Impact of miR-29c-3p in the Nucleus Accumbens on Methamphetamine-Induced Behavioral Sensitization and Neuroplasticity-Related Proteins
Abstract
1. Introduction
2. Results
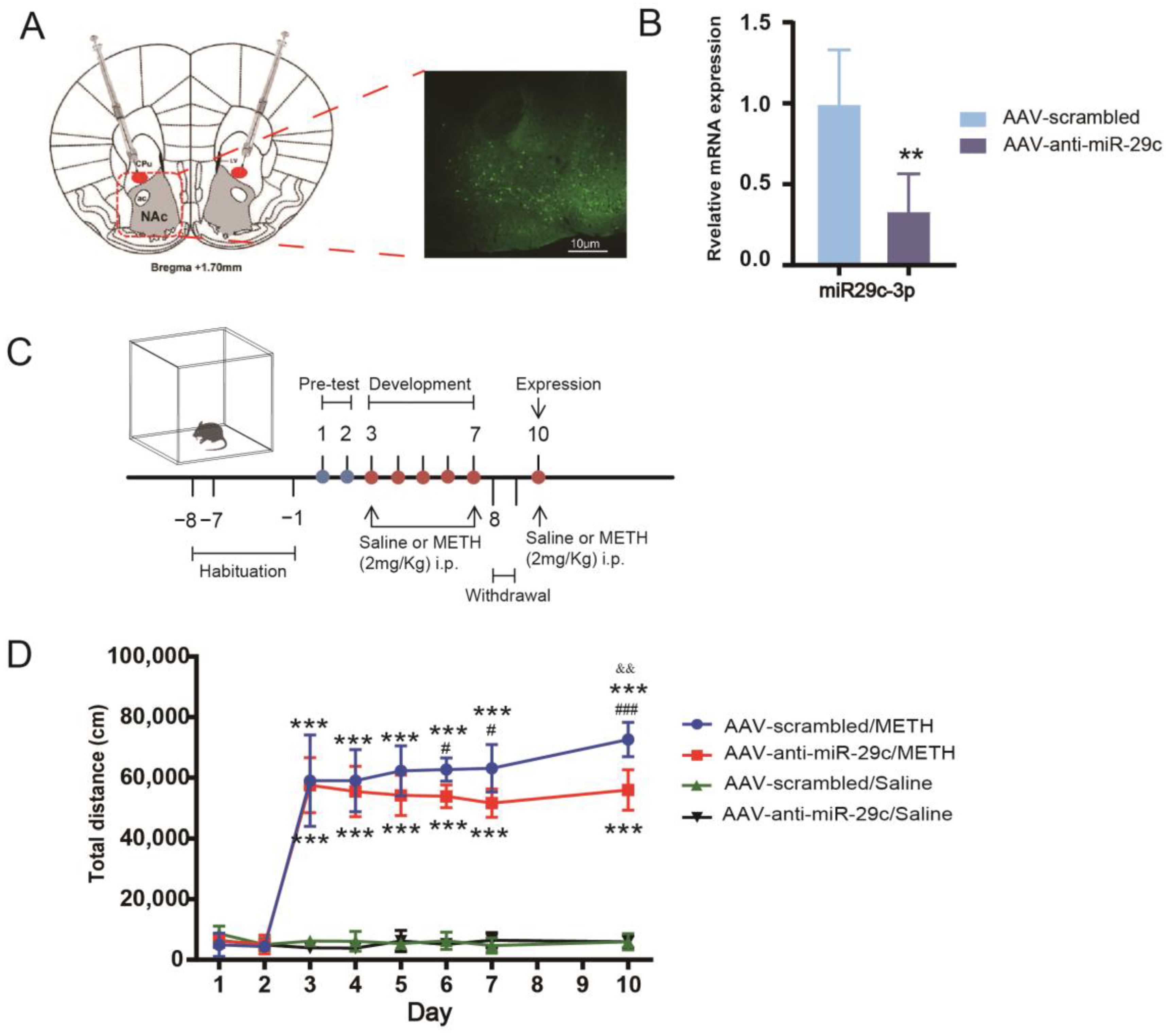
2.1. Inhibition of miR-29c-3p in the NAc Diminished the Properties of METH
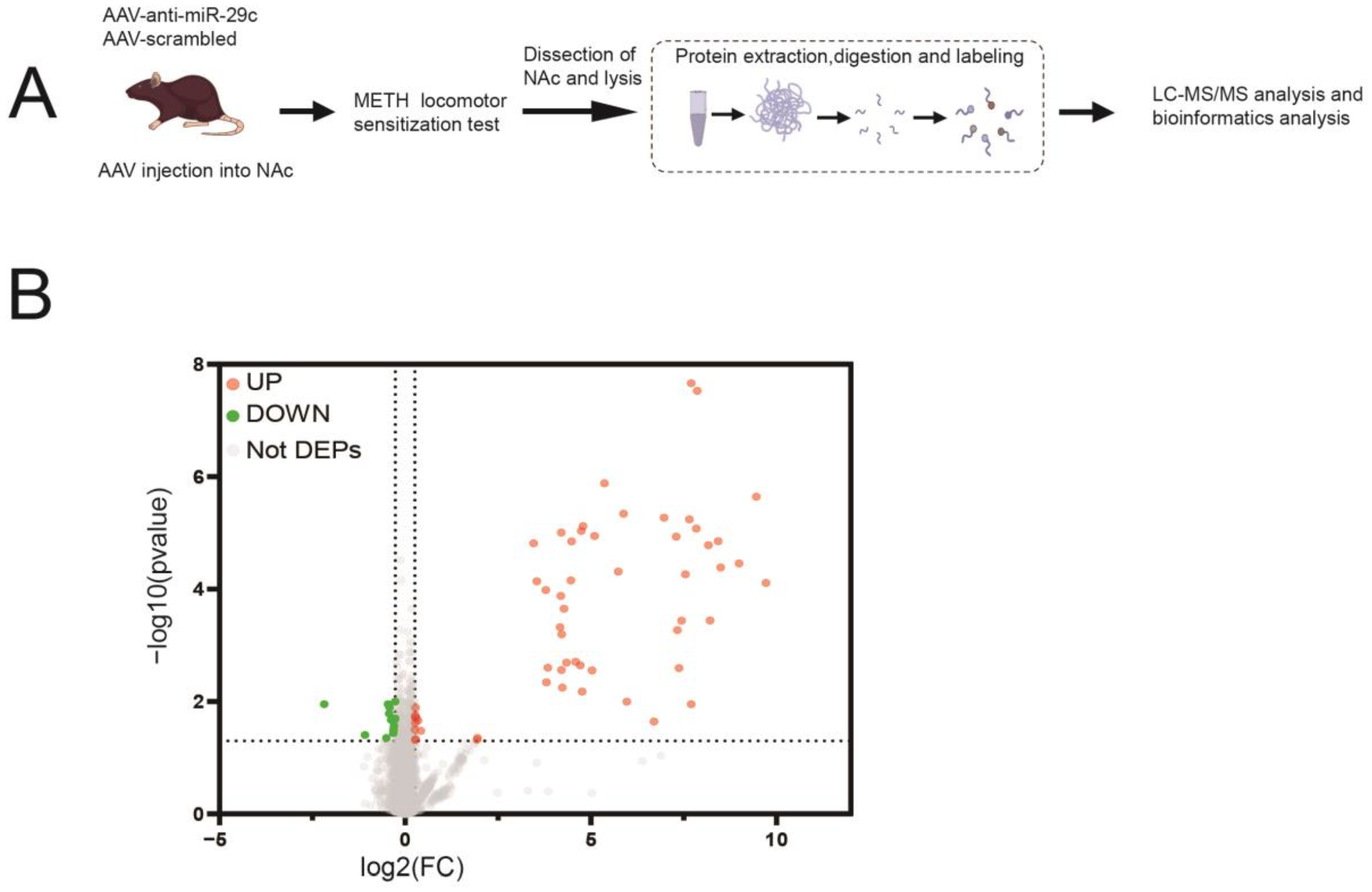
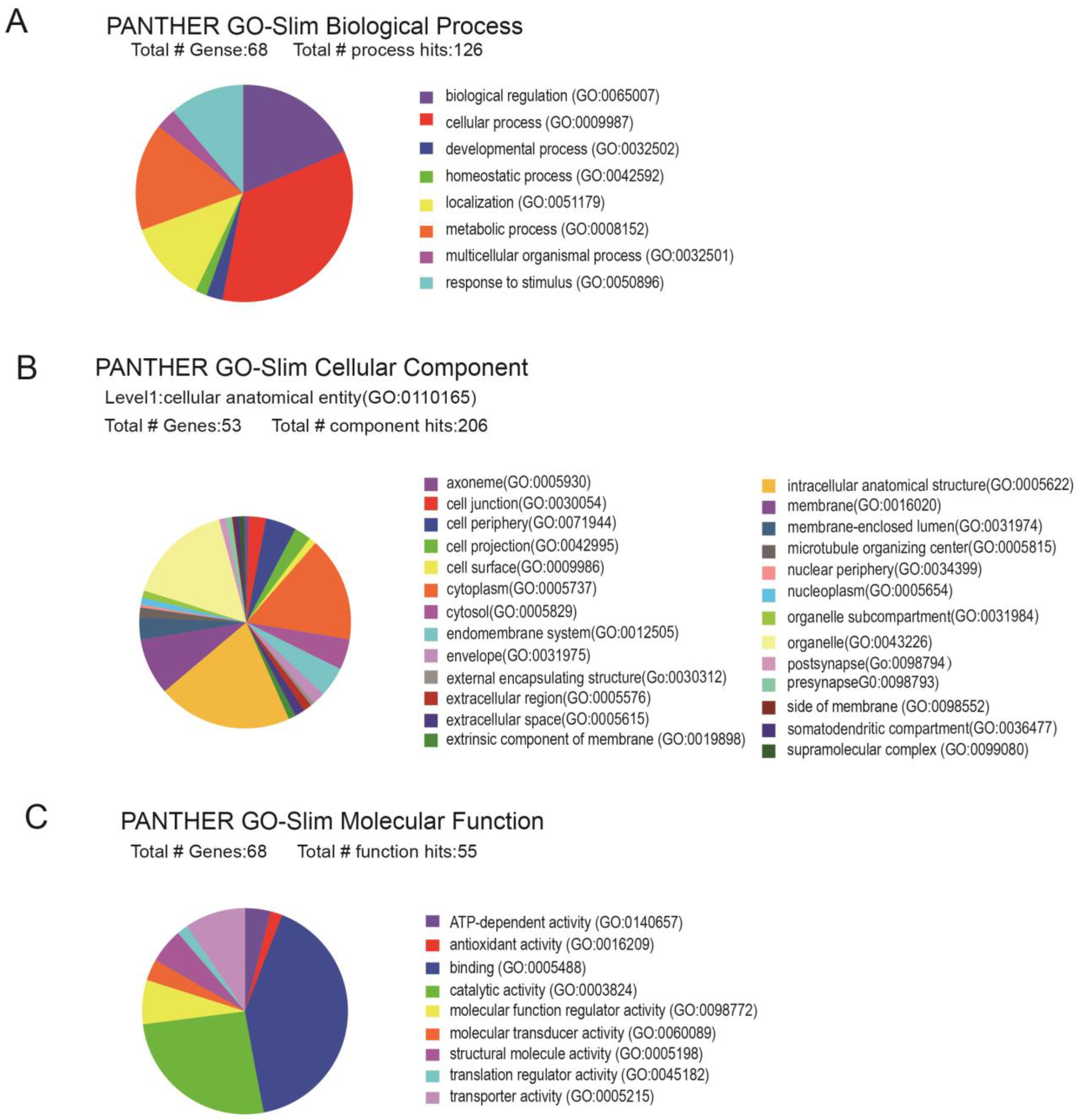
2.2. Protein Expression Profile Involved in miR-29c-3p-Dependent METH-Induced Behavior Sensitization
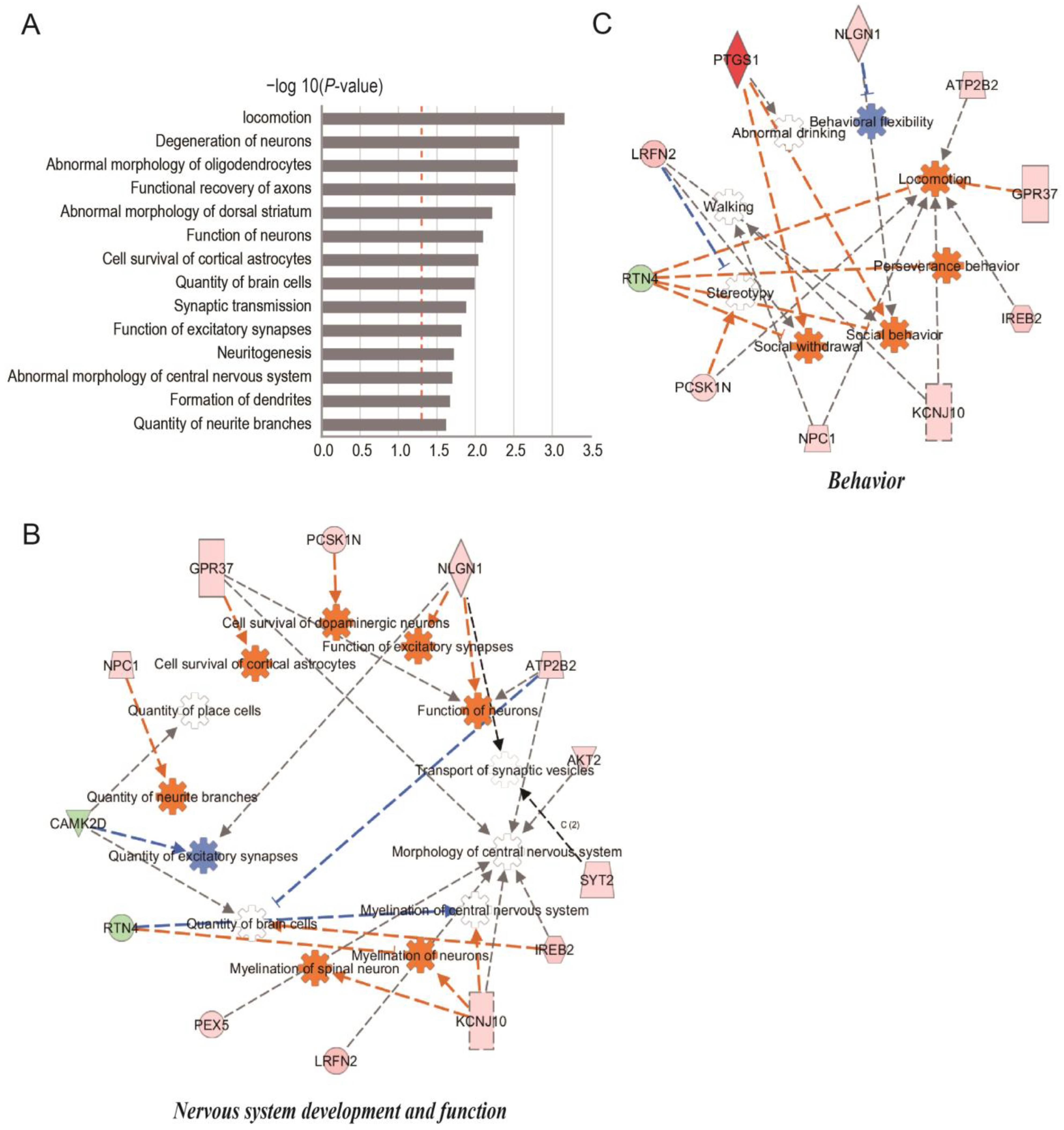
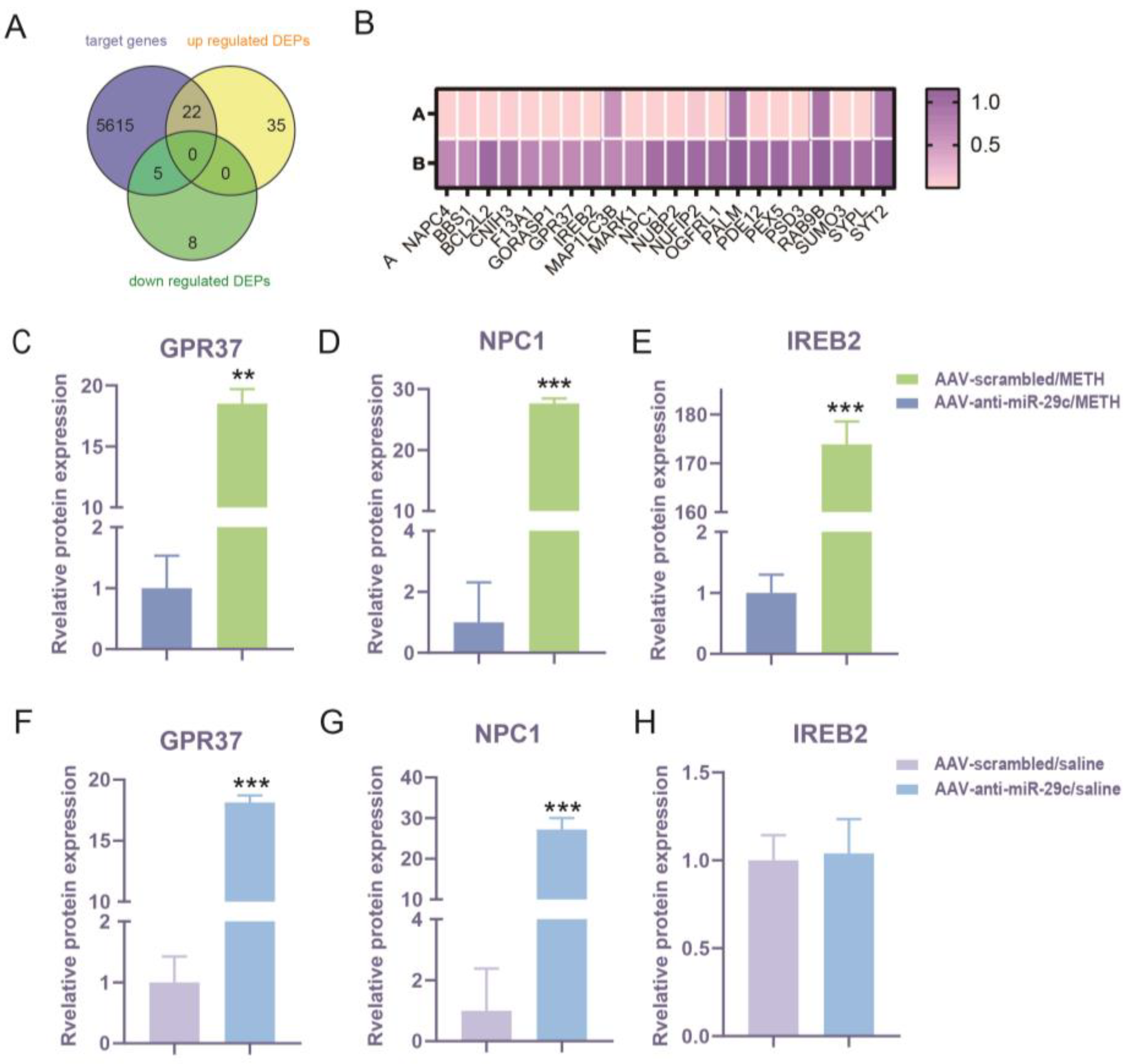
2.3. Comparison of miR-29c-3p Predicted Target Gene Interactions with DEPs
2.4. Effects of miR-29c-3p on the Expression Changes of GPR37, NPC1, and IREB2 in METH-Induced Behavioral Sensitization
2.5. Effects of METH on the Expression of Gpr37, Npc1, and Ireb2
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Drugs
4.3. Adeno-Associated Virus (AAV) eGFP Expression
4.4. Surgery with Stereotaxy
4.5. Animal Groups
4.6. Open Field Test
4.7. RNA Extraction and Reverse Transcription
4.8. Quantitative Real-Time PCR Analyses (qRT-PCR)
4.9. Protein Extraction
4.10. iTRAQ
4.10.1. Protein Labeling
4.10.2. Strong Cation Exchange (SCX) Chromatography and LC-MS/MS Analysis
4.10.3. Proteomic Analysis and Characterization of Protein Function
4.11. IPA
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Honeywell, K.M.; Doren, E.V.; Szumlinski, K.K. Selective inhibition of PDE4B reduces methamphetamine reinforcement in two c57bl/6 substrains. Int. J. Mol. Sci. 2022, 23, 4872. [Google Scholar] [CrossRef] [PubMed]
- Mullen, J.M.; Richards, J.R.; Crawford, A.T. Amphetamine Related Psychiatric Disorders; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Limanaqi, F.; Gambardella, S.; Biagioni, F.; Busceti, C.L.; Fornai, F. Epigenetic effects induced by methamphetamine and methamphetamine-dependent oxidative stress. Oxidative Med. Cell. Longev. 2018, 2018, 4982453. [Google Scholar] [CrossRef] [PubMed]
- Crime UNOO. World Drug Report 2023. [Internet]. Available online: https://www.unodc.org/unodc/en/data-and-analysis/world-drug-report-2023.html (accessed on 23 October 2023).
- Kim, Y.; Won, S.D.; Kwon, H.; Han, C. The ratio of second and fourth digit length: A biomarker for methamphetamine dependence? Clin. Psychopharmacol. Neurosci. 2022, 20, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.J.; Ren, Z.X.; Shi, Z.F.; Wan, C.L.; Han, C.J.; Zhu, L.S.; Li, N.N.; Waddington, J.L.; Zhen, X.C. Dysregulation of iron homeostasis and methamphetamine reward behaviors in Clk1-deficient mice. Acta Pharmacol Sin. 2022, 43, 1686–1698. [Google Scholar] [CrossRef] [PubMed]
- Salem, B.E.; Brecht, M.L.; Ekstrand, M.L.; Faucette, M.; Nyamathi, A.M. Correlates of physical, psychological, and social frailty among formerly incarcerated, homeless women. Health Care Women Int. 2019, 40, 788–812. [Google Scholar] [CrossRef] [PubMed]
- Darke, S.; Kaye, S.; McKetin, R.; Duflou, J. Major physical and psychological harms of methamphetamine use. Drug Alcohol Rev. 2008, 27, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Blum, K.; Cadet, J.L.; Gold, M.S. Psychostimulant use disorder emphasizing methamphetamine and the opioid-dopamine connection: Digging out of a hypodopaminergic ditch. J. Neurol. Sci. 2021, 420, 117252. [Google Scholar] [CrossRef] [PubMed]
- Christoffel, D.J.; Walsh, J.J.; Hoerbelt, P.; Heifets, B.D.; Llorach, P.; Lopez, R.C.; Ramakrishnan, C.; Deisseroth, K.; Malenka, R.C. Selective filtering of excitatory inputs to nucleus accumbens by dopamine and serotonin. Proc. Natl. Acad. Sci. USA 2021, 118, e2106648118. [Google Scholar] [CrossRef]
- Fischer, K.D.; Knackstedt, L.A.; Rosenberg, P.A. Glutamate homeostasis and dopamine signaling: Implications for psychostimulant addiction behavior. Neurochem. Int. 2021, 144, 104896. [Google Scholar] [CrossRef]
- Lewis, D.; Kenneally, M.; van DenHeuvel, C.; Byard, R.W. Methamphetamine deaths: Changing trends and diagnostic issues. Med. Sci.Law 2021, 61, 130–137. [Google Scholar] [CrossRef]
- Shi, S.; Chen, T.; Zhao, M. The crosstalk between neurons and glia in methamphetamine-induced neuroinflammation. Neurochem. Res. 2022, 47, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J. Molecular basis of long-term plasticity underlying addiction. Nat. Rev. Neurosci. 2001, 2, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Nan, J.; Lan, Y. The nucleus accumbens: A common target in the comorbidity of depression and addiction. Front. Neural Circuits 2020, 14, 37. [Google Scholar] [CrossRef]
- Buck, S.A.; Torregrossa, M.M.; Logan, R.W.; Freyberg, Z. Roles of dopamine and glutamate co-release in the nucleus accumbens in mediating the actions of drugs of abuse. FEBS J. 2021, 288, 1462–1474. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Fayette, N.; Heinsbroek, J.A.; Ford, C.P. Cocaine shifts dopamine D2 receptor sensitivity to gate conditioned behaviors. Neuron 2021, 109, 3421–3435. [Google Scholar] [CrossRef]
- Sharifi, A.; Karimi-Haghighi, S.; Shabani, R.; Asgari, H.R.; Ahadi, R.; Haghparast, A. Cannabidiol impairs the rewarding effects of methamphetamine: Involvement of dopaminergic receptors in the nucleus accumbens. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2022, 113, 110458. [Google Scholar] [CrossRef]
- Zhu, L.; Wu, F.; Yan, Z.; He, L.; Wang, S.; Hu, H.; Goh, E.; Zhu, Y.; Guan, F.; Chen, T. A novel microRNA, novel-m009c, regulates methamphetamine rewarding effects. Mol. Psychiatry 2022, 27, 3885–3897. [Google Scholar] [CrossRef]
- Li, J.; Zhu, L.; Su, H.; Liu, D.; Yan, Z.; Ni, T.; Wei, H.; Goh, E.L.K.; Chen, T. Regulation of miR-128 in the nucleus accumbens affects methamphetamine-induced behavioral sensitization by modulating proteins involved in neuroplasticity. Addict. Biol. 2021, 26, e12881. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, J.; Duan, Z.; Gong, T.; Chen, W.; Wang, S.; Xu, H. MiR-27b-3p and miR-607 cooperatively regulate BLM gene expression by directly targeting the 3’-utr in pc3 cells. Mol. Med. Rep. 2019, 19, 4819–4831. [Google Scholar] [CrossRef]
- Song, M. miRNAs-dependent regulation of synapse formation and function. Genes Genom. 2020, 42, 837–845. [Google Scholar] [CrossRef]
- Dong, X.; Cong, S. The emerging role of microRNAs in polyglutamine diseases. Front. Mol. Neurosci. 2019, 12, 156. [Google Scholar] [CrossRef] [PubMed]
- Rashidi, S.K.; Kalirad, A.; Rafie, S.; Behzad, E.; Dezfouli, M.A. The role of microRNAs in neurobiology and pathophysiology of the hippocampus. Front. Mol. Neurosci. 2023, 16, 1226413. [Google Scholar] [CrossRef] [PubMed]
- Sanacora, G.; Yan, Z.; Popoli, M. The stressed synapse 2.0: Pathophysiological mechanisms in stress-related neuropsychiatric disorders. Nat. Rev. Neurosci. 2022, 23, 86–103. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, M.L.; Prochnik, A.; Wald, M.R.; Genaro, A.M. Chronic stress and glucocorticoid receptor resistance in asthma. Clin. Ther. 2020, 42, 993–1006. [Google Scholar] [CrossRef]
- Allen, L.; Dwivedi, Y. MicroRNA mediators of early life stress vulnerability to depression and suicidal behavior. Mol. Psychiatry 2020, 25, 308–320. [Google Scholar] [CrossRef]
- Chen, Y.; Yao, S.Y.; Shu, X.; Wang, Y.J.; Liu, J.G. Changes in mRNA and miRNA expression in the prelimbic cortex related to depression-like syndrome induced by chronic social defeat stress in mice. Behav. Brain Res. 2023, 438, 114211. [Google Scholar] [CrossRef]
- Su, H.; Zhu, L.; Li, J.; Wang, R.; Liu, D.; Han, W.; Cadet, J.L.; Chen, T. Regulation of microRNA-29c in the nucleus accumbens modulates methamphetamine-induced locomotor sensitization in mice. Neuropharmacology 2019, 148, 160–168. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. Panther version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive api. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Kehl, T.; Kern, F.; Backes, C.; Fehlmann, T.; Stöckel, D.; Meese, E.; Lenhof, H.P.; Keller, A. Mirpathdb 2.0: A novel release of the mirna pathway dictionary database. Nucleic Acids Res. 2020, 48, D142–D147. [Google Scholar] [CrossRef]
- Paraskevopoulou, M.D.; Georgakilas, G.; Kostoulas, N.; Vlachos, I.S.; Vergoulis, T.; Reczko, M.; Filippidis, C.; Dalamagas, T.; Hatzigeorgiou, A.G. Diana-microt web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013, 41, W169–W173. [Google Scholar] [CrossRef]
- Xu, Z.J.; Zhang, P.Y.; Li, Z.Q.; Zhu, H.P.; Tan, W.L.; Ren, P.H. LncRNA ac125982.2 regulates apoptosis of cardiomyocytes through mir-450b-3p/atg4b axis in a rat model with myocardial infarction. Heliyon 2023, 9, e22467. [Google Scholar] [CrossRef] [PubMed]
- Segaran, R.C.; Chan, L.Y.; Wang, H.; Sethi, G.; Tang, F.R. Neuronal development-related miRNAs as biomarkers for Alzheimer’s disease, depression, schizophrenia and ionizing radiation exposure. Curr. Med. Chem. 2021, 28, 19–52. [Google Scholar] [CrossRef]
- Zong, Y.; Yu, P.; Cheng, H.; Wang, H.; Wang, X.; Liang, C.; Zhu, H.; Qin, Y.; Qin, C. MiR-29c regulates nav3 protein expression in a transgenic mouse model of Alzheimer’s disease. Brain Res. 2015, 1624, 95–102. [Google Scholar] [CrossRef]
- Rial, D.; Morató, X.; Real, J.I.; Gonçalves, F.Q.; Stagljar, I.; Pereira, F.C.; Fernández-Dueñas, V.; Cunha, R.A.; Ciruela, F. Parkinson’s disease-associated GPR37 receptor regulates cocaine-mediated synaptic depression in corticostriatal synapses. Neurosci. Lett. 2017, 638, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Kim, E. Alterations in striatal microRNA-mRNA networks contribute to neuroinflammation in multiple system atrophy. Mol. Neurobiol. 2019, 56, 7003–7021. [Google Scholar] [CrossRef] [PubMed]
- Kane, C.J.M.; Douglas, J.C.; Rafferty, T.; Johnson, J.W.; Niedzwiedz Massey, V.M.; Phelan, K.D.; Majewska, A.K.; Drew, P.D. Ethanol modulation of cerebellar neuroinflammation in a postnatal mouse model of fetal alcohol spectrum disorders. J. Neurosci. Res. 2021, 99, 1986–2007. [Google Scholar] [CrossRef]
- Marazziti, D.; Di Pietro, C.; Mandillo, S.; Golini, E.; Matteoni, R.; Tocchini Valentini, G.P. Absence of the GPR37/PAEL receptor impairs striatal AKT and ERK2 phosphorylation, δfosb expression, and conditioned place preference to amphetamine and cocaine. FASEB J. 2011, 25, 2071–2081. [Google Scholar] [CrossRef]
- Marazziti, D.; Golini, E.; Mandillo, S.; Magrelli, A.; Witke, W.; Matteoni, R.; Tocchini-Valentini, G.P. Altered dopamine signaling and MPTP resistance in mice lacking the Parkinson’s disease-associated GPR37/parkin-associated endothelin-like receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 10189–10194. [Google Scholar] [CrossRef]
- Xie, C.; Gong, X.; Luo, J.; Li, B.; Song, B. AAV9-NPC1 significantly ameliorates Purkinje cell death and behavioral abnormalities in mouse NPC disease. J. Lipid Res. 2017, 58, 512–518. [Google Scholar] [CrossRef]
- Ye, Z.; Mo, C.; Liu, S.; Hatch, K.S.; Gao, S.; Ma, Y.; Hong, L.E.; Thompson, P.M.; Jahanshad, N.; Acheson, A.; et al. White matter integrity and nicotine dependence: Evaluating vertical and horizontal pleiotropy. Front. Neurosci. 2021, 15, 738037. [Google Scholar] [CrossRef]
- Zumbrennen-Bullough, K.B.; Becker, L.; Garrett, L.; Holter, S.M.; Calzada-Wack, J.; Mossbrugger, I.; Quintanilla-Fend, L.; Racz, I.; Rathkolb, B.; Klopstock, T.; et al. Abnormal brain iron metabolism in Irp2 deficient mice is associated with mild neurological and behavioral impairments. PLoS ONE 2014, 9, e98072. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.P.; Morató, X.; Souza, C.; Pinhal, C.; Machado, N.J.; Canas, P.M.; Silva, H.B.; Stagljar, I.; Gandía, J.; Fernández-Dueñas, V.; et al. The role of Parkinson’s disease-associated receptor GPR37 In the hippocampus: Functional interplay with the adenosinergic system. J. Neurochem. 2015, 134, 135–146. [Google Scholar] [CrossRef]
- Morató, X.; Gonçalves, F.Q.; Lopes, J.P.; Jauregui, O.; Soler, C.; Fernández-Dueñas, V.; Cunha, R.A.; Ciruela, F. Chronic adenosine A2A receptor blockade induces locomotor sensitization and potentiates striatal LTD in GPR37-deficient mice. J. Neurochem. 2019, 148, 796–809. [Google Scholar] [CrossRef]
- Mitroi, D.N.; Pereyra Gómez, G.; Soto Huelin, B.; Senovilla, F.; Kobayashi, T.; Esteban, J.A.; Ledesma, M.D. NPC1 enables cholesterol mobilization during long-term potentiation that can be restored in Niemann–pick disease type c by CYP46A1 activation. EMBO Rep. 2019, 20, e48143. [Google Scholar] [CrossRef] [PubMed]
- Karten, B.; Campenot, R.B.; Vance, D.E.; Vance, J.E. The Niemann-pick C1 protein in recycling endosomes of presynaptic nerve terminals. J. Lipid Res. 2006, 47, 504–514. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, J.K.; Bae, Y.C.; Yang, S.H.; Okino, N.; Schuchman, E.H.; Yamashita, T.; Bae, J.; Jin, H.K. Inhibition of GM3 synthase attenuates neuropathology of Niemann-pick disease type c by affecting sphingolipid metabolism. Mol. Cells 2014, 37, 161–171. [Google Scholar] [CrossRef]
- D Arcangelo, G.; Grossi, D.; Racaniello, M.; Cardinale, A.; Zaratti, A.; Rufini, S.; Cutarelli, A.; Tancredi, V.; Merlo, D.; Frank, C. Miglustat reverts the impairment of synaptic plasticity in a mouse model of NPC disease. Neural Plast. 2016, 2016, 3830424. [Google Scholar]
- Minicã, C.C.; Mbarek, H.; Pool, R.; Dolan, C.V.; Boomsma, D.I.; Vink, J.M. Pathways to smoking behaviours: Biological insights from the tobacco and genetics consortium meta-analysis. Mol. Psychiatry 2017, 22, 82–88. [Google Scholar] [CrossRef]
- Mayo, L.M.; Paul, E.; DeArcangelis, J.; Van Hedger, K.; and de Wit, H. Gender differences in the behavioral and subjective effects of methamphetamine in healthy humans. Psychopharmacology 2019, 236, 2413–2423. [Google Scholar] [CrossRef]
- Daiwile, A.P.; Jayanthi, S.; Cadet, J.L. Sex differences in methamphetamine use disorder perused from pre-clinical and clinical studies: Potential therapeutic impacts. Neurosci. Biobehav. Rev. 2022, 137, 104674. [Google Scholar] [CrossRef]
- Dluzen, D.E.; Liu, B. Gender differences in methamphetamine use and responses: A review. Gend. Med. 2008, 5, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Ujike, H.; Sato, M. Clinical features of sensitization to methamphetamine observed in patients with methamphetamine dependence and psychosis. Ann. N. Y. Acad. Sci. 2004, 1025, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.B.J.; Paxinos, G. The mouse brain. In Stereotaxic Coordinates; Academic Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, H.; Zhu, L.; Su, L.; Li, M.; Wang, R.; Zhu, J.; Chen, Y.; Chen, T. Impact of miR-29c-3p in the Nucleus Accumbens on Methamphetamine-Induced Behavioral Sensitization and Neuroplasticity-Related Proteins. Int. J. Mol. Sci. 2024, 25, 942. https://doi.org/10.3390/ijms25020942
Su H, Zhu L, Su L, Li M, Wang R, Zhu J, Chen Y, Chen T. Impact of miR-29c-3p in the Nucleus Accumbens on Methamphetamine-Induced Behavioral Sensitization and Neuroplasticity-Related Proteins. International Journal of Molecular Sciences. 2024; 25(2):942. https://doi.org/10.3390/ijms25020942
Chicago/Turabian StyleSu, Hang, Li Zhu, Linlan Su, Min Li, Rui Wang, Jie Zhu, Yanjiong Chen, and Teng Chen. 2024. "Impact of miR-29c-3p in the Nucleus Accumbens on Methamphetamine-Induced Behavioral Sensitization and Neuroplasticity-Related Proteins" International Journal of Molecular Sciences 25, no. 2: 942. https://doi.org/10.3390/ijms25020942
APA StyleSu, H., Zhu, L., Su, L., Li, M., Wang, R., Zhu, J., Chen, Y., & Chen, T. (2024). Impact of miR-29c-3p in the Nucleus Accumbens on Methamphetamine-Induced Behavioral Sensitization and Neuroplasticity-Related Proteins. International Journal of Molecular Sciences, 25(2), 942. https://doi.org/10.3390/ijms25020942





