K.; Wang, J.-J.; Zhong, W.-T.; Fang, Y.; Li, Z.; Zhang, Q.-S.; Yan, D.-C. Transcriptome analysis of P. clarkii infected with infectious hypodermal and haematopoietic necrosis virus. Fish Shellfish Immunol. 2020, 98, 766–772. [Google Scholar] [CrossRef]
- Rodríguez-Moro, G.; Román-Hidalgo, C.; Ramírez-Acosta, S.; Aranda-Merino, N.; Gómez-Ariza, J.L.; Abril, N.; Bello-López, M.A.; Fernández-Torres, R.; García-Barrera, T. Targeted and untargeted metabolomic analysis of P. clarkii exposed to a “chemical cocktail” of heavy metals and diclofenac. Chemosphere 2022, 293, 133410. [Google Scholar] [CrossRef]
- Fernández-Cisnal, R.; García-Sevillano, M.A.; García-Barrera, T.; Gómez-Ariza, J.L.; Abril, N. Metabolomic alterations and oxidative stress are associated with environmental pollution in P. clarkii. Aquat. Toxicol. 2018, 205, 76–88. [Google Scholar] [CrossRef]
- Yan, B.; Sun, Y.; Fu, K.; Zhang, Y.; Lei, L.; Men, J.; Guo, Y.; Wu, S.; Han, J.; Zhou, B. Effects of glyphosate exposure on gut-liver axis: Metabolomic and mechanistic analysis in grass carp (Ctenopharyngodon idellus). Sci. Total Environ. 2023, 902, 166062. [Google Scholar] [CrossRef]
- Sulukan, E.; Baran, A.; Kankaynar, M.; Kızıltan, T.; Bolat, İ.; Yıldırım, S.; Ceyhun, H.A.; Ceyhun, S.B. Global warming and glyphosate toxicity (II): Offspring zebrafish modelling with behavioral, morphological and immunohistochemical approaches. Sci. Total Environ. 2023, 856, 158903. [Google Scholar] [CrossRef]
- Telahigue, K.; Rabeh, I.; Mhadhbi, L.; Nechi, S.; Chelbi, E.; Ali, M.B.; Hedfi, A.; Al-Harbi, M.S.; Hajji, T. Glyphosate exposure modulates lipid composition, histo-architecture and oxidative stress status and induces neurotoxicity in the smooth scallop Flexopecten glaber. Pestic. Biochem. Physiol. 2022, 184, 105099. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chang, L.; Zhao, T.; Liu, L.; Zhang, M.; Li, C.; Xie, F.; Jiang, J.; Zhu, W. Metabolic switch in energy metabolism mediates the sublethal effects induced by glyphosate-based herbicide on tadpoles of a farmland frog Microhyla fissipes. Ecotoxicol. Environ. Saf. 2019, 186, 109794. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhu, J.; Wang, W.; Ruan, P.; Rajeshkumar, S.; Li, X. Biochemical and molecular impacts of glyphosate-based herbicide on the gills of common carp. Environ. Pollut. 2019, 252, 1288–1300. [Google Scholar] [CrossRef] [PubMed]
- Jiao, T.; Yang, T.-T.; Wang, D.; Gao, Z.-Q.; Wang, J.-L.; Tang, B.-P.; Liu, Q.-N.; Zhang, D.-Z.; Dai, L.-S. Characterization and expression analysis of immune-related genes in the red swamp crayfish, Procambarus clarkii in response to lipopolysaccharide challenge. Fish Shellfish Immunol. 2019, 95, 140–150. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.-Y.; Kang, C.-Q.; Dang, W.; Melvin, S.-D.; Lu, H.-L. Minor metabolomic disturbances induced by glyphosate-isopropylammonium exposure at environmentally relevant concentrations in an aquatic turtle, Pelodiscus sinensis. Aquat. Toxicol. 2023, 256, 106415. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Adineh, H.; Reverter, M.; Hamidi, M.K.; Vatnikov, Y.A.; Kulikov, E.V.; Hoseinifar, S.H.; Van Doan, H. Protective effects of black seed (Nigella sativa) diet supplementation in common carp (Cyprinus carpio) against immune depression, oxidative stress and metabolism dysfunction induced by glyphosate. Fish Shellfish Immunol. 2021, 109, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Lucia, R.M.; Huang, W.-L.; Pathak, K.V.; McGilvrey, M.; David-Dirgo, V.; Alvarez, A.; Goodman, D.; Masunaka, I.; Odegaard, A.O.; Ziogas, A.; et al. Association of Glyphosate Exposure with Blood DNA Methylation in a Cross-Sectional Study of Postmenopausal Women. Environ. Health Perspect. 2022, 130, 47001. [Google Scholar] [CrossRef] [PubMed]
- Velisek, J.; Stara, A.; Zuskova, E.; Kubec, J.; Buric, M.; Kouba, A. Chronic toxicity of metolachlor OA on growth, ontogenetic development, antioxidant biomarkers and histopathology of early life stages of marbled crayfish. Sci. Total Environ. 2018, 643, 1456–1463. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Harshitha, R.; Arunraj, D.R. Real-time quantitative PCR: A tool for absolute and relative quantification. Biochem. Mol. Biol. Educ. 2021, 49, 800–812. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, T.; Gan, J.; Yuan, K.; He, L.; Yu, Y.; Liu, Z.; Zhou, Y.; Wu, G. Effects of Aminomethylphosphonic Acid on the Transcriptome and Metabolome of Red Swamp Crayfish, Procambarus clarkii. Int. J. Mol. Sci. 2024, 25, 943. https://doi.org/10.3390/ijms25020943
Mao T, Gan J, Yuan K, He L, Yu Y, Liu Z, Zhou Y, Wu G. Effects of Aminomethylphosphonic Acid on the Transcriptome and Metabolome of Red Swamp Crayfish, Procambarus clarkii. International Journal of Molecular Sciences. 2024; 25(2):943. https://doi.org/10.3390/ijms25020943
Chicago/Turabian StyleMao, Tao, Jinhua Gan, Keping Yuan, Li He, Yali Yu, Ziduo Liu, Yuntao Zhou, and Gaobing Wu. 2024. "Effects of Aminomethylphosphonic Acid on the Transcriptome and Metabolome of Red Swamp Crayfish, Procambarus clarkii" International Journal of Molecular Sciences 25, no. 2: 943. https://doi.org/10.3390/ijms25020943
APA StyleMao, T., Gan, J., Yuan, K., He, L., Yu, Y., Liu, Z., Zhou, Y., & Wu, G. (2024). Effects of Aminomethylphosphonic Acid on the Transcriptome and Metabolome of Red Swamp Crayfish, Procambarus clarkii. International Journal of Molecular Sciences, 25(2), 943. https://doi.org/10.3390/ijms25020943




