Analysis of Receptor-Type Protein Tyrosine Phosphatase Extracellular Regions with Insights from AlphaFold
Abstract
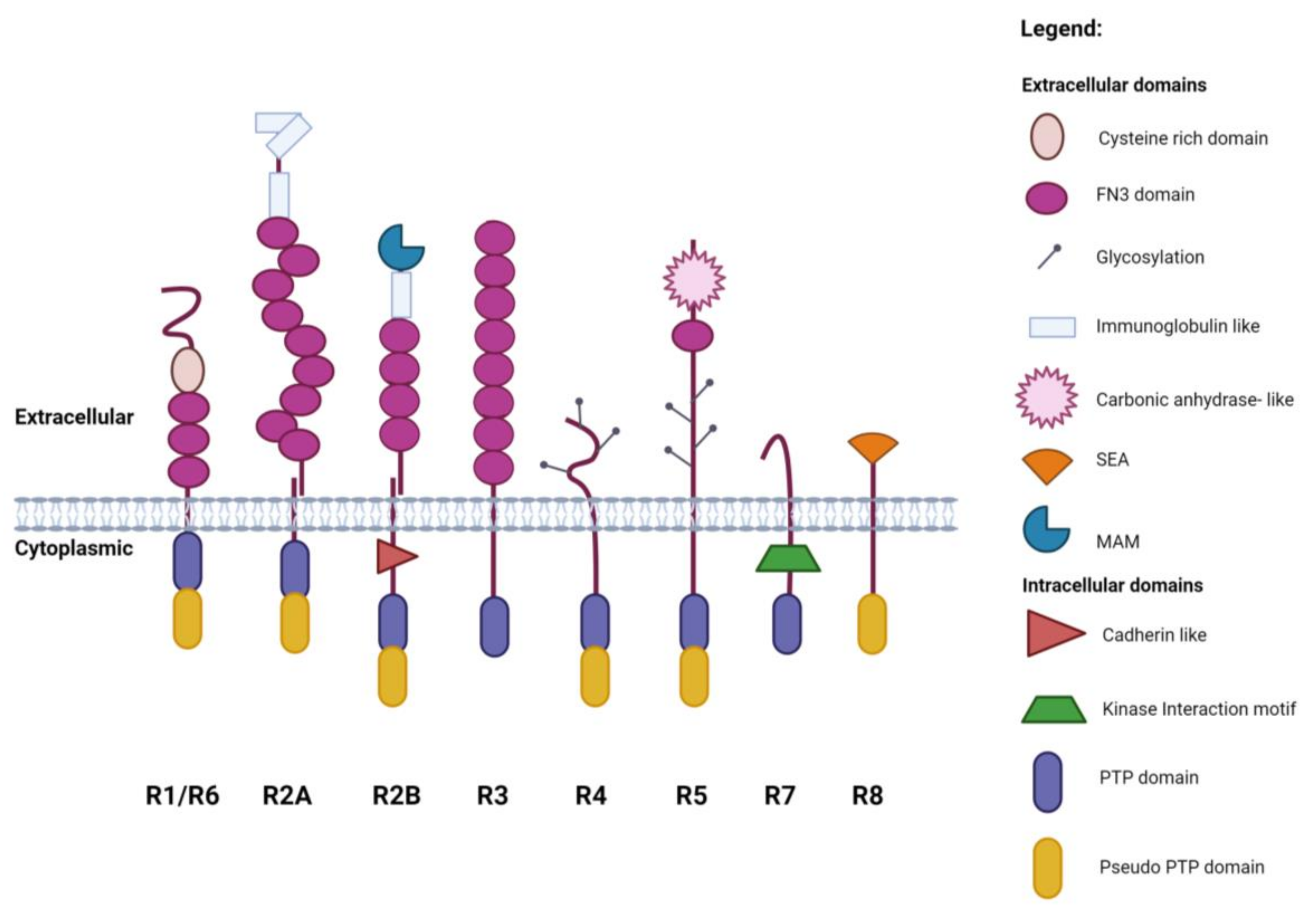
1. Introduction
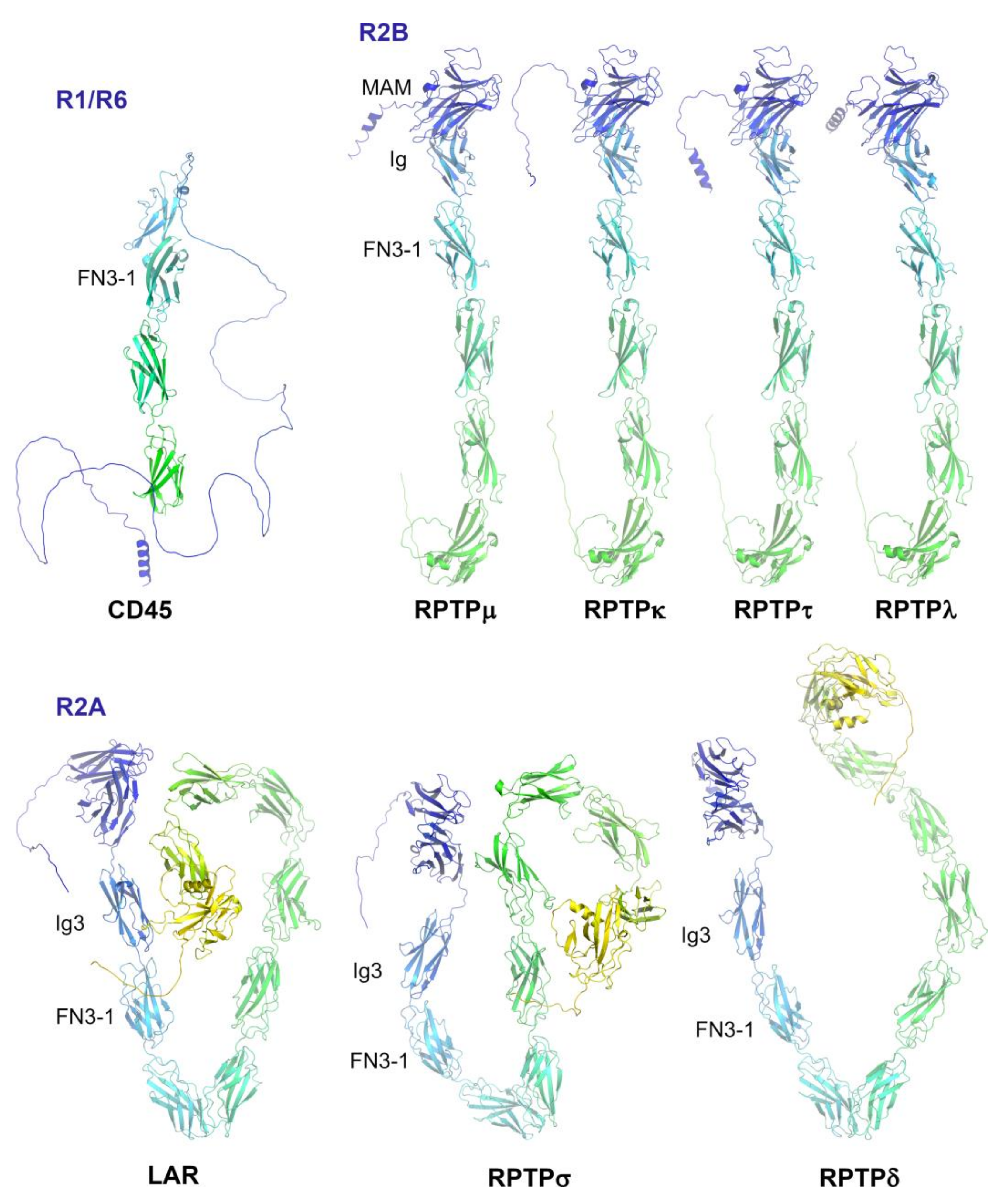
2. Subgroup R1/R6 (CD45)
3. Subgroup R2A (RPTPσ, RPTPδ and Leukocyte Common Antigen-Related (LAR))
4. Subgroup R2B (RPTPμ, RPTPρ, RPTPκ, and PCP2/RPTPλ)
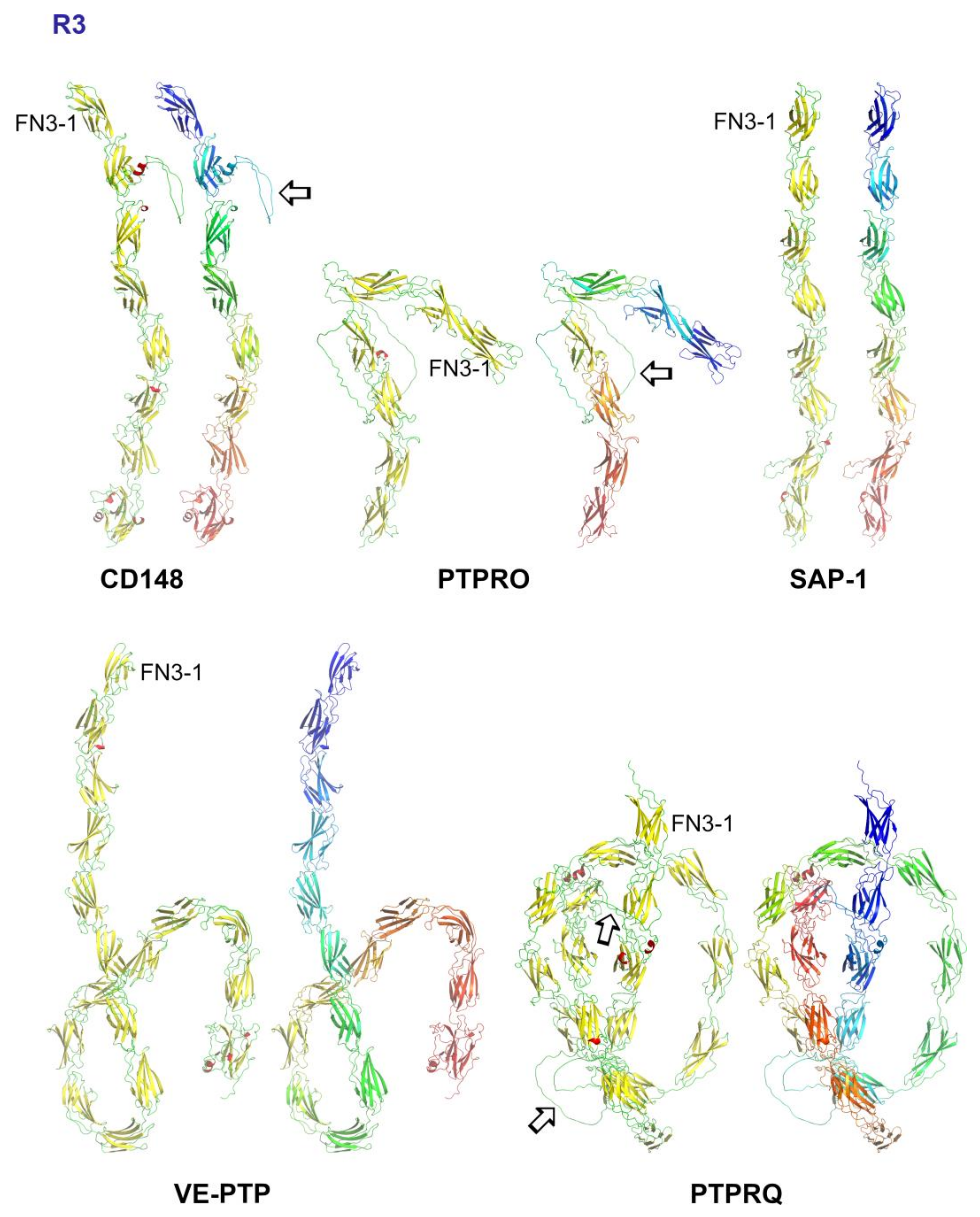
5. Subgroup R3 (CD148, VE-PTP, GLEPP1, SAP-1 and PTPRQ)
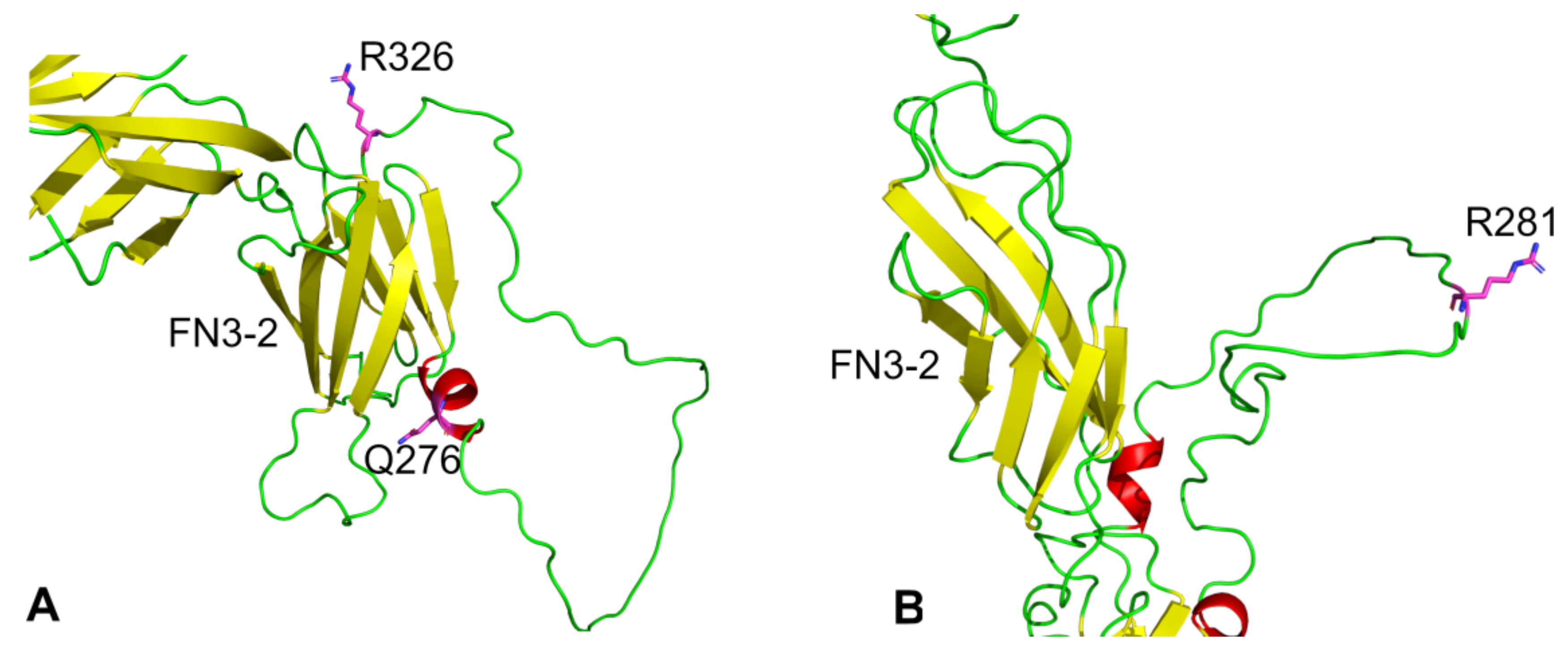
5.1. Ligands for R3 PTPs and Interactions Mediated via the Extracellular Domain
5.2. R3-PTPs: A Role for Size Exclusion
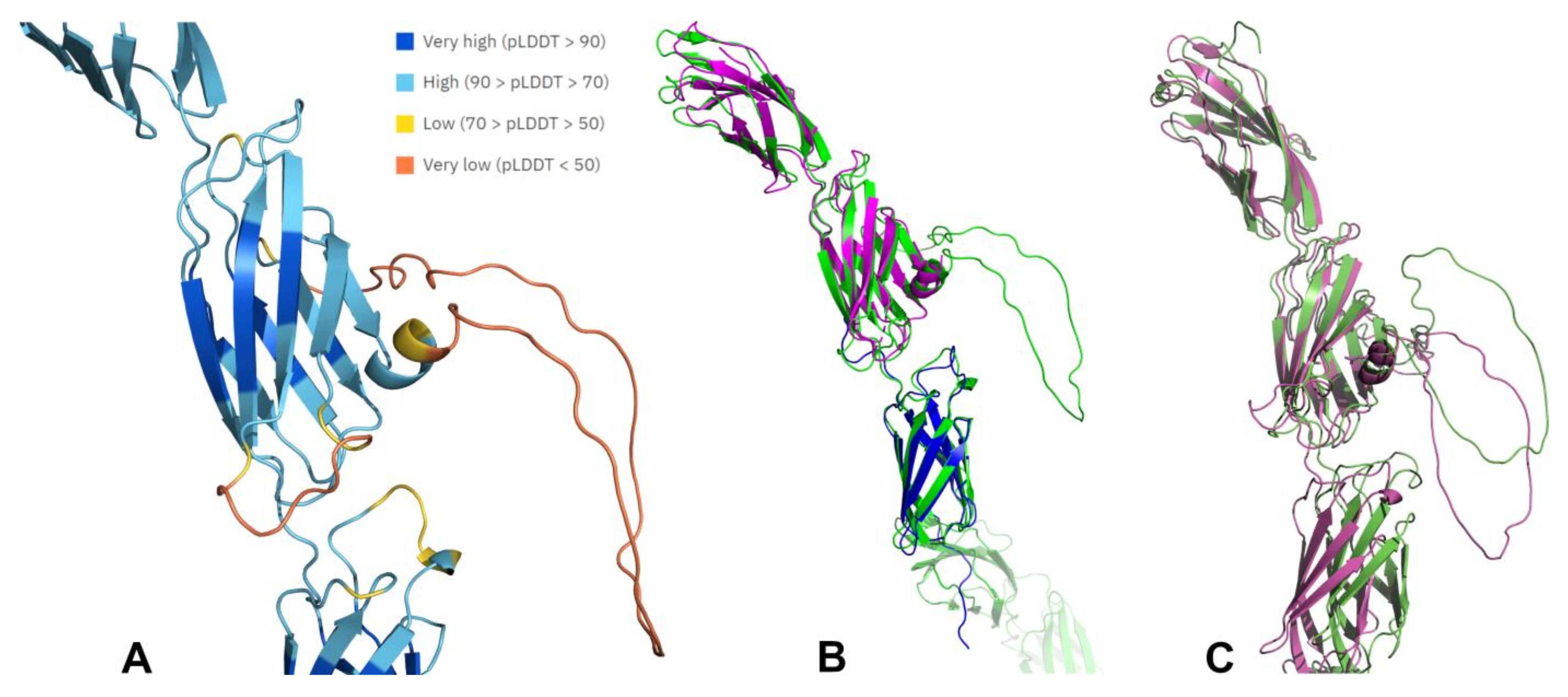
5.3. Structural Analysis of R3-PTPs Extracellular Domains
5.4. Genetic Variants of R3 Subgroup PTPs
6. Subgroup R4 (RPTPα, RPTPε)
7. Subgroup R5 (RPTPγ, RPTPζ)
8. Subgroup R7 (STEP, PTPRR)
9. Subgroup R8 (IA-2, IA2β)
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hunter, T. Tyrosine phosphorylation: Thirty years and counting. Curr. Opin. Cell Biol. 2009, 21, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, R.; Neel, B.G.; Rapoport, T.A. Mathematical models of protein kinase signal transduction. Mol. Cell 2002, 9, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.N.; Mortensen, O.H.; Peters, G.H.; Drake, P.G.; Iversen, L.F.; Olsen, O.H.; Jansen, P.G.; Andersen, H.S.; Tonks, N.K.; Moller, N.P. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell Biol. 2001, 21, 7117–7136. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Den Hertog, J.; Ostman, A.; Bohmer, F. Protein tyrosine phosphatases: Regulatory mechanisms. FEBS J. 2008, 275, 831–847. [Google Scholar] [CrossRef] [PubMed]
- Mohebiany, A.N.; Nikolaienko, R.M.; Bouyain, S.; Harroch, S. Receptor-type tyrosine phosphatase ligands: Looking for the needle in the haystack. FEBS J. 2013, 280, 388–400. [Google Scholar] [CrossRef]
- Pao, L.I.; Badour, K.; Siminovitch, K.A.; Neel, B.G. Nonreceptor protein-tyrosine phosphatases in immune cell signaling. Annu. Rev. Immunol. 2007, 25, 473–523. [Google Scholar] [CrossRef]
- Coles, C.H.; Jones, E.Y.; Aricescu, A.R. Extracellular regulation of type IIa receptor protein tyrosine phosphatases: Mechanistic insights from structural analyses. Semin. Cell Dev. Biol. 2015, 37, 98–107. [Google Scholar] [CrossRef]
- Senis, Y.A.; Barr, A.J. Targeting Receptor-Type Protein Tyrosine Phosphatases with Biotherapeutics: Is Outside-in Better than Inside-Out? Molecules 2018, 23, 569. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Zidek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; van der Merwe, P.A. The kinetic-segregation model: TCR triggering and beyond. Nat. Immunol. 2006, 7, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y. Pre-organized landscape of T cell surface. Front. Immunol. 2023, 14, 1264721. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.T.; Fernandes, R.A.; Ganzinger, K.A.; Lee, S.F.; Siebold, C.; McColl, J.; Jonsson, P.; Palayret, M.; Harlos, K.; Coles, C.H.; et al. Initiation of T cell signaling by CD45 segregation at ‘close contacts’. Nat. Immunol. 2016, 17, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Reily, C.; Stewart, T.J.; Renfrow, M.B.; Novak, J. Glycosylation in health and disease. Nat. Rev. Nephrol. 2019, 15, 346–366. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.G.; Tenney, A.P.; Ghose, A.; Duckworth, A.M.; Higashi, M.E.; Parfitt, K.; Marcu, O.; Heslip, T.R.; Marsh, J.L.; Schwarz, T.L.; et al. The HSPGs Syndecan and Dallylike bind the receptor phosphatase LAR and exert distinct effects on synaptic development. Neuron 2006, 49, 517–531. [Google Scholar] [CrossRef]
- Kwon, S.; Woo, J.; Kim, S.; Kim, H.; Kim, E. Trans-synaptic adhesions between netrin-G ligand-3 (NGL-3) and receptor tyrosine phosphatases LAR, protein-tyrosine phosphatase delta (PTPdelta), and PTPsigma via specific domains regulate excitatory synapse formation. J. Biol. Chem. 2010, 285, 13966–13978. [Google Scholar] [CrossRef]
- Woo, J.; Kwon, S.; Choi, S.; Kim, S.; Lee, J.; Dunah, A.W.; Sheng, M.; Kim, E. Trans-synaptic adhesion between NGL-3 and LAR regulates the formation of excitatory synapses. Nat. Neurosci. 2009, 12, 428–437. [Google Scholar] [CrossRef]
- Coles, C.H.; Shen, Y.; Tenney, A.P.; Siebold, C.; Sutton, G.C.; Lu, W.; Gallagher, J.T.; Jones, E.Y.; Flanagan, J.G.; Aricescu, A.R. Proteoglycan-specific molecular switch for RPTPsigma clustering and neuronal extension. Science 2011, 332, 484–488. [Google Scholar] [CrossRef]
- Coles, C.H.; Mitakidis, N.; Zhang, P.; Elegheert, J.; Lu, W.; Stoker, A.W.; Nakagawa, T.; Craig, A.M.; Jones, E.Y.; Aricescu, A.R. Structural basis for extracellular cis and trans RPTPsigma signal competition in synaptogenesis. Nat. Commun. 2014, 5, 5209. [Google Scholar] [CrossRef]
- Qian, Z.; Song, D.; Ipsaro, J.J.; Bautista, C.; Joshua-Tor, L.; Yeh, J.T.; Tonks, N.K. Manipulating PTPRD function with ectodomain antibodies. Genes. Dev. 2023, 37, 743–759. [Google Scholar] [CrossRef] [PubMed]
- Ensslen-Craig, S.E.; Brady-Kalnay, S.M. Receptor protein tyrosine phosphatases regulate neural development and axon guidance. Dev. Biol. 2004, 275, 12–22. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hay, I.M.; Fearnley, G.W.; Rios, P.; Kohn, M.; Sharpe, H.J.; Deane, J.E. The receptor PTPRU is a redox sensitive pseudophosphatase. Nat. Commun. 2020, 11, 3219. [Google Scholar] [CrossRef]
- Aricescu, A.R.; Siebold, C.; Choudhuri, K.; Chang, V.T.; Lu, W.; Davis, S.J.; van der Merwe, P.A.; Jones, E.Y. Structure of a tyrosine phosphatase adhesive interaction reveals a spacer-clamp mechanism. Science 2007, 317, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- Hay, I.M.; Shamin, M.; Caroe, E.R.; Mohammed, A.S.A.; Svergun, D.I.; Jeffries, C.M.; Graham, S.C.; Sharpe, H.J.; Deane, J.E. Determinants of receptor tyrosine phosphatase homophilic adhesion: Structural comparison of PTPRK and PTPRM extracellular domains. J. Biol. Chem. 2023, 299, 102750. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.G.; Dube, N.; Hardy, S.; Tremblay, M.L. Inside the human cancer tyrosine phosphatome. Nat. Rev. Cancer 2011, 11, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Matozaki, T.; Murata, Y.; Mori, M.; Kotani, T.; Okazawa, H.; Ohnishi, H. Expression, localization, and biological function of the R3 subtype of receptor-type protein tyrosine phosphatases in mammals. Cell Signal 2010, 22, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Oganesian, A.; Poot, M.; Daum, G.; Coats, S.A.; Wright, M.B.; Seifert, R.A.; Bowen-Pope, D.F. Protein tyrosine phosphatase RQ is a phosphatidylinositol phosphatase that can regulate cell survival and proliferation. Proc. Natl. Acad. Sci. USA 2003, 100, 7563–7568. [Google Scholar] [CrossRef]
- Bilotta, A.; Dattilo, V.; D’Agostino, S.; Belviso, S.; Scalise, S.; Bilotta, M.; Gaudio, E.; Paduano, F.; Perrotti, N.; Florio, T.; et al. A novel splice variant of the protein tyrosine phosphatase PTPRJ that encodes for a soluble protein involved in angiogenesis. Oncotarget 2017, 8, 10091–10102. [Google Scholar] [CrossRef]
- Aguiar, R.C.; Yakushijin, Y.; Kharbanda, S.; Tiwari, S.; Freeman, G.J.; Shipp, M.A. PTPROt: An alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood 1999, 94, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Dong, H.; Zhang, H. Regulatory Functions of Protein Tyrosine Phosphatase Receptor Type O in Immune Cells. Front. Immunol. 2021, 12, 783370. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Mernaugh, R.L.; Friedman, D.B.; Weller, R.; Tsuboi, N.; Yamashita, H.; Quaranta, V.; Takahashi, T. Thrombospondin-1 acts as a ligand for CD148 tyrosine phosphatase. Proc. Natl. Acad. Sci. USA 2012, 109, 1985–1990. [Google Scholar] [CrossRef] [PubMed]
- Whiteford, J.R.; Xian, X.; Chaussade, C.; Vanhaesebroeck, B.; Nourshargh, S.; Couchman, J.R. Syndecan-2 is a novel ligand for the protein tyrosine phosphatase receptor CD148. Mol. Biol. Cell 2011, 22, 3609–3624. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.S.; Gutierrez, J. Thrombospondin 1 in Metabolic Diseases. Front. Endocrinol. 2021, 12, 638536. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Sumarriva, K.; Kim, R.; Jiang, R.; Brantley-Sieders, D.M.; Chen, J.; Mernaugh, R.L.; Takahashi, T. Determination of the CD148-Interacting Region in Thrombospondin. PLoS ONE 2016, 11, e0154916. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Takahashi, K.; Pasic, L.; Narui, C.; Ellinger, P.; Grundmann, M.; Takahashi, T. The effects of CD148 Q276P/R326Q polymorphisms in A431D epidermoid cancer cell proliferation and epidermal growth factor receptor signaling. Cancer Rep. 2022, 5, e1566. [Google Scholar] [CrossRef]
- Tsoyi, K.; Liang, X.; De Rossi, G.; Ryter, S.W.; Xiong, K.; Chu, S.G.; Liu, X.; Ith, B.; Celada, L.J.; Romero, F.; et al. CD148 Deficiency in Fibroblasts Promotes the Development of Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2021, 204, 312–325. [Google Scholar] [CrossRef]
- Vestweber, D. Vascular Endothelial Protein Tyrosine Phosphatase Regulates Endothelial Function. Physiology 2021, 36, 84–93. [Google Scholar] [CrossRef]
- Nawroth, R.; Poell, G.; Ranft, A.; Kloep, S.; Samulowitz, U.; Fachinger, G.; Golding, M.; Shima, D.T.; Deutsch, U.; Vestweber, D. VE-PTP and VE-cadherin ectodomains interact to facilitate regulation of phosphorylation and cell contacts. EMBO J. 2002, 21, 4885–4895. [Google Scholar] [CrossRef]
- Cordoba, S.; Choudhuri, K.; Zhang, H.; Bridge, M.; Basat, A.B.; Dustin, M.L.; van der Merwe, P.A. The large ectodomains of CD45 and CD148 regulate their segregation from and inhibition of ligated T-cell receptor. Blood 2013, 121, 4295–4302. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Roy, S.; Niculcea, J.; Gould, K.; Adams, E.J.; van der Merwe, P.A.; Choudhuri, K. Ligand-induced segregation from large cell-surface phosphatases is a critical step in γδ TCR triggering. bioRxiv 2023. [Google Scholar] [CrossRef]
- Goodridge, H.S.; Reyes, C.N.; Becker, C.A.; Katsumoto, T.R.; Ma, J.; Wolf, A.J.; Bose, N.; Chan, A.S.H.; Magee, A.S.; Danielson, M.E.; et al. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 2011, 472, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Montague, S.J.; Patel, P.; Martin, E.M.; Slater, A.; Quintanilla, L.G.; Perrella, G.; Kardeby, C.; Nagy, M.; Mezzano, D.; Mendes, P.M.; et al. Platelet activation by charged ligands and nanoparticles: Platelet glycoprotein receptors as pattern recognition receptors. Platelets 2021, 32, 1018–1030. [Google Scholar] [CrossRef] [PubMed]
- Ruff, K.M.; Pappu, R.V. AlphaFold and Implications for Intrinsically Disordered Proteins. J. Mol. Biol. 2021, 433, 167208. [Google Scholar] [CrossRef]
- Mirdita, M.; Schutze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016, 44, 1185–1200. [Google Scholar] [CrossRef]
- Leahy, D.J.; Aukhil, I.; Erickson, H.P. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 1996, 84, 155–164. [Google Scholar] [CrossRef]
- Hendriks, W.J.A.J.; Pulido, R. Protein tyrosine phosphatase variants in human hereditary disorders and disease susceptibilities. Biochim. Biophys. Acta 2013, 1832, 1673–1696. [Google Scholar] [CrossRef]
- Rollin, J.; Pouplard, C.; Gratacap, M.; Leroux, D.; May, M.; Aupart, M.; Gouilleux-Gruart, V.; Payrastre, B.; Gruel, Y. Polymorphisms of protein tyrosine phosphatase CD148 influence FcgammaRIIA-dependent platelet activation and the risk of heparin-induced thrombocytopenia. Blood 2012, 120, 1309–1316. [Google Scholar] [CrossRef]
- Laczmanska, I.; Sasiadek, M.M. Meta-analysis of association between Arg326Gln (rs1503185) and Gln276Pro (rs1566734) polymorphisms of PTPRJ gene and cancer risk. J. Appl. Genet. 2019, 60, 57–62. [Google Scholar] [CrossRef]
- Schraders, M.; Oostrik, J.; Huygen, P.L.M.; Strom, T.M.; van Wijk, E.; Kunst, H.P.M.; Hoefsloot, L.H.; Cremers, C.W.R.J.; Admiraal, R.J.C.; Kremer, H. Mutations in PTPRQ are a cause of autosomal-recessive nonsyndromic hearing impairment DFNB84 and associated with vestibular dysfunction. Am. J. Hum. Genet. 2010, 86, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Pulido, R.; Stoker, A.W.; Hendriks, W.J.A.J. PTPs emerge as PIPs: Protein tyrosine phosphatases with lipid-phosphatase activities in human disease. Hum. Mol. Genet. 2013, 22, 66. [Google Scholar] [CrossRef] [PubMed]
- Chien, Y.; Wang, Y.; Sridharan, D.; Kuo, C.; Chien, C.; Uchihashi, T.; Kato, K.; Angata, T.; Meng, T.; Hsu, S.D.; et al. High Density of N- and O-Glycosylation Shields and Defines the Structural Dynamics of the Intrinsically Disordered Ectodomain of Receptor-type Protein Tyrosine Phosphatase Alpha. JACS Au 2023, 3, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Zhang, Y.; Huang, C.; Lu, R.; Yu, J.; Chen, R.; Wang, Y.; Zhao, X.; Yu, J.; Huang, J. Glucose-mediated N-glycosylation of RPTPalpha affects its subcellular localization and Src activation. Oncogene 2023, 42, 1058–1071. [Google Scholar] [CrossRef] [PubMed]
- Sendo, S.; Kiosses, W.B.; Yang, S.; Wu, D.J.; Lee, D.W.K.; Liu, L.; Aschner, Y.; Vela, A.J.; Downey, G.P.; Santelli, E.; et al. Clustering of phosphatase RPTPalpha promotes Src signaling and the arthritogenic action of synovial fibroblasts. Sci. Signal 2023, 16, eabn8668. [Google Scholar] [CrossRef] [PubMed]
- Nagai, K.; Fujii, M.; Kitazume, S. Protein Tyrosine Phosphatase Receptor Type Z in Central Nervous System Disease. Int. J. Mol. Sci. 2022, 23, 4414. [Google Scholar] [CrossRef] [PubMed]
- Boni, C.; Laudanna, C.; Sorio, C. A Comprehensive Review of Receptor-Type Tyrosine-Protein Phosphatase Gamma (PTPRG) Role in Health and Non-Neoplastic Disease. Biomolecules 2022, 12, 84. [Google Scholar] [CrossRef]
- Barr, A.J.; Ugochukwu, E.; Lee, W.H.; King, O.N.; Filippakopoulos, P.; Alfano, I.; Savitsky, P.; Burgess-Brown, N.; Muller, S.; Knapp, S. Large-scale structural analysis of the classical human protein tyrosine phosphatome. Cell 2009, 136, 352–363. [Google Scholar] [CrossRef]
- Fujikawa, A.; Sugawara, H.; Tanga, N.; Ishii, K.; Kuboyama, K.; Uchiyama, S.; Suzuki, R.; Noda, M. A head-to-toe dimerization has physiological relevance for ligand-induced inactivation of protein tyrosine receptor type Z. J. Biol. Chem. 2019, 294, 14953–14965. [Google Scholar] [CrossRef]
- Hendriks, W.J.A.J.; Dilaver, G.; Noordman, Y.E.; Kremer, B.; Fransen, J.A.M. PTPRR protein tyrosine phosphatase isoforms and locomotion of vesicles and mice. Cerebellum 2009, 8, 80–88. [Google Scholar] [CrossRef][Green Version]
- Won, S.; Roche, K.W. Regulation of glutamate receptors by striatal-enriched tyrosine phosphatase 61 (STEP(61)). J. Physiol. 2021, 599, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Caromile, L.A.; Oganesian, A.; Coats, S.A.; Seifert, R.A.; Bowen-Pope, D.F. The neurosecretory vesicle protein phogrin functions as a phosphatidylinositol phosphatase to regulate insulin secretion. J. Biol. Chem. 2010, 285, 10487–10496. [Google Scholar] [CrossRef] [PubMed]
- Noguera, M.E.; Primo, M.E.; Jakoncic, J.; Poskus, E.; Solimena, M.; Ermacora, M.R. X-ray structure of the mature ectodomain of phogrin. J. Struct. Funct. Genom. 2015, 16, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Buel, G.R.; Walters, K.J. Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 2022, 29, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Liebschner, D.; Croll, T.I.; Williams, C.J.; McCoy, A.J.; Poon, B.K.; Afonine, P.V.; Oeffner, R.D.; Richardson, J.S.; Read, R.J.; et al. AlphaFold predictions are valuable hypotheses and accelerate but do not replace experimental structure determination. Nat. Methods 2023. [Google Scholar] [CrossRef] [PubMed]

| Name | Gene Name | UniProt | UniProt (Prosite) FN3 Domains | AlphaFold FN3 Domains | Loops, Length and Position |
|---|---|---|---|---|---|
| CD148 | PTPRJ | Q12913 | 9 | 8 | N278–D327 (49aa) at FN3-2 |
| VE-PTP | PTPRB | P23467 | 17 | 17 | none |
| GLEPP-1 | PTPRO | Q16827 | 8 | 7 | N234–P342 (108aa) at FN3-3 |
| SAP-1 | PTPRH | Q9HD43 | 8 | 8 | none |
| PTPRQ | PTPRQ | Q9UMZ3 | 18 | 18 | L254–N298 (44aa) at FN3-3 T491–E569 (78aa) at FN3-5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Badaoui, L.; Barr, A.J. Analysis of Receptor-Type Protein Tyrosine Phosphatase Extracellular Regions with Insights from AlphaFold. Int. J. Mol. Sci. 2024, 25, 820. https://doi.org/10.3390/ijms25020820
El Badaoui L, Barr AJ. Analysis of Receptor-Type Protein Tyrosine Phosphatase Extracellular Regions with Insights from AlphaFold. International Journal of Molecular Sciences. 2024; 25(2):820. https://doi.org/10.3390/ijms25020820
Chicago/Turabian StyleEl Badaoui, Lina, and Alastair J. Barr. 2024. "Analysis of Receptor-Type Protein Tyrosine Phosphatase Extracellular Regions with Insights from AlphaFold" International Journal of Molecular Sciences 25, no. 2: 820. https://doi.org/10.3390/ijms25020820
APA StyleEl Badaoui, L., & Barr, A. J. (2024). Analysis of Receptor-Type Protein Tyrosine Phosphatase Extracellular Regions with Insights from AlphaFold. International Journal of Molecular Sciences, 25(2), 820. https://doi.org/10.3390/ijms25020820





