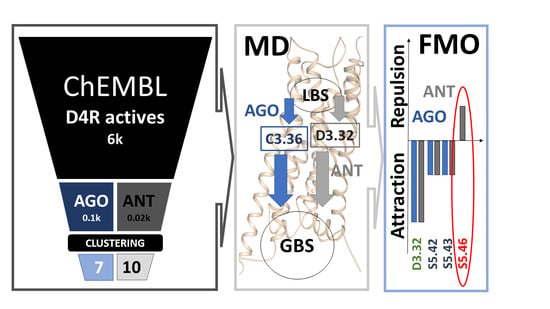
The Pivotal Distinction between Antagonists’ and Agonists’ Binding into Dopamine D4 Receptor—MD and FMO/PIEDA Studies
Abstract
1. Introduction
2. Results and Discussion
2.1. Docking
2.2. MD Simulations
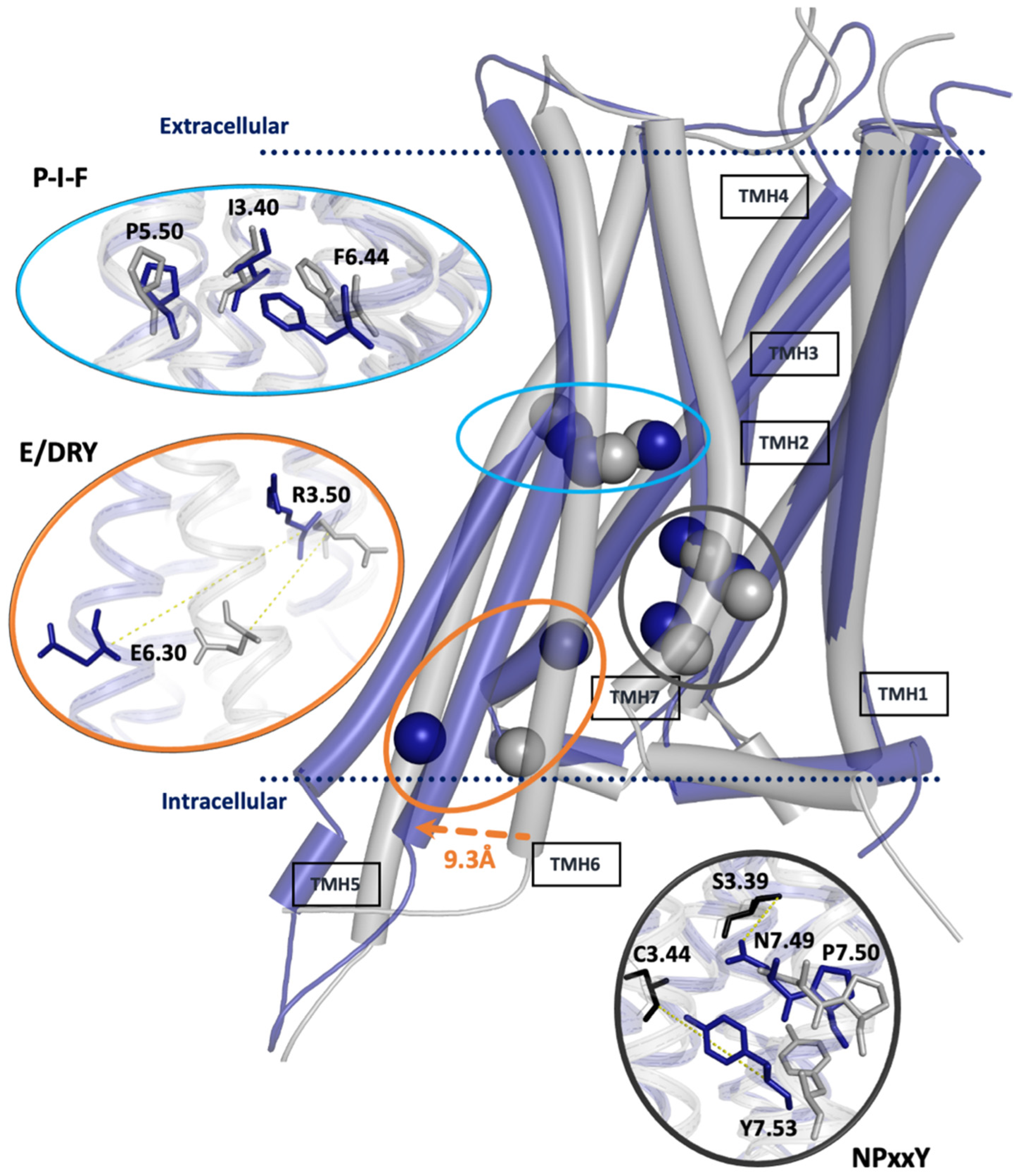
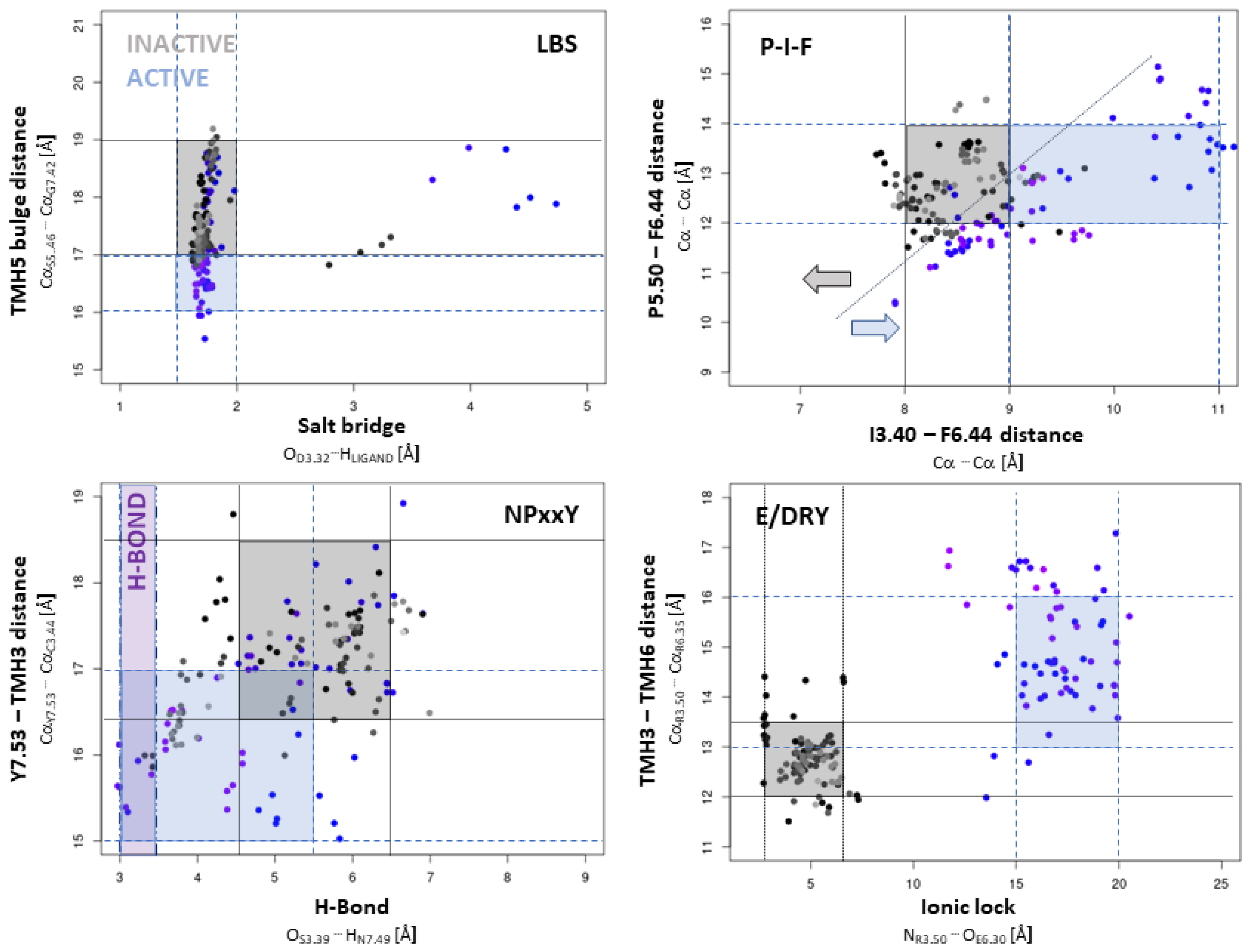
2.2.1. Influence of Agonists/Antagonists on Receptor Dynamics
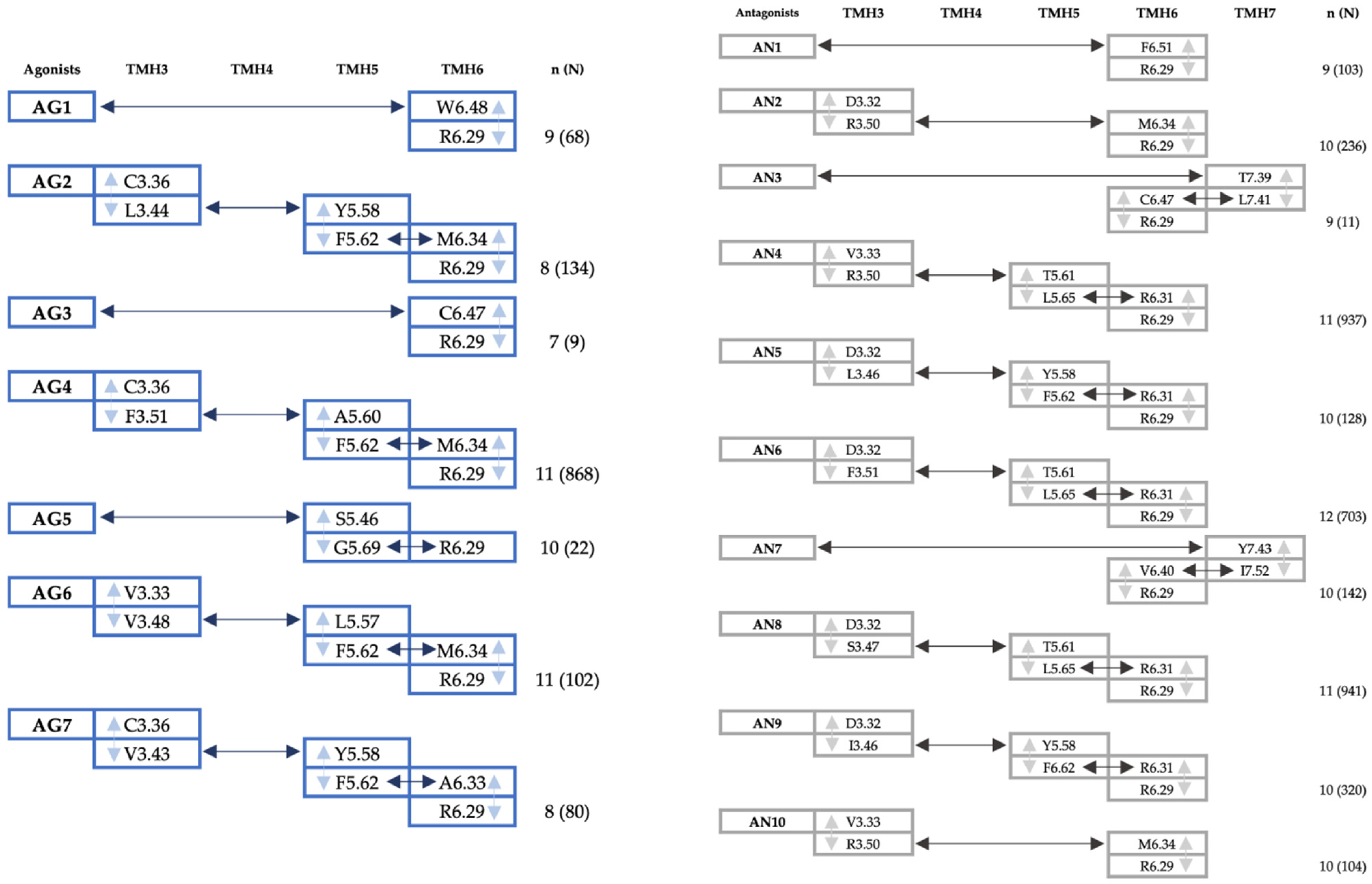
2.2.2. Signal Transfer Based on the Dynamical Network Analysis
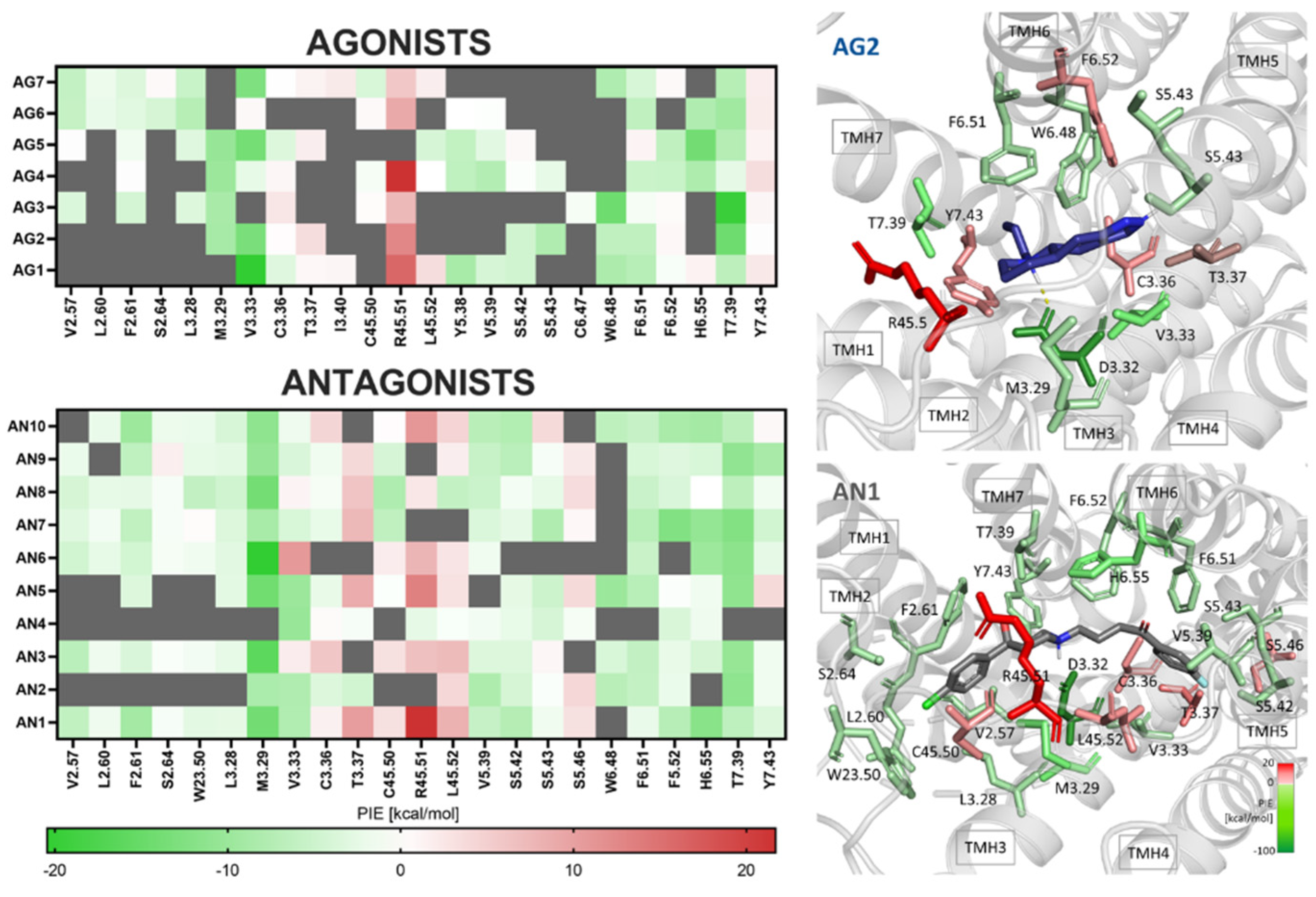
2.3. The Importance of Repulsive or Attractive Forces in Activating D4R—FMO/PIEDA Calculations
3. Materials and Methods
3.1. Protein and Agonists/Antagonists Set Preparations
3.2. Docking
3.3. Molecular Dynamic and Analysis
3.4. FMO/PIEDA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ramachandraiah, C.T.; Subramaniam, N.; Tancer, M. The Story of Antipsychotics: Past and Present. Indian J. Psychiatry 2009, 51, 324–326. [Google Scholar] [CrossRef] [PubMed]
- Ban, T.A. Fifty Years Chlorpromazine: A Historical Perspective. Neuropsychiatr. Dis. Treat. 2007, 3, 495–500. [Google Scholar] [PubMed]
- Mishra, A.; Singh, S.; Shukla, S. Physiological and Functional Basis of Dopamine Receptors and Their Role in Neurogenesis: Possible Implication for Parkinson’s Disease. J. Exp. Neurosci. 2018, 12, 1179069518779829. [Google Scholar] [CrossRef] [PubMed]
- Kebabian, J.W.; Petzold, G.L.; Greengard, P. Dopamine-Sensitive Adenylate Cyclase in Caudate Nucleus of Rat Brain, and Its Similarity to the “Dopamine Receptor”. Proc. Natl. Acad. Sci. USA 1972, 69, 2145–2149. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Snyder, G.L.; Vanover, K.E. Dopamine Targeting Drugs for the Treatment of Schizophrenia: Past, Present and Future. Curr. Top. Med. Chem. 2016, 16, 3385–3403. [Google Scholar] [CrossRef] [PubMed]
- Roth, B.L.; Tandra, S.; Burgess, L.H.; Sibley, D.R.; Meltzer, H.Y. D4 Dopamine Receptor Binding Affinity Does Not Distinguish between Typical and Atypical Antipsychotic Drugs. Psychopharmacology 1995, 120, 365–368. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.E. Neuroleptic Drug-Induced Extrapyramidal Syndromes and Tardive Dyskinesia. Schizophr. Res. 1991, 4, 109–120. [Google Scholar] [CrossRef]
- Kramer, M.S.; Last, B.; Getson, A.; Reines, S.A. The Effects of a Selective D4 Dopamine Receptor Antagonist (L-745,870) in Acutely Psychotic Inpatients with Schizophrenia. D4 Dopamine Antagonist Group. Arch. Gen. Psychiatry 1997, 54, 567–572. [Google Scholar] [CrossRef]
- Corrigan, M.H.; Gallen, C.C.; Bonura, M.L.; Merchant, K.M. Effectiveness of the Selective D4 Antagonist Sonepiprazole in Schizophrenia: A Placebo-Controlled Trial. Biol. Psychiatry 2004, 55, 445–451. [Google Scholar] [CrossRef]
- Dolma, S.; Selvadurai, H.J.; Lan, X.; Lee, L.; Kushida, M.; Voisin, V.; Whetstone, H.; So, M.; Aviv, T.; Park, N.; et al. Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells. Cancer Cell 2016, 29, 859–873. [Google Scholar] [CrossRef]
- Sebastianutto, I.; Maslava, N.; Hopkins, C.R.; Cenci, M.A. Validation of an Improved Scale for Rating L-DOPA-Induced Dyskinesia in the Mouse and Effects of Specific Dopamine Receptor Antagonists. Neurobiol. Dis. 2016, 96, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Huot, P.; Johnston, T.H.; Koprich, J.B.; Aman, A.; Fox, S.H.; Brotchie, J.M. L-745,870 Reduces L-DOPA-Induced Dyskinesia in the 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine-Lesioned Macaque Model of Parkinson’s Disease. J. Pharmacol. Exp. Ther. 2012, 342, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Botticelli, L.; Micioni Di Bonaventura, E.; del Bello, F.; Giorgioni, G.; Piergentili, A.; Romano, A.; Quaglia, W.; Cifani, C.; Micioni Di Bonaventura, M.V. Underlying Susceptibility to Eating Disorders and Drug Abuse: Genetic and Pharmacological Aspects of Dopamine D4 Receptors. Nutrients 2020, 12, 2288. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.B.; Paulus, M.P.; Hartman, D.S.; Godel, T.; Geyer, M.A. RO-10-5824 Is a Selective Dopamine D4 Receptor Agonist That Increases Novel Object Exploration in C57 Mice. Neuropharmacology 2003, 44, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, M.; Neugebauer, N.M.; Meltzer, H.Y. Dopamine D4 Receptor Stimulation Contributes to Novel Object Recognition: Relevance to Cognitive Impairment in Schizophrenia. J. Psychopharmacol. 2017, 31, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Brioni, J.D.; Moreland, R.B.; Cowart, M.; Hsieh, G.C.; Stewart, A.O.; Hedlund, P.; Donnelly-Roberts, D.L.; Nakane, M.; Lynch, J.J.; Kolasa, T.; et al. Activation of Dopamine D4 Receptors by ABT-724 Induces Penile Erection in Rats. Proc. Natl. Acad. Sci. USA 2004, 101, 6758–6763. [Google Scholar] [CrossRef]
- Melis, M.R.; Succu, S.; Mascia, M.S.; Argiolas, A. PD-168077, a Selective Dopamine D4 Receptor Agonist, Induces Penile Erection When Injected into the Paraventricular Nucleus of Male Rats. Neurosci. Lett. 2005, 379, 59–62. [Google Scholar] [CrossRef]
- Ladefoged, K.L.; Koch, R.; Biggin, P.C.; Schiøtt, B. Binding and Activation of Serotonergic G-Protein Coupled Receptors by the Multimodal Antidepressant Vortioxetine. ACS Chem. Neurosci. 2022, 13, 1129–1142. [Google Scholar] [CrossRef]
- Latorraca, N.R.; Venkatakrishnan, A.J.; Dror, R.O. GPCR Dynamics: Structures in Motion. Chem. Rev. 2017, 117, 139–155. [Google Scholar] [CrossRef]
- Jatana, N.; Thukral, L.; Latha, N. Structure and Dynamics of DRD4 Bound to an Agonist and an Antagonist Using in Silico Approaches. Proteins: Struct. Funct. Bioinform. 2014, 83, 867–880. [Google Scholar] [CrossRef]
- Sethi, A.; Eargle, J.; Black, A.A.; Luthey-Schulten, Z. Dynamical Networks in TRNA:Protein Complexes. Proc. Natl. Acad. Sci. USA 2009, 106, 6620–6625. [Google Scholar] [CrossRef] [PubMed]
- Girvan, M.; Newman, M.E.J. Community Structure in Social and Biological Networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; Trani, G.; Aldeghi, M.; MacKinnon, C.H.; McEwan, P.A.; Brookfield, F.A.; Chudyk, E.I.; Bodkin, M.; Pei, Z.; Burch, J.D.; et al. Fragment Molecular Orbital Method Applied to Lead Optimization of Novel Interleukin-2 Inducible T-Cell Kinase (ITK) Inhibitors. J. Med. Chem. 2016, 59, 4352–4363. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, D.G.; Nagata, T.; Kitaura, K. Exploring Chemistry with the Fragment Molecular Orbital Method. Phys. Chem. Chem. Phys. 2012, 14, 7562–7577. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, D.G.; Kitaura, K. Pair Interaction Energy Decomposition Analysis. J. Comput. Chem. 2007, 28, 222–237. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, D.G.; Kitaura, K. Extending the Power of Quantum Chemistry to Large Systems with the Fragment Molecular Orbital Method. J. Phys. Chem. A 2007, 111, 6904–6914. [Google Scholar] [CrossRef] [PubMed]
- Kurczab, R.; Kucwaj-Brysz, K.; Śliwa, P. The Significance of Halogen Bonding in Ligand–Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor. Molecules 2019, 25, 91. [Google Scholar] [CrossRef]
- Preikša, J.; Śliwa, P. Application of Fragment Molecular Orbital Method to Investigate Dopamine Receptors. Sci. Technol. Innov. 2019, 6, 24–33. [Google Scholar] [CrossRef]
- Śliwa, P.; Kurczab, R.; Rafał, K.; Drabczyk, A.; Jaśkowska, J. Recognition of Repulsive and Attractive Regions of Selected Serotonin Receptor Binding Site Using FMO-EDA Approach. J. Mol. Model. 2019, 25, 114. [Google Scholar] [CrossRef]
- Zaręba, P.; Śliwa, P.; Satała, G.; Zajdel, P.; Latacz, G.; Jaśkowska, J. New N-Aryl-N′-Aryl-/(Thio)Ureido-/Sulfamoylamino-Derivatives of Alkyl/Alkylcarbamoyl Piperazines: Effect of Structural Modifications on Selectivity over 5-HT1A Receptor. Eur. J. Med. Chem. 2022, 235, 114319. [Google Scholar] [CrossRef]
- Śliwa, P.; Kurczab, R.; Bojarski, A.J. ONIOM and FMO-EDA Study of Metabotropic Glutamate Receptor 1: Quantum Insights into the Allosteric Binding Site. Int. J. Quantum Chem. 2018, 118, e25617. [Google Scholar] [CrossRef]
- Matalińska, J.; Lipiński, P.F.J.; Kosson, P.; Kosińska, K.; Misicka, A. In Vivo, in Vitro and in Silico Studies of the Hybrid Compound Aa3266, an Opioid Agonist/NK1R Antagonist with Selective Cytotoxicity. Int. J. Mol. Sci. 2020, 21, 7738. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; Aldeghi, M.; Chudyk, E.I.; Fedorov, D.G.; Bodkin, M.J.; Biggin, P.C. Using the Fragment Molecular Orbital Method to Investigate Agonist-Orexin-2 Receptor Interactions. Biochem. Soc. Trans. 2016, 44, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Paudel, P.; Seong, S.H.; Jung, H.A.; Choi, J.S. Characterizing Fucoxanthin as a Selective Dopamine D3/D4 Receptor Agonist: Relevance to Parkinson’s Disease. Chem. Biol. Interact. 2019, 310, 108757. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, M.; Nadri, H.; Moradi, A.; Sakhteman, A. Homology Modeling, Molecular Dynamic Simulation, and Docking Based Binding Site Analysis of Human Dopamine (D4) Receptor. J. Mol. Model. 2015, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Wang, S.; Balius, T.E.; Singh, I.; Levit, A.; Moroz, Y.S.; O’Meara, M.J.; Che, T.; Algaa, E.; Tolmachova, K.; et al. Ultra-Large Library Docking for Discovering New Chemotypes. Nature 2019, 566, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wacker, D.; Levit, A.; Che, T.; Betz, R.M.; McCorvy, J.D.; Venkatakrishnan, A.J.; Huang, X.P.; Dror, R.O.; Shoichet, B.K.; et al. D4 Dopamine Receptor High-Resolution Structures Enable the Discovery of Selective Agonists. Science 2017, 358, 381–386. [Google Scholar] [CrossRef]
- Yin, J.; Chen, K.Y.M.; Clark, M.J.; Hijazi, M.; Kumari, P.; Bai, X.C.; Sunahara, R.K.; Barth, P.; Rosenbaum, D.M. Structure of a D2 Dopamine Receptor–G-Protein Complex in a Lipid Membrane. Nature 2020, 584, 125–129. [Google Scholar] [CrossRef]
- Jatana, N.; Thukral, L.; Latha, N. Structural Signatures of DRD4 Mutants Revealed Using Molecular Dynamics Simulations: Implications for Drug Targeting. J. Mol. Model. 2016, 22, 14. [Google Scholar] [CrossRef]
- Kurczab, R.; Śliwa, P.; Rataj, K.; Kafel, R.; Bojarski, A.J. Salt Bridge in Ligand–Protein Complexes—Systematic Theoretical and Statistical Investigations. J. Chem. Inf. Model. 2018, 58, 2224–2238. [Google Scholar] [CrossRef]
- Dror, R.O.; Arlow, D.H.; Maragakis, P.; Mildorf, T.J.; Pan, A.C.; Xu, H.; Borhani, D.W.; Shaw, D.E.; Research, D.E.S.; York, N.; et al. Activation Mechanism of the β 2-Adrenergic Receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 18684–18689. [Google Scholar] [CrossRef] [PubMed]
- Kohlhoff, K.J.; Shukla, D.; Lawrenz, M.; Bowman, G.R.; Konerding, D.E.; Belov, D.; Altman, R.B.; Pande, V.S. Cloud-Based Simulations on Google Exacycle Reveal Ligand-Modulation of GPCR Activation Pathways HHS Public Access Author Manuscript. Nat. Chem. 2014, 6, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Manglik, A.; Kim, T.H.; Masureel, M.; Altenbach, C.; Yang, Z.; Hilger, D.; Lerch, M.T.; Kobilka, T.S.; Thian, F.S.; Hubbell, W.L.; et al. Structural Insights into the Dynamic Process of Β2-Adrenergic Receptor Signaling. Cell 2015, 161, 1101–1111. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.G.F.; Choi, H.J.; Fung, J.J.; Pardon, E.; Casarosa, P.; Chae, P.S.; Devree, B.T.; Rosenbaum, D.M.; Thian, F.S.; Kobilka, T.S.; et al. Structure of a Nanobody-Stabilized Active State of the Β2 Adrenoceptor. Nature 2011, 469, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.G.F.; Devree, B.T.; Zou, Y.; Kruse, A.C.; Chung, K.Y.; Kobilka, T.S.; Thian, F.S.; Chae, P.S.; Pardon, E.; Calinski, D.; et al. Crystal Structure of the Β2 Adrenergic Receptor–Gs Protein Complex. Nature 2011, 477, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; Chudyk, E.; Gleave, L.; Aldeghi, M.; Cherezov, V.; Fedorov, D.G.; Biggin, P.C.; Bodkin, M. The Fragment Molecular Orbital Method Reveals New Insight into the Chemical Nature of GPCR-Ligand Interactions. J. Chem. Inf. Model. 2016, 56, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Isberg, V.; Vroling, B.; van der Kant, R.; Li, K.; Vriend, G.; Gloriam, D. GPCRdb: An Information System for G Protein-Coupled Receptors. Nucleic Acids Res. 2014, 42, D422–D425. [Google Scholar] [CrossRef]
- Isberg, V.; Mordalski, S.; Munk, C.; Rataj, K.; Harpsøe, K.; Hauser, A.S.; Vroling, B.; Bojarski, A.J.; Vriend, G.; Gloriam, D.E. GPCRdb: An Information System for G Protein-Coupled Receptors. Nucleic Acids Res. 2016, 44, D356–D364. [Google Scholar] [CrossRef]
- Isberg, V.; De Graaf, C.; Bortolato, A.; Cherezov, V.; Katritch, V.; Marshall, F.H.; Mordalski, S.; Pin, J.P.; Stevens, R.C.; Vriend, G.; et al. Generic GPCR Residue Numbers—Aligning Topology Maps While Minding the Gaps. Trends Pharmacol. Sci. 2015, 36, 22–31. [Google Scholar] [CrossRef]
- Kooistra, A.J.; Mordalski, S.; Pándy-Szekeres, G.; Esguerra, M.; Mamyrbekov, A.; Munk, C.; Keserű, G.M.; Gloriam, D.E. GPCRdb in 2021: Integrating GPCR Sequence, Structure and Function. Nucleic Acids Res. 2021, 49, D335–D343. [Google Scholar] [CrossRef] [PubMed]
- Pandy-Szekeres, G.; Caroli, J.; Mamyrbekov, A.; Kermani, A.A.; Keseru, G.M.; Kooistra, A.J.; Gloriam, D.E. GPCRdb in 2023: State-Specific Structure Models Using AlphaFold2 and New Ligand Resources. Nucleic Acids Res. 2023, 51, D395–D402. [Google Scholar] [CrossRef] [PubMed]
- Willighagen, E.L.; Waagmeester, A.; Spjuth, O.; Ansell, P.; Williams, A.J.; Tkachenko, V.; Hastings, J.; Chen, B.; Wild, D.J. The ChEMBL Database as Linked Open Data. J. Cheminform 2013, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.; Nowotka, M.; Papadatos, G.; Dedman, N.; Gaulton, A.; Atkinson, F.; Bellis, L.; Overington, J.P. ChEMBL Web Services: Streamlining Access to Drug Discovery Data and Utilities. Nucleic Acids Res. 2015, 43, W612. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Dixon, S.L.; Lowrie, J.F.; Sherman, W. Analysis and Comparison of 2D Fingerprints: Insights into Database Screening Performance Using Eight Fingerprint Methods. J. Mol. Graph. Model. 2010, 29, 157–170. [Google Scholar] [CrossRef]
- Sastry, M.; Lowrie, J.F.; Dixon, S.L.; Sherman, W. Large-Scale Systematic Analysis of 2D Fingerprint Methods and Parameters to Improve Virtual Screening Enrichments. J. Chem. Inf. Model. 2010, 50, 771–784. [Google Scholar] [CrossRef] [PubMed]
- LigPrep, version 3.7; Schrödinger, LLC: New York, NY, USA, 2016.
- Epik, Version 3.5; Schrödinger, LLC: New York, NY, USA, 2016.
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef]
- Vonommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone φ, ψ and Side-Chain Χ1 and Χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Ribeiro, J.V.; Bernardi, R.C.; Rudack, T.; Stone, J.E.; Phillips, J.C.; Freddolino, P.L.; Schulten, K. QwikMD—Integrative Molecular Dynamics Toolkit for Novices and Experts. Sci. Rep. 2016, 6, 26536. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K.; Vert, J.-P.; Ballesteros, J.; Pardo, L.; Kobilka, B.; Kuhn, P.; Weis, W.; Kobilka, B.; et al. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hou, T. CaFE: A Tool for Binding Affinity Prediction Using End-Point Free Energy Methods. Bioinformatics 2016, 32, 2216–2218. [Google Scholar] [CrossRef] [PubMed]
- Jurrus, E.; Engel, D.; Star, K.; Monson, K.; Brandi, J.; Felberg, L.E.; Brookes, D.H.; Wilson, L.; Chen, J.; Liles, K.; et al. Improvements to the APBS Biomolecular Solvation Software Suite. Protein Sci. 2018, 27, 112–128. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Glykos, N.M. Software News and Updates. Carma: A Molecular Dynamics Analysis Program. J. Comput. Chem. 2006, 27, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Luthey-Schulten, Z.; Sethi, A.; Eargle, J.; Black, A. Subopt 200. Available online: https://luthey-schulten.chemistry.illinois.edu/software/networkTools/ (accessed on 15 February 2023).
- Hout, R.F.; Levi, B.A.; Hehre, W.J. Effect of Electron Correlation on Theoretical Vibrational Frequencies. J. Comput. Chem. 1982, 3, 234–250. [Google Scholar] [CrossRef]
- Fedorov, D.G. The Fragment Molecular Orbital Method: Theoretical Development, Implementation in GAMESS, and Applications. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2017, 7, e1322. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. General Atomic and Molecular Electronic Structure System. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Suenaga, M. Facio: New Computational Chemistry Environment for PC GAMESS. J. Comput. Chem. Jpn. 2005, 4, 25–32. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Śliwa, P.; Dziurzyńska, M.; Kurczab, R.; Kucwaj-Brysz, K. The Pivotal Distinction between Antagonists’ and Agonists’ Binding into Dopamine D4 Receptor—MD and FMO/PIEDA Studies. Int. J. Mol. Sci. 2024, 25, 746. https://doi.org/10.3390/ijms25020746
Śliwa P, Dziurzyńska M, Kurczab R, Kucwaj-Brysz K. The Pivotal Distinction between Antagonists’ and Agonists’ Binding into Dopamine D4 Receptor—MD and FMO/PIEDA Studies. International Journal of Molecular Sciences. 2024; 25(2):746. https://doi.org/10.3390/ijms25020746
Chicago/Turabian StyleŚliwa, Paweł, Magdalena Dziurzyńska, Rafał Kurczab, and Katarzyna Kucwaj-Brysz. 2024. "The Pivotal Distinction between Antagonists’ and Agonists’ Binding into Dopamine D4 Receptor—MD and FMO/PIEDA Studies" International Journal of Molecular Sciences 25, no. 2: 746. https://doi.org/10.3390/ijms25020746
APA StyleŚliwa, P., Dziurzyńska, M., Kurczab, R., & Kucwaj-Brysz, K. (2024). The Pivotal Distinction between Antagonists’ and Agonists’ Binding into Dopamine D4 Receptor—MD and FMO/PIEDA Studies. International Journal of Molecular Sciences, 25(2), 746. https://doi.org/10.3390/ijms25020746