Abstract
Microtia-atresia is a rare type of congenital craniofacial malformation causing severe damage to the appearance and hearing ability of affected individuals. The genetic factors associated with microtia-atresia have not yet been determined. The AMER1 gene has been identified as potentially pathogenic for microtia-atresia in two twin families. An amer1 mosaic knockdown zebrafish model was constructed using CRISPR/Cas9. The phenotype and the development process of cranial neural crest cells of the knockdown zebrafish were examined. Components of the Wnt/β-catenin pathway were examined by qPCR, Western blotting, and immunofluorescence assay. IWR-1-endo, a reversible inhibitor of the Wnt/β-catenin pathway, was applied to rescue the abnormal phenotype. The present study showed that the development of mandibular cartilage in zebrafish was severely compromised by amer1 knockdown using CRISPR/Cas9. Specifically, amer1 knockdown was found to affect the proliferation and apoptosis of cranial neural crest cells, as well as their differentiation to chondrocytes. Mechanistically, amer1 exerted an antagonistic effect on the Wnt/β-catenin pathway. The application of IWR-1-endo could partially rescue the abnormal phenotype. We demonstrated that amer1 was essential for the craniofacial development of zebrafish by interacting with the Wnt/β-catenin pathway. These findings provide important insight into the role of amer1 in zebrafish mandibular development and the pathology of microtia-atresia caused by AMER1 gene mutations in humans.
1. Introduction
Microtia-atresia is a rare type of craniofacial developmental malformation characterized by shrinkage or deformity of the auricle and frequently accompanied by atresia of the external ear canal. Its incidence has been reported to range from 0.67 to 3.06 per 10,000 births [1,2]. As a manifestation of craniofacial maldevelopment, it can occur either unilaterally (70–90%) or bilaterally (10–30%), with or without middle ear malformation, and either independently (40–80%) or as a part of clinical developmental syndrome (20–60%) [3]. Etiologically, this condition is regarded as multifactorial, including both environmental and genetic factors. Although the causative genetic variations have been identified in patients with several types of syndromic microtia, the genetic variations in most patients with sporadic isolated microtia-atresia have not yet been determined.
To identify the genetic causes in patients with sporadic microtia-atresia, six families with monozygotic twins carrying discordant microtia phenotypes were subjected to whole-exome sequencing (WES) [4]. Five genes with recurrent mutations were identified, with a missense mutation (c.61C>T: p.R21C) in AMER1, the gene encoding APC membrane recruitment protein 1, detected in two families and predicted to be “Damaging” by both SIFT and Polyphen-2. AMER1, which is located on the X chromosome in humans, specifically at Xq11, has been shown to be the pathological gene responsible for osteopathia striata with cranial sclerosis (OSCS, MIM: 300373) [5], an X-linked dominant developmental disease manifesting mainly in females as macrocephaly, cleft palate, mild learning disabilities, sclerosis of the long bones and skull, and longitudinal striations visible on radiographs of the bones [6]. The disorder is usually considered lethal in males. The phenotype of microtia-atresia, however, has not been previously associated with either OSCS or AMER1.
Tissue-tissue interactions mediated by the Tgf-β, Wnt, Fgf, and Hh signaling molecules have been found to play key regulatory roles during the early stages of craniofacial development [7]. The Wnt signaling pathway has been shown to contribute to the development and homeostasis of craniofacial tissues [8], as well as being involved in the morphogenesis of craniofacial cartilage [9]. The protein encoded by AMER1 was found to exert an antagonistic effect on the Wnt/β-catenin pathway [4,10]. This protein, however, is also required for the phosphorylation of the LRP6 receptor [11]. The association of the dichotomic nature of this complex interaction with craniofacial development, however, has not yet been analyzed [12,13].
Taken together, the above findings suggested that AMER1 may play an important pathogenic role in the development of microtia-atresia. Although amer1 has been reported to be dispensable in the development of zebrafish [14], amer1 may be associated with early craniofacial development in the zebrafish. The present study, therefore, assessed the effects of amer1 knockdown using CRISPR/Cas9 on abnormal phenotypes of inbred mutant offspring and on the association of these phenotypes with dysregulation of the Wnt pathway. These findings suggested that amer1 may regulate key developmental processes and chondrogenic differentiation of cranial neural crest cells (CNCCs), and may interact with the Wnt/β-catenin pathway, showing that amer1 may play a crucial regulatory role in early craniofacial development in zebrafish.
2. Results
2.1. Homology and Expression Profile of AMER1 in Zebrafish
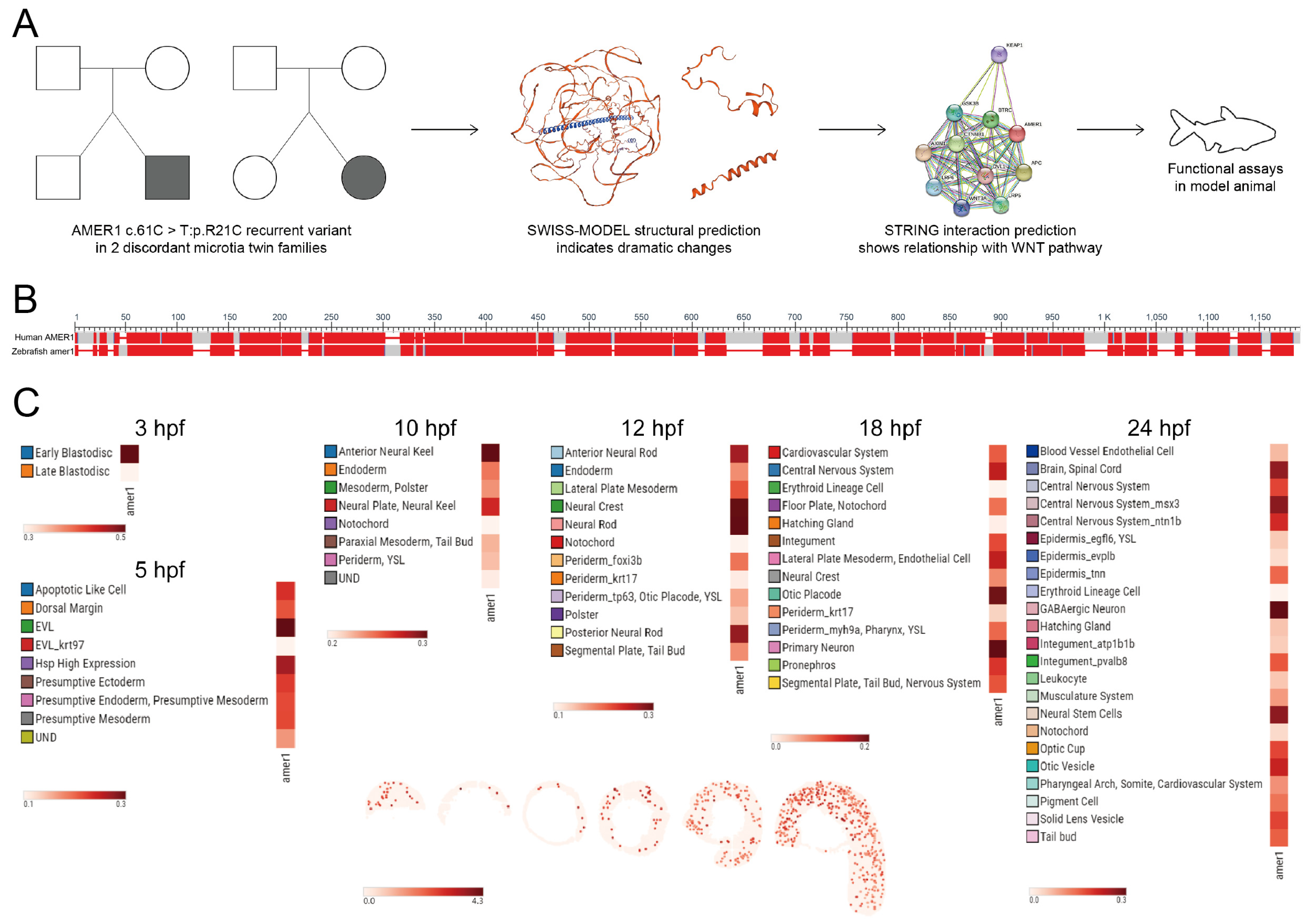
Evaluation of two families having twins with microtia showed that the AMER1 (c.61C>T: p.R21C) variant was present in both sets of twins, with both SIFT and Polyphen-2 predicting that this variant was “Damaging” and suggesting that AMER1 may be potentially pathogenic (Figure 1A) [4]. Bioinformatics analyses also showed structural alterations in this gene and its functional association with the Wnt pathway. The protein encoded by AMER1 was found to consist of 1135 amino acids, containing a region with six conserved block sequences and a highly divergent region. Its functional domains include those involved in phospholipid binding, APC interaction, and β-catenin binding. Evaluation of genes homologous to human AMER1 in zebrafish by NCBI BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 3 December 2022)) (Figure 1B) identified a zebrafish gene encoding a 953-amino acid protein. The human and zebrafish protein sequences were 35.91% identical, with an expected value of 2 × 10−70, indicating a high degree of homology.
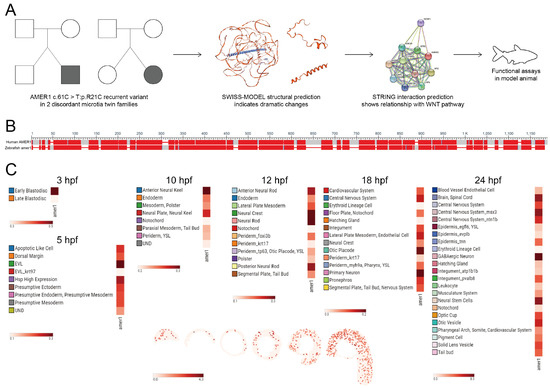
Figure 1.
Identification of AMER1 and its homologous gene in zebrafish. (A) Schematic diagram of AMER1 sequences in families of patients with dichotomous monozygotic twins with microtia. The AMER1 variant (c.61C>T: p.R21C) was detected in two patients with sporadic microtia (shaded), as well as in their normal twins and parents (blank). The predicted protein structure showed dramatic changes from wildtype, and pathway prediction showed a relationship between AMER1 and the Wnt pathway. (B) BLAST analysis of sequences homologous to human AMER1 in zebrafish. (C) Expression profile of zebrafish amer1, as determined using the Spatial Transcript Omics DataBase.
The spatiotemporal pattern of amer1 expression during zebrafish embryonic development was examined using two approaches. The expression of amer1 during the early stages of zebrafish embryonic development was evaluated initially using the recently established ZESTA database (https://db.cngb.org/stomics/zesta/ (accessed on 19 December 2022)) (Figure 1C, Supplementary Figure S1). Examination of the scRNA data showed that, at 10 h post-fertilization (hpf), amer1 was expressed in the anterior neural keel and neural plate. At 12 hpf, amer1 was mainly expressed in the neural crest and neural rod, whereas, at 18 and 24 hpf, amer1 was expressed in the otic placode, otic cup, and otic vesical at 18 and 24 hpf (Figure 1C). Evaluation of the stereoseq data at six time points (3, 5.25, 10, 12, 18, 24 hpf) yielded results similar to those of the scRNA data, with amer1 mainly expressed in the otic vesicle at 18 and 24 hpf (Supplementary Figure S1).
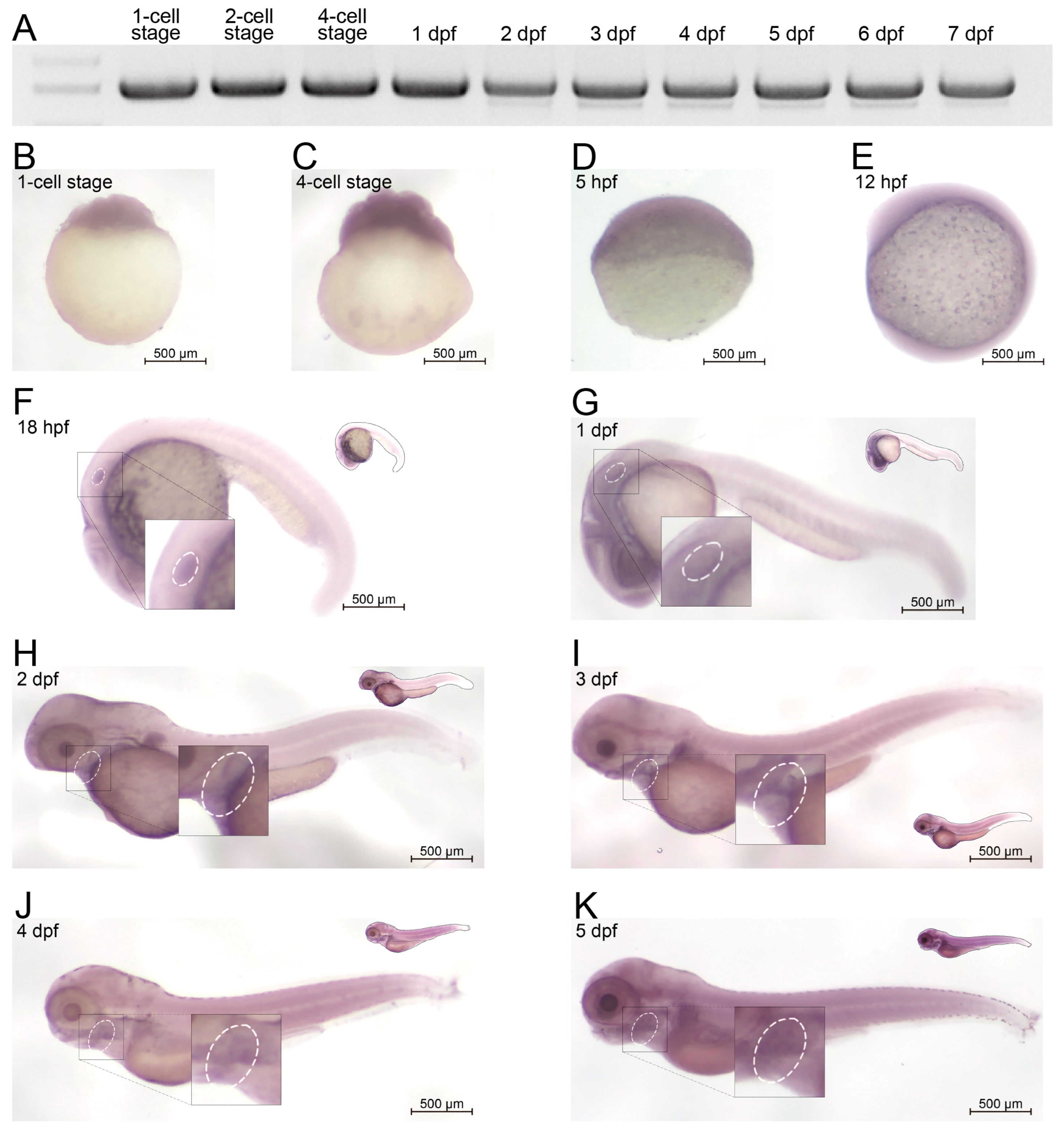
To evaluate the in vivo expression profile of amer1 in zebrafish, RNA was extracted from zebrafish embryos at different developmental stages and cDNA libraries were constructed. The expression of amer1 could be detected beginning at the one-cell stage up to 7 days post-fertilization (dpf) (Figure 2A). To confirm these findings and to broaden the time window for expression, embryos during the first 6 days of development were subjected to whole-mount in situ hybridization (Figure 2B–K). Expression of amer1 was detected in zebrafish ova, indicating maternal expression of this gene, similar to findings in the ZESTA database (Figure 2B). At 18 hpf, amer1 expression could be observed in the otic vesicle (Figure 2F). At 1 dpf, amer1 was detected in the pharyngeal arches (Figure 2G), with this gene being mainly expressed in the pharyngeal arches beginning at 48 hpf (Figure 2H–K), indicating that this gene plays a role in pharyngeal development.
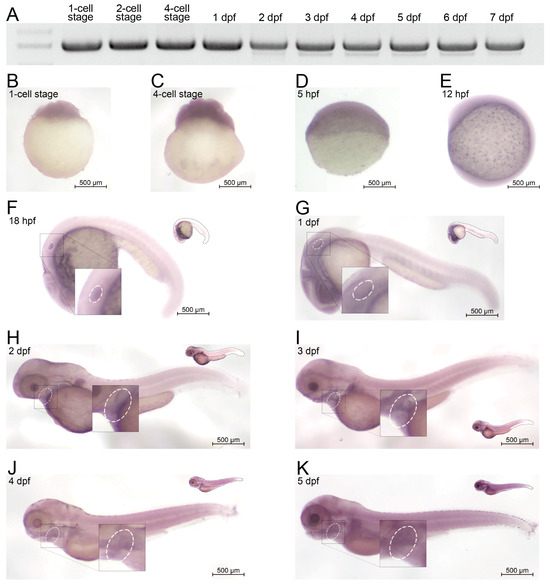
Figure 2.
Expression of amer1 in zebrafish. (A) RT-PCR analysis, showing that amer1 was constitutively expressed in zebrafish embryos, beginning at the 1-cell stage to 7 dpf. (B–K) In situ hybridization of amer1, showing that (F) amer1 was expressed in otic vesicles (white circle) at 18 hpf; and that (H–K) concentrated expression of amer1 in the mandible region (white circle) was observed beginning at 2 dpf.
2.2. Knockdown of amer1 Results in Severe Pharyngeal Malformation
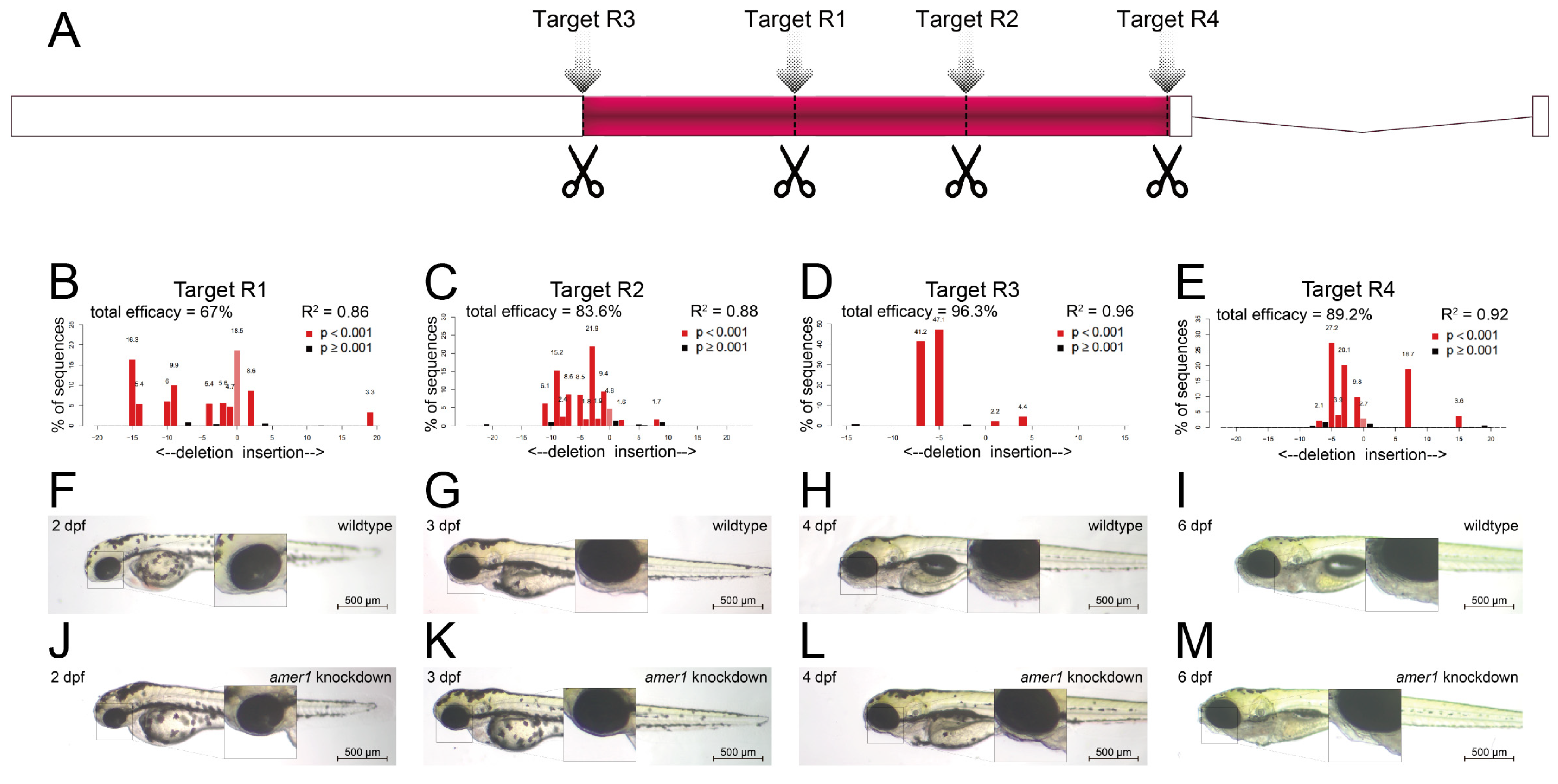
To assess the effects of amer1 variants on pharyngeal development, amer1 expression was knocked down in zebrafish using the CRISPR/Cas9 technique [15]. Four gRNAs were designed and validated (Figure 3A–E, Supplementary Table S2), with each co-injected with Cas9 into one-cell embryos. Evaluation of representative amer1 knockdown embryos at 1 dpf showed that the four gRNAs had efficacies of 67.0%, 83.6%, 96.3%, and 89.2%, respectively.
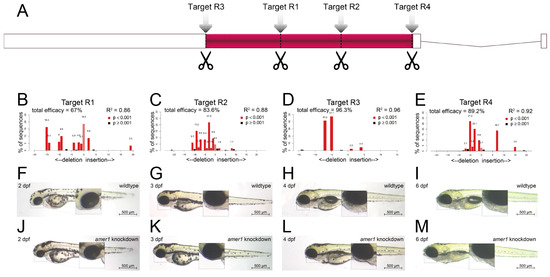
Figure 3.
Knockdown of amer1 and phenotypic changes. (A) Schematic diagram showing the four amer1 gRNA targets of the CRISPR/Cas9 knockdown system. (B–E) PCR analysis, showing representative amer1 knockdown efficiencies of the four gRNA targets. (F-M) Brightfield images of embryos of (F–I) wildtype and (J–M) amer1 knockdown zebrafish at (F,J) 2, (G,K) 3, (H,L) 4, and (I,M) 6 dpf, showing malformed mandibles in the amer1 knockdown embryos.
The gRNA (R3) with the highest knockdown efficacy was selected for the construction of a zebrafish amer1 knockdown model. The phenotypes of the wildtype and amer1 mosaic knockdown zebrafish were compared (Figure 3F–M). The amer1 knockdown embryos were observed to have no obvious phenotype anomalies during the early developmental stages (Figure 3F,J). At 3 dpf, however, when the wildtype embryos began to form the structure of the lower jaw, the amer1 knockdown embryos abnormally lacked visible signs of mandible development (Figure 3G,K). These differences were even more apparent after 4 dpf, as the bladder began to inflate and the lower jaw continued to protrude forward in wildtype embryos, whereas neither the bladder nor the lower jaw showed signs of development in the amer1 knockdown embryos (Figure 3H,L). The differences persisted until 6 dpf, with all abnormal zebrafish dying of breathing and/or swallowing difficulties (Figure 3I,M). The otocysts and otoliths of wildtype and amer1 knockdown embryos were essentially the same, however, indicating that the differences in lower jaw and bladder development were not caused by overall developmental delays, but by defects in the development of these specific tissues.
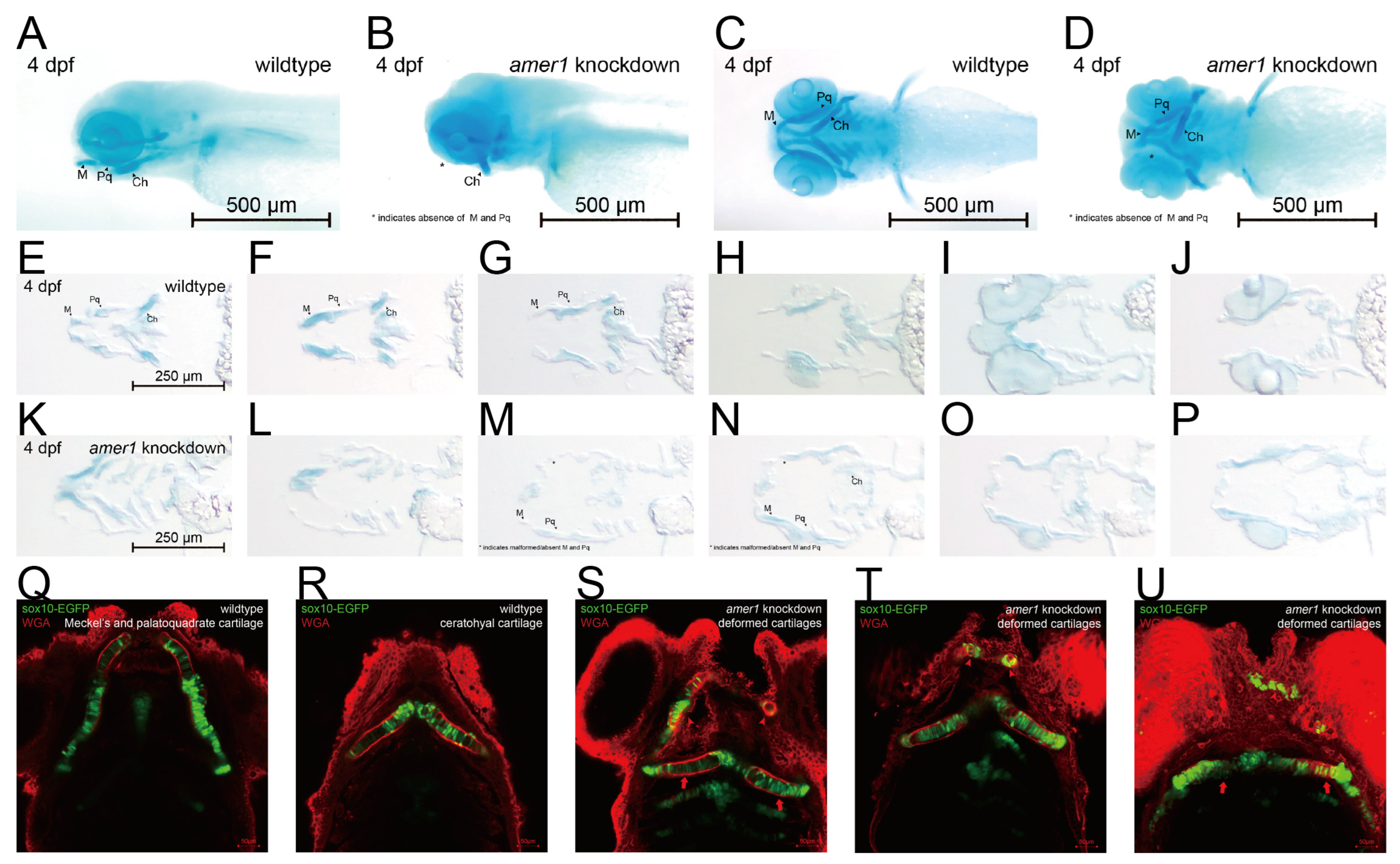
Differences in mandible development between wildtype and amer1 knockdown embryos were further demonstrated by Alcian staining of pharyngeal arch cartilage from complete and sliced embryos at 4 dpf (Figure 4A–P). Partial loss and malformation of Meckel’s and the palatoquadrate cartilage were observed in the amer1 knockdown embryos (Figure 4A–D). Staining of the slices more clearly demonstrated the deformed cartilages (Figure 4M–O). To further examine the structure of pharyngeal chondrocytes at a cellular level, cartilage samples were also stained with wheat germ agglutinin (WGA) (Figure 4Q–U). Chondrocytes in Meckel’s cartilage of amer1 knockdown embryos were swollen and their alignment was disoriented (Figure 4S–U), indicating severe defects in cartilage development. To further validate that loss of amer1 is the cause of the abnormal pharyngeal arch development, we performed a rescue experiment with amer1 mRNA and observed a complete reversal of the abnormal phenotype (Supplementary Figure S2).
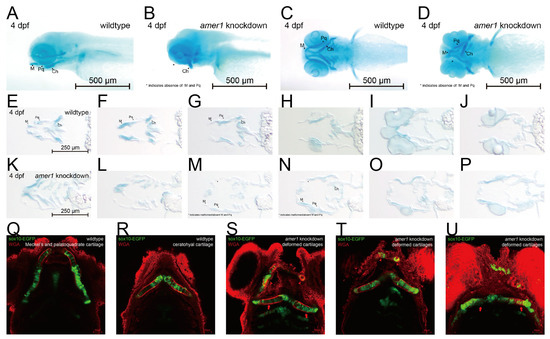
Figure 4.
Phenotypic changes induced by amer1 knockdown, as shown by Alcian and WGA staining. (A–D) Alcian staining of embryos of (A,C) wildtype and (B,D) amer1 knockdown embryos at 4 dpf, showing malformation of Meckel’s and palatoquadrate cartilages in the amer1 knockdown embryos. (E–P) Alcian staining of tissue sections of embryos of (E–J) wildtype and (K–P) amer1 knockdown zebrafish at 4 dpf, showing that Meckel’s cartilage was severely deformed in amer1 knockdown embryos (red rectangles in (M–O)). (Q,R) WGA staining of wildtype embryos, showing regularly distributed, well-shaped chondrocytes, with Meckel’s cartilage and ceratohyal cartilage normally not observed in the same plane. (S–U) WGA staining of amer1 knockdown embryos, showing Meckel’s cartilage (red arrowheads) and palatoquadrate and ceratohyal cartilage (long red arrows) in the same plane. All cartilage was deformed and the arrangement of chondrocytes was disordered.
2.3. Inbred Homozygous Zebrafish Demonstrates Similar Phenotype after Excluding Potential Compensation
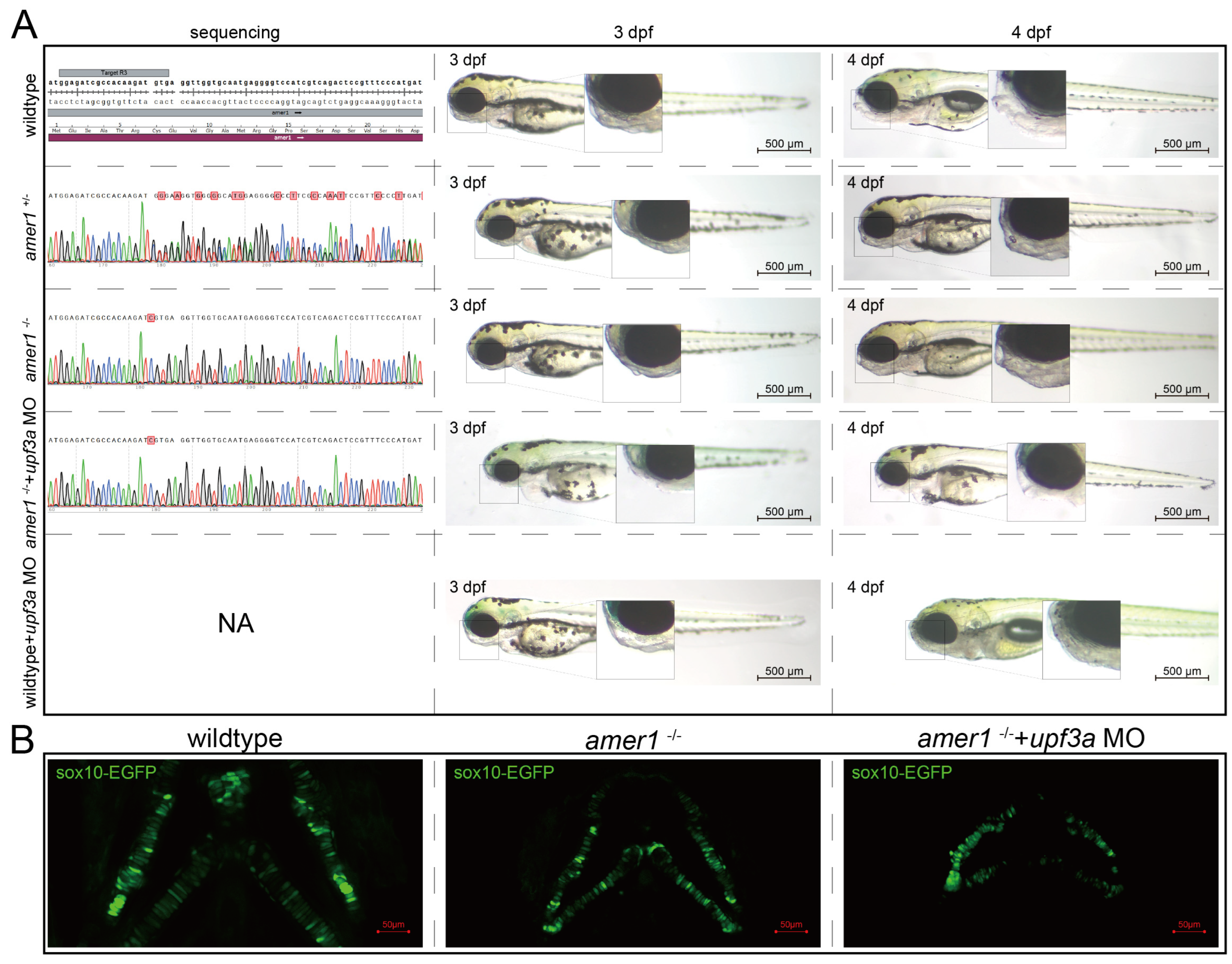
To determine if the abnormal pharyngeal trait could be passed on, F0 mosaic knockdown zebrafishes were mated to obtain stable heterozygotic amer1+/− F1 fishes. These fishes showed no apparent developmental anomalies, and their genotypes were validated. F1 males and females with the same genotype were mated to obtain amer1−/− F2 fishes, with genotyping showing that approximately 1/4 of these F2 fishes were homozygotic amer1−/−. However, these amer1−/− fishes did not demonstrate visible malformation during early developmental stages (0 hpf to 6 dpf) (Figure 5A). WGA staining of their cartilage also showed no apparent deformation or disorderly arrangement of their chondrocytes (Figure 5B). Examination of the altered DNA sequence “TCGTGA” showed that an extra C was present in the encoded mRNA, causing a shift of the reading frame and a pre-mature “UGA” termination codon. Because a compensation effect may have been triggered [16], Upf3a morpholino was injected into amer1−/− embryos to mitigate potential genetic compensatory effects. Sole injection of upf3a morpholino into wildtype embryos did not induce apparent anomalies. After injection, the offspring demonstrated a 3:1 phenotypic segregation of lower jaw deformity vs. normal fishes (Figure 5A). Fluorescence imaging revealed swallowed and misaligned chondrocytes in amer1−/− embryos treated with upf3a morpholino (Figure 5B). Unfortunately, amer1−/− zebrafish could not stably pass on this genotype, as these fish died of breathing and swallowing difficulties after 6 dpf.
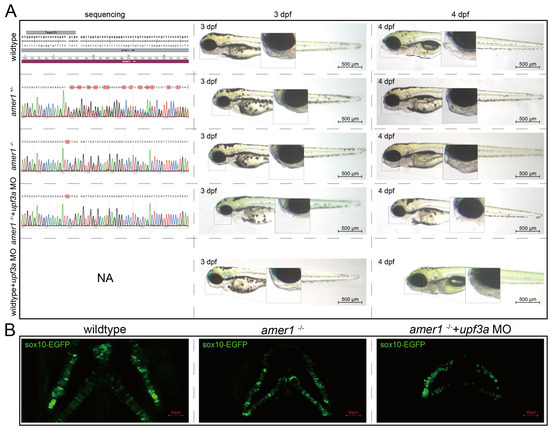
Figure 5.
Genotypes and phenotypes of F2 zebrafish. (A) Sequencing results and brightfield images of F2 zebrafishes at 3 and 4 dpf with five different genotypes/treatments: wildtype, amer1+/−, amer1−/−, and amer1−/− + upf3a MO, wildtype + upf3a MO. amer1−/− F2 embryos treated with upf3a MO showed malformation of the mandible. Red suqares indicate mutated sites. NA: not applicable. (B) Fluorescence staining showing that chondrocytes were regularly distributed in wildtype and amer1−/− F2 embryos, but were disarranged in amer1−/− F2 embryos treated with upf3a MO.
2.4. Knockdown of amer1 Affects Chondrocyte Differentiation, Proliferation, and Apoptosis
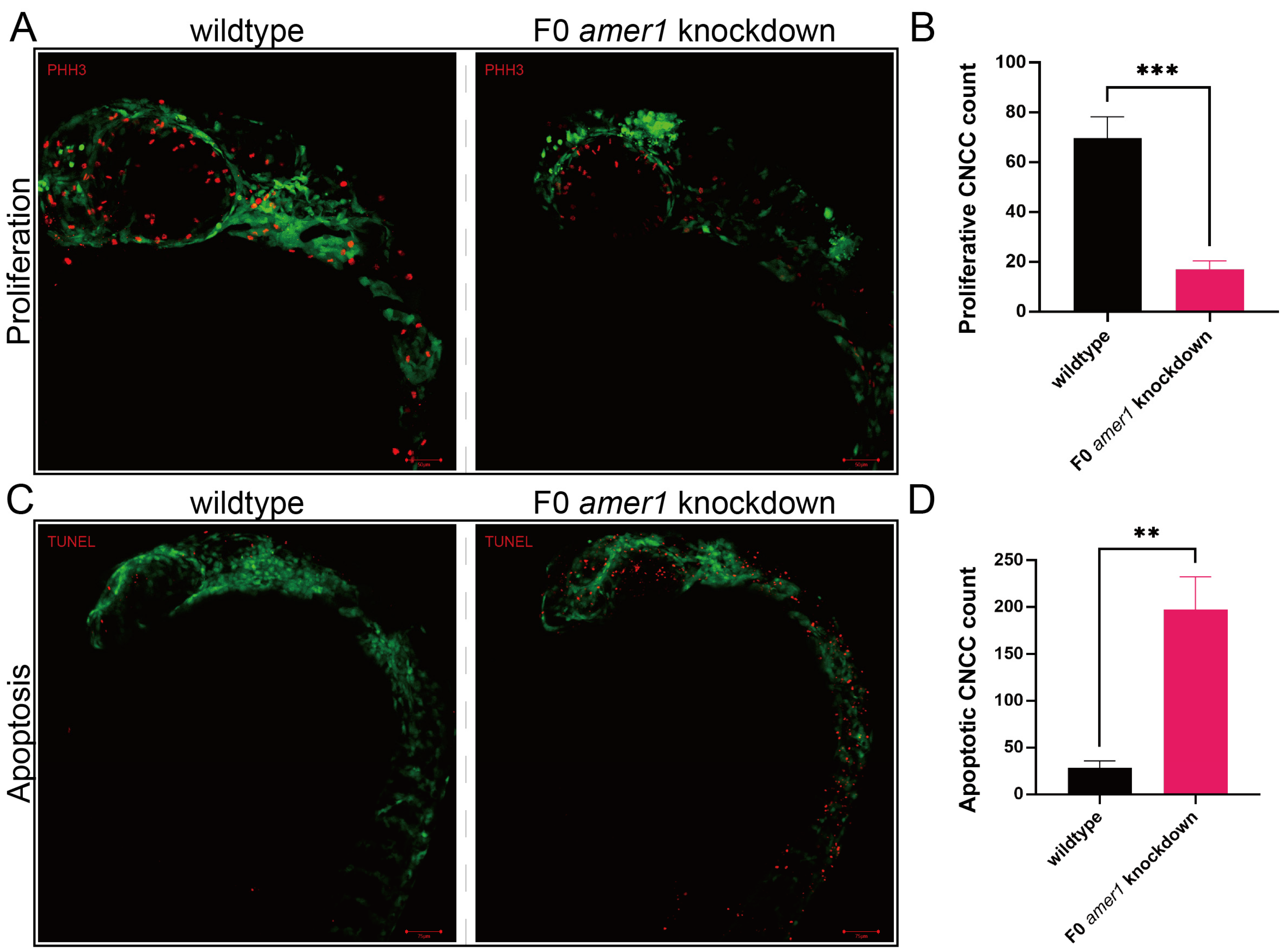
Because malformation of multiple cartilages was observed in amer1 knockdown embryos, the process by which chondrocytes develop from CNCCs was evaluated in zebrafish embryos. Determination of the numbers of CNCCs in wildtype and F0 amer1 knockdown embryos at 24 hpf showed that the proliferation of CNCCs, as determined by staining for PH3, was significantly lower in amer1 knockdown than in wildtype embryos (17.00 ± 3.46 vs. 69.67 ± 8.62 cells per fish, p = 0.0006) (Figure 6). Conversely, the number of cells that had undergone apoptosis, as determined by TUNEL staining, was significantly higher in amer1 knockdown than in wildtype embryos (197.33 ± 35.36 vs. 28.33 ± 7.50 cells per fish, p = 0.0013) (Figure 6).
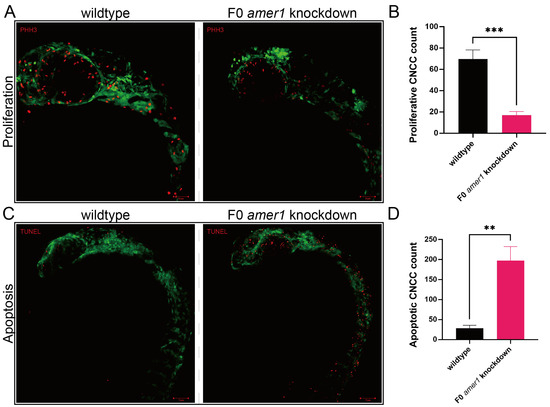
Figure 6.
Proliferation and apoptosis of CNCCs in zebrafish embryos. (A) Proliferation of CNCCs, as shown by PHH3 staining, in wildtype and F0 amer1 knockdown embryos. (B) Quantitation of CNCC proliferation in wildtype and amer1 knockdown embryos (n = 3 each). (C) Apoptosis of CNCCs, as shown by TUNEL staining. (D) Quantitation of CNCC apoptosis in wildtype and amer1 knockdown embryos (n = 3 each). ** p < 0.01; *** p < 0.001.
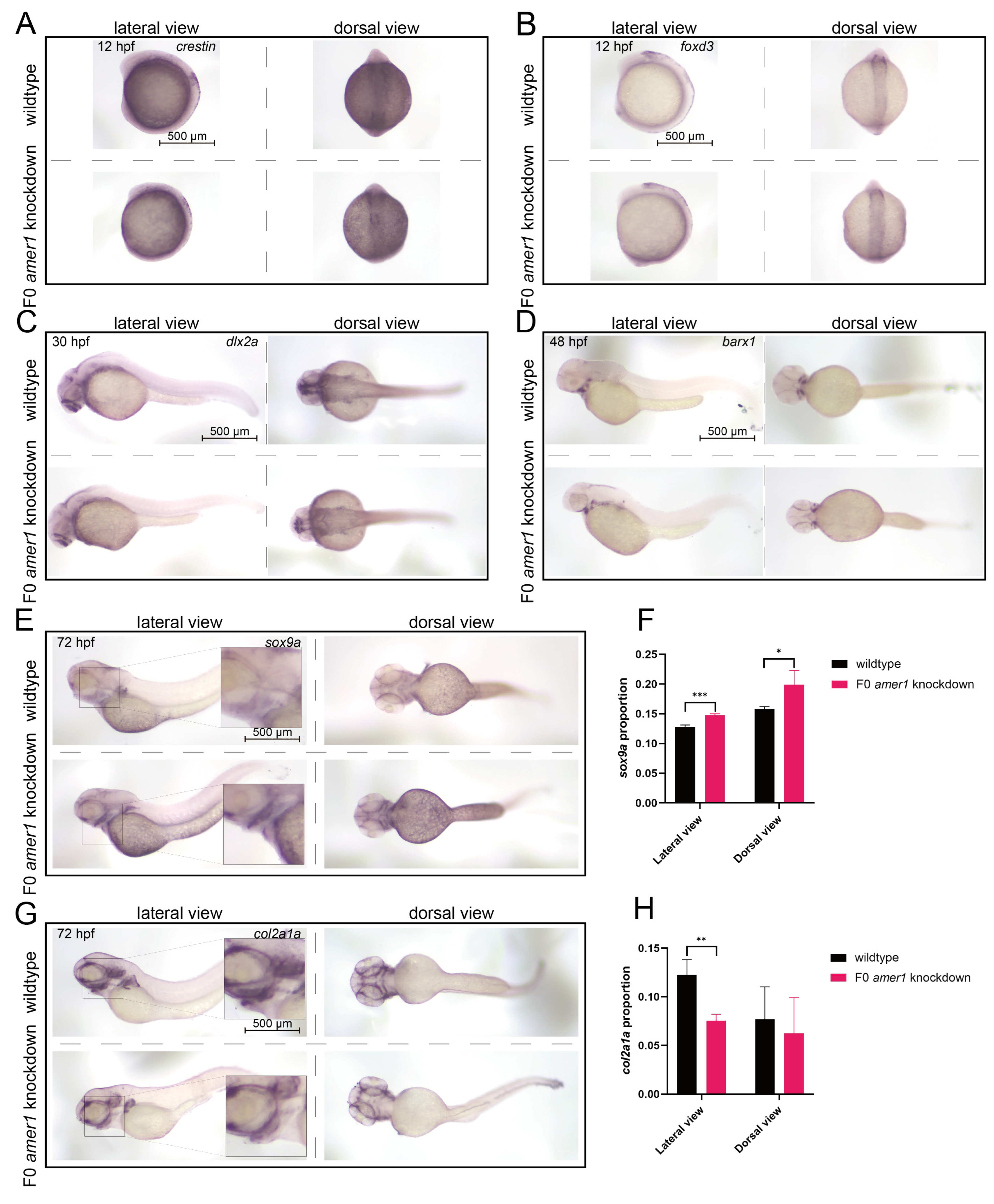
To evaluate the effects of amer1 knockdown on the specific developmental stages from CNCCs to chondrocytes, marker genes for CNCC development were assayed in zebrafish embryos of various developmental stages by in situ hybridization. The levels of expression of crestin and foxd3 were similar in wildtype and F0 amer1 knockdown embryos at 12 hpf, indicating that CNCC formation was not affected by amer1 knockdown (Figure 7A,B). The levels of expression of dlx2a in wildtype and F0 amer1 knockdown embryos at 30 hpf also showed no apparent differences, suggesting that amer1 was not necessary for the specialization of CNCCs to pharyngeal arch CNCCs (Figure 7C). The area of expression of barx1 was smaller in amer1 knockdown than in wildtype embryos at 48 hpf, although their expression patterns were similar, indicating that amer1 knockdown had a slight effect on the number of CNCCs rather than on the coagulation of mesenchymal cells (Figure 7D). Expression of the chondrogenic markers sox9a and col2a1a in wildtype and F0 amer1 knockdown embryos at 72 hpf differed significantly, with sox9 being significantly upregulated (p = 0.0007 for lateral view and p = 0.0441 for dorsal view) and col2a1a significantly downregulated (p = 0.0092 for lateral view and p = 0.6436 for dorsal view) in F0 amer1 knockdown embryos (Figure 7E–H). Taken together, these results showed that chondrocyte differentiation and collagen formation were partially blocked by amer1 knockdown.
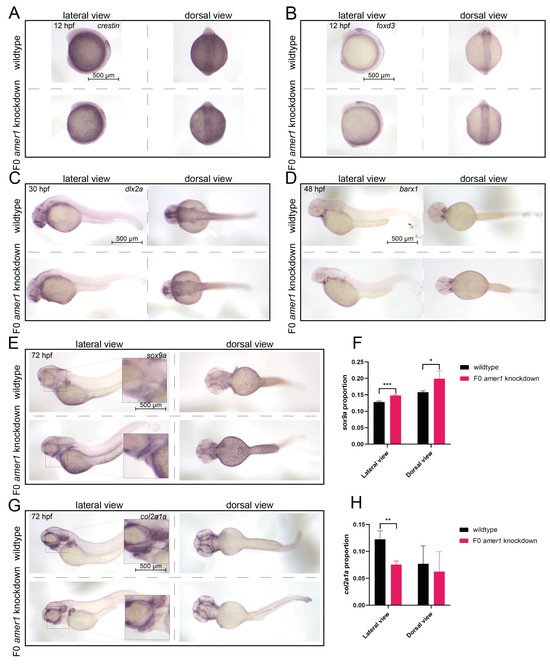
Figure 7.
Expression of CNCC related markers in zebrafish embryos. In situ hybridization, showing the expression of (A) crestin and (B) foxd3 at 12 hpf, (C) dlx2a at 30 hpf, and (D) barx1 at 48 hpf in wildtype and F0 amer1 knockdown embryos. (A–C) The levels of expression of crestin, foxd3, and dlx2a showed no apparent differences in wildtype and amer1 knockdown embryos. (D) The expression pattern of barx1 was smaller in amer1 knockdown than in wildtype embryos. (E,F) The expression of sox9a at 72 hpf was increased in amer1 knockdown than in wildtype embryos (for quantitative analysis, n = 3 each). (G,H) The expression of col2a1a at 72 hpf was decreased in amer1 knockdown than in wildtype embryos (for quantitative analysis, n = 3 each). * p < 0.05; ** p < 0.01; *** p < 0.001.
2.5. Amer1 Exerts Its Effects via the Wnt/β-Catenin Signaling Pathway
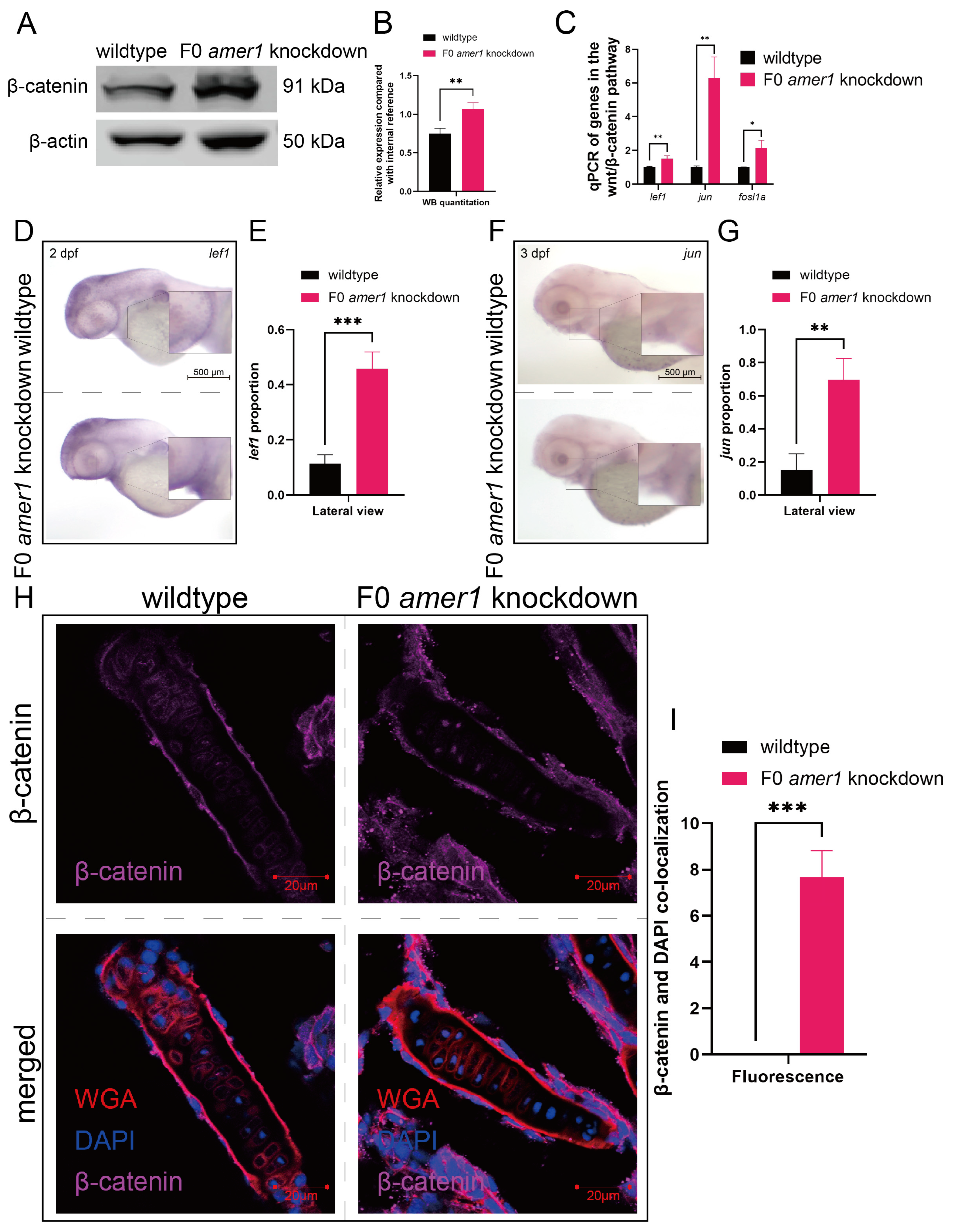
Because the amer1 gene has been associated with the Wnt/β-catenin pathway, a vital regulatory pathway in early craniofacial and pharyngeal arch development, amer1 knockdown may cause malformations in amer knockdown embryos by interfering with the wnt/β-catenin pathway. Western blotting of full-embryo proteins extracted from wildtype and F0 amer1 knockdown embryos at 4 dpf showed that β-catenin expression was significantly higher in amer1 knockdown embryos (Figure 8A,B). Evaluation by qPCR of genes acting downstream of β-catenin showed that the levels of expression of several genes, including lef1, jun, and fosl1a, were significantly higher (p = 0.0069 for lef1, p = 0.0020 for jun, p = 0.0113 for fosl1a) in amer1 knockdown embryos (Figure 8C). Moreover, in situ hybridization showed that lef1 and jun expression was upregulated in the mandible region of amer1 knockdown embryos at 2 and 3 dpf (Figure 8D–G).
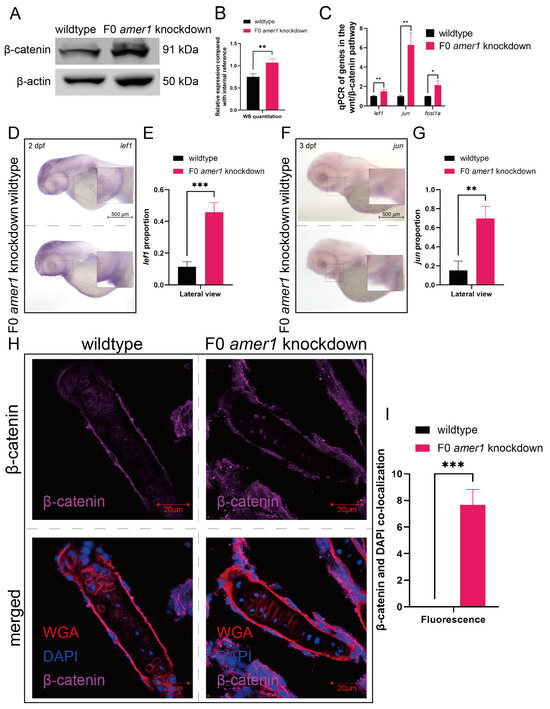
Figure 8.
Expression and localization of components of the wnt pathway in zebrafish embryos. (A,B) Western blotting and quantitation showing that β-catenin levels were higher in amer1 knockdown than in wildtype embryos at 4 dpf. (C) qPCR showing that the levels of expression of lef1, jun, and fosl1a were significantly higher in amer1 knockdown embryos. (D–G) Validation of the expression of (D) lef1 and (F) jun by in situ hybridization and their quantitation (E,G) (for quantitative analysis, n = 3 each). (H) Immunofluorescence staining of the ceratohyal cartilage using WGA, β-catenin, and DAPI in wildtype and amer1 knockdown embryo sections at 4 dpf. (I) Manual counting of co-localization of β-catenin with cell nuclei (n = 3 each). * p < 0.05; ** p < 0.01; *** p < 0.001.
Upregulation of these canonical TCF/β-catenin target genes suggested the possible translocation of β-catenin. Immunofluorescence staining of tissue sections from wildtype and amer1 knockdown embryo sections of the ceratohyal cartilage plane at 4 dpf showed that β-catenin was ubiquitously expressed in the cytoplasm of cells of the ceratohyal cartilage of wildtype embryos, but co-localized with the nucleus of mutant embryos (Figure 8H,I).
2.6. An Inhibitor of the Wnt/β-Catenin Signaling Pathway Rescues the Malformation of amer1 Knockdown Embryos
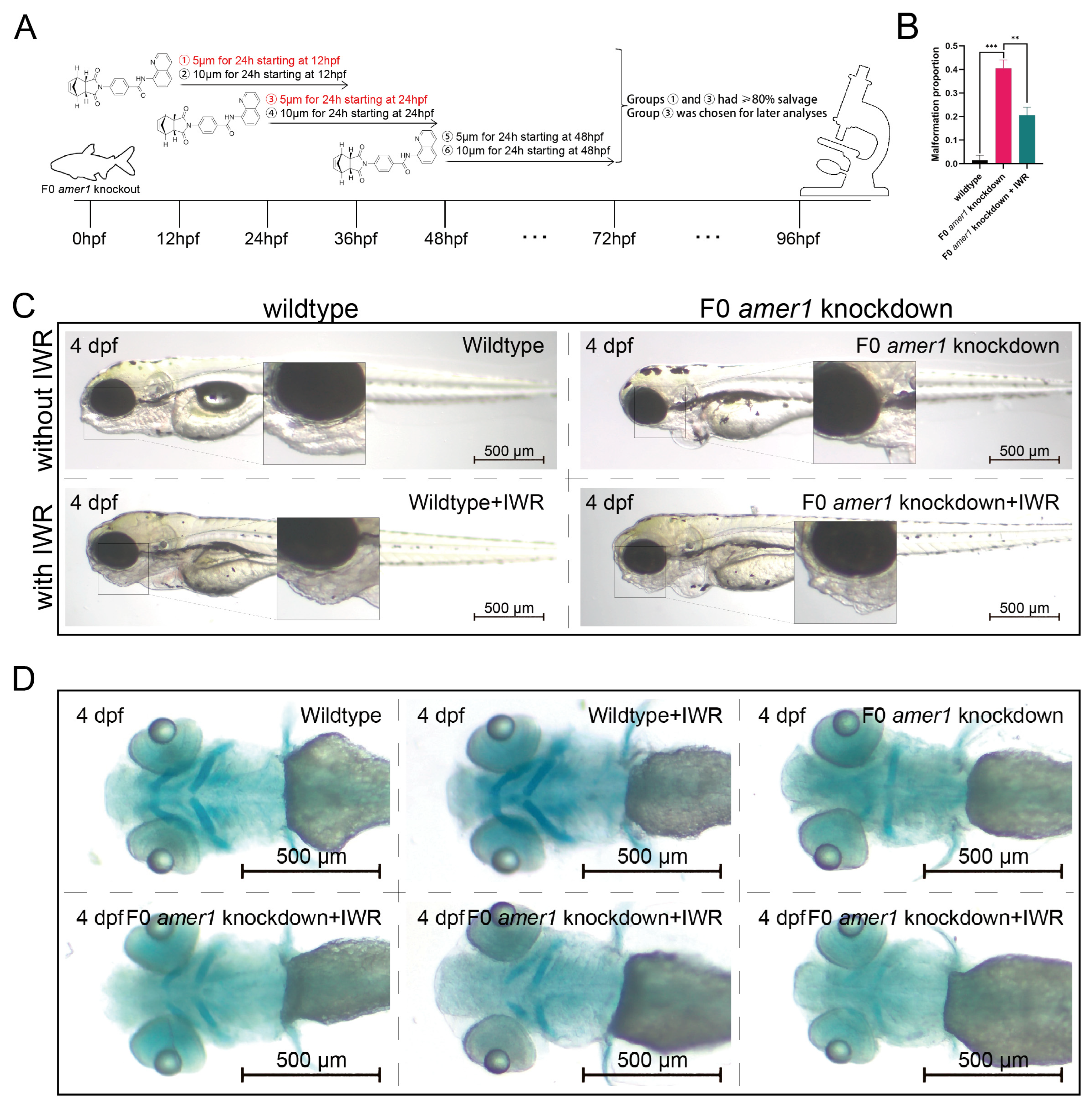
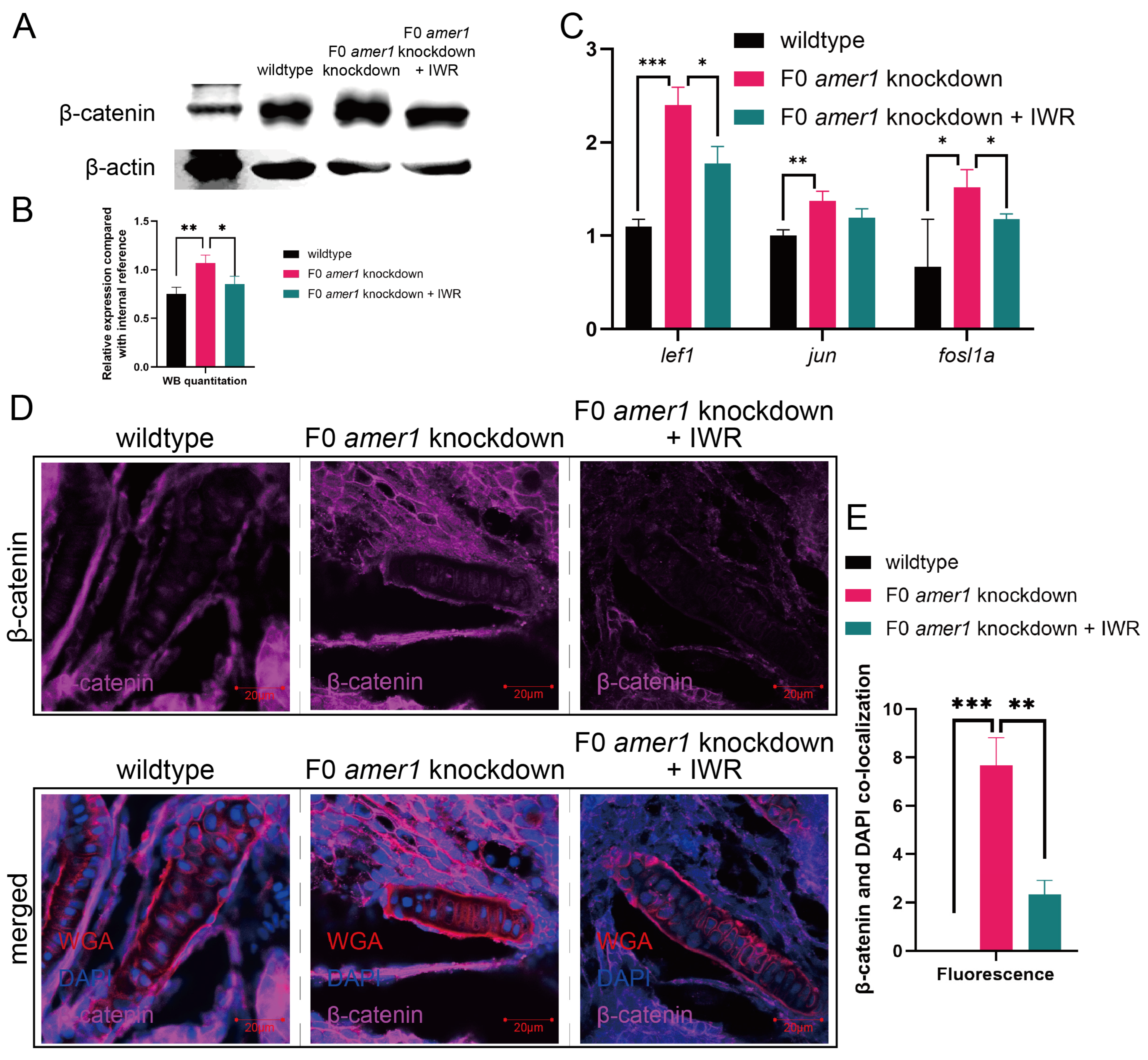
To further explore the causative role of dysregulation of the Wnt/β-catenin signaling pathway in the pathogenicity of amer1 mutation, F0 amer1 knockdown embryos were treated with IWR-1-endo, a reversible inhibitor that promotes β-catenin phosphorylation by interacting with axin, and observed whether IWR-1-endo could rescue embryos from the malformed phenotype. Mutant embryos at 12, 24, and 48 hpf were treated with 5 and 10 μM IWR-1-endo for 24 h; the inhibitor was removed; and the embryos were fixed and observed at 96 hpf (Figure 9A). Treatment of embryos at 12 and 24 hpf with 5 μM IWR-1-endo for 24 h resulted in phenotypic salvage of more than 80% of embryos, with application starting at 24 hpf being the most effective dose with the lowest toxicity. The malformation rate of amer1 knockdown embryos was significantly lower in the presence that in the absence of IWR-1-endo (20.50% vs. 40.50%, p = 0.009), indicating successful rescue (Figure 9B), with similar results observed on representative brightfield images (Figure 9C). Alcian staining of the pharyngeal arch cartilage in 100 amer1 knockdown embryos treated with IWR-1-endo at 4 dpf showed that cartilage development could be divided into five categories: normal (n = 53), mild dysplasia (n = 21), partial loss (n = 15), complete loss (n = 3), and others (including dead embryos) (n = 8) (Figure 9D). Western blotting of β-catenin and qPCR of canonical TCF/β-catenin target genes demonstrated that the expression levels of β-catenin, lef1, jun, and fosl1a were all reduced by IWR-1-endo treatment, to concentrations similar to those in wildtype embryos (Figure 10A–C). Immunofluorescence staining demonstrated that co-localization of β-catenin with the nucleus in amer1 knockdown embryos was lower in the presence than in the absence of IWR-1-endo (Figure 10D,E).
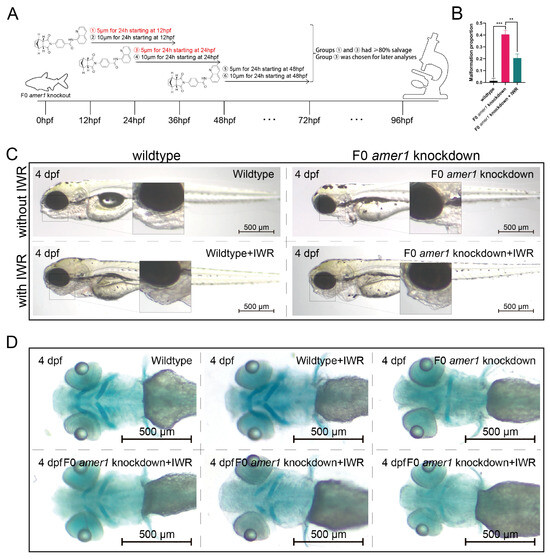
Figure 9.
Phenotypic rescue of F0 amer1 knockdown embryos using IWR-1-endo. (A) Schematic diagram of the rescue and observation process. Different numbers indicate different treatment. (B) Proportions of malformed wildtype, amer1 knockdown and IWR-treated amer1 knockdown embryos, showing that the proportion of malformed IWR-treated amer1 knockdown was significantly lower than that of amer1 knockdown embryos. (C) Representative brightfield images of wildtype, amer1 knockdown embryos, and IWR-treated amer1 knockdown embryos. (D) Representative Alcian staining images of wildtype (first row, left), IWR-treated wildtype (first row, middle), amer1 knockdown (first row, right), and IWR-treated amer1 knockdown embryos showing normal development of pharyngeal arch cartilage (second row, left), partial loss (second row, middle), and complete loss (second row, right). ** p < 0.01; *** p < 0.001.
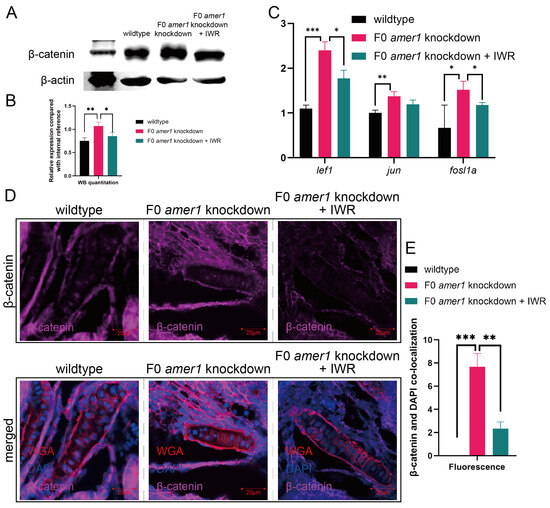
Figure 10.
Effects of IWR-1-endo on components of the Wnt pathway in F0 amer1 knockdown embryos. (A,B) Western blotting and quantitation showing that β-catenin expression was lower in IWR-treated amer1 knockdown embryos than in amer1 knockdown embryos at 4 dpf. (C) qPCR showing that the levels of expression of lef1, jun, and fosl1a were lower in IWR-treated than in untreated amer1 knockdown embryos. (D) Co-localization of β-catenin with the cell nucleus in the ceratohyal cartilage was reduced in amer1 knockdown embryos treated with IWR. (E) Manual counting of co-localization of β-catenin with cell nuclei in embryos with different treatments (n = 3 each). * p < 0.05; ** p < 0.01; *** p < 0.001.
3. Discussion
Because both environmental and genetic factors may contribute to the etiology of microtia, it is difficult to investigate these factors separately, especially in patients with sporadic microtia and no apparent family history. This issue was previously addressed by recruiting six families with monozygotic twins having discordant microtia/normal phenotypes [4]. These twins grew in the same environment and their genotypes were nearly identical, enabling isolation of a genetic component contributing to the etiology of microtia. The findings of that study suggested that AMER1 (c.61C>T: p.R21C) may be a pathogenic genetic variant of AMER1, as not only was this variant observed in more than one sporadic family, but it resulted in both structural alteration and predicted functional damage.
Previous studies have assessed the consequences of the loss-of-function of AMER1 in zebrafish and mice, but these results were inconsistent [14,17,18]. Knockdown of Amer1 in mice resulted in neonatal lethality, somatic overgrowth, and deficiencies in multiple mesenchyme-derived tissues [17], and a previous study of morpholino-based amer1 knockdown in zebrafish revealed abnormal phenotypes, including anterior truncations, epiboly defects, headless malformations, and small eyes [18]. In contrast, another study in zebrafish based on morpholino knockdown and TALEN knockdown techniques found no apparent abnormal phenotypes, suggesting that amer1 was dispensable in zebrafish [14]. The present study used the CRISPR/Cas9 strategy and four gRNAs to knock down amer1 expression, with the gRNA having the highest efficiency being chosen for further evaluation. Morpholino-based knockdown of amer1 and inbred fish lines were both utilized to validate phenotypic findings, with the abnormal phenotype in zebrafish being homologous to the malformations observed in patients with microtia [4]. These findings suggest that amer1 can regulate embryonic and craniofacial development in zebrafish.
Three major hypotheses to date have been associated with microtia-atresia and its related craniofacial malformations: vascular abnormality, damage to Meckel’s cartilage, and disruption of CNCC development [19]. Amer1 knockdown in mice has been reported to affect multiple mesenchyme-derived tissues, suggesting that this gene affects the development of CNCCs, which form a mesenchymal core between the ectoderm and endoderm along with mesoderm cells and play an important role in the development of cartilage [17]. PH3 and TUNEL staining differed significantly between amer1 knockdown and control zebrafish, suggesting that amer1 knockdown reduced the numbers of CNCCs by reducing cell proliferation and enhancing apoptosis. In situ hybridization assessing the expression of marker genes during each step of CNCC development showed that the expression of sox9a was significantly upregulated and that of col2a1a was downregulated in amer1 knockdown zebrafish. During chondrogenesis, sox9a and col2a1 are normally expressed in parallel, with Col2a1 acting directly downstream of sox9a [20,21]. The findings of the present study suggest that amer1 knockdown affected the differentiation of CNCCs to chondrocytes, with the upregulation of sox9a likely caused by a negative feedback mechanism.
Studies have evaluated the relationship between AMER1 and the Wnt/β-catenin pathway. For example, AMER1 was found to inhibit the Wnt pathway through its assembly of the β-catenin destruction complex at the plasma membrane, resulting in the induction of β-catenin degradation, with in vitro knockdown of AMER1 in cell lines resulting in the activation of Wnt target genes [22]. AMER1, however, has also been shown to act as a scaffold protein to stimulate LRP6 phosphorylation, a key step in Wnt/β-catenin signaling, with knockdown of AMER1 reducing Wnt-induced LRP6 phosphorylation [23]. These dichotomic regulatory roles of AMER1 provide an answer to its enigmatic relationship with the Wnt/β-catenin pathway [11].
Although interactions between AMER1 and the Wnt/β-catenin pathway have been explored during the process of tumorigenesis, less is known about this relationship during embryonic and craniofacial development. Defective cell fate determination caused by Amer1 deficiency in mice was shown to be due to aberrant β-catenin activation [17], and silencing of amer1 in zebrafish was found to activate Wnt/β-catenin reporter genes [18]. The current study assessed whether malformations observed in zebrafish had similar underlying mechanisms. Many of the genes in the Wnt/β-catenin pathway, including lef1, jun, and fosl1a, were found to be significantly upregulated in amer1 mutant zebrafish. A previous study in mice concerning Lmp1 found that Amer1 was downregulated, accompanied by the cytoplasmic enrichment and endonuclear accumulation of β-catenin [13]. This finding was in agreement with the results of the present study, showing that decreased amer1 expression in zebrafish hindered the destruction of β-catenin, leading to its accumulation and translocation from the cytoplasm to the cell nucleus. Taken together, these findings support the hypothesis that amer1 negatively regulates the Wnt/β-catenin pathway during the early stages of embryonic development and amer1 mutation aberrantly activates this pathway, resulting in craniofacial malformation.
To further show that the abnormal phenotype induced by amer1 mutation was due to abnormal activation of the Wnt/β-catenin pathway, two types of rescue analyses, both targeting this pathway, were performed. Treatment of zebrafish embryos with IWR-1-endo inhibitor or expression of its mRNA downregulated the Wnt/β-catenin pathway to near normal levels, as well as reducing the percentage of abnormal embryos. In addition to showing that amer1 regulates the Wnt/β-catenin pathway, these findings support the significance of this pathway in the development of microtia and suggest potential therapeutic targets.
4. Materials and Methods
4.1. Zebrafish and Embryos
Adult zebrafish were maintained under standard conditions [24], and embryos were morphologically staged using standard methods [25]. Tuebingen and the transgenic fish line, Tg (sox10:EGFP) (ID:CZ156, ba2Tg/C) [26], were acquired from China Zebrafish Resource Center (CZRC) (http://www.zfish.cn/ (accessed on 1 May 2021)). All zebrafish experiments were approved by the Animal Ethics Committee of Peking University (protocol code: LSC-ZhangB-3, date of approval: 1 September 2019) and conformed to institutional, local, and national rules and guidelines. All phenotypic experiments were carried out within 5 days post fertilization (dpf).
4.2. Photography and Image Processing
A Stemi 2000-C optical stereomicroscope (Zeiss, Oberkochen, Germany) was used for all brightfield observations, including in situ hybridization. Brightfield images were taken with a Zeiss AxioCam MRc5 camera. Immunofluorescence images were taken with a Zeiss LSM 710 NLO laser scanning confocal microscope and Duoscan System. Images were processed and analyzed by Zeiss Zen 2009.
4.3. Bioinformatic Prediction of amer1 Expression Profile in Zebrafish
The expression of amer1 during the early stages of zebrafish embryonic development was evaluated in the Spatial Transcript Omics DataBase (STOmics DB) (https://db.cngb.org/stomics/ (accessed on 19 December 2022)) [27], both in single-cell RNA (scRNA) sequencing and stereo-sequencing (stereoseq) data. Bin annotation was applied for analysis of stereoseq data.
4.4. Whole-Mount In Situ Hybridization
RNA probes were transcribed from linearized plasmids using the proper RNA polymerase (Roche Diagnostics, Tokyo, Japan; Stratagene Japan, Tokyo, Japan). Whole-mount in situ hybridization and immunostaining were performed as described in [28]. Embryos were fixed overnight in 4% paraformaldehyde, followed by dehydration in methanol and storage in methanol at −20 °C. Primers for in situ hybridization included those for amer1 (forward, 5′-GTACCCAACAGTGACGAGGG-3′; reverse, 5′-GGGCCAAAGGGAGAACTTGA-3′), and for crestin, foxd3, dlx2a, barx1, sox9, col2a1a, fgf3, tbx1, and nkx2.3 (Supplementary Table S1).
4.5. Genetic Manipulation of amer1
Knockdown of amer1 was performed using the CRISPR/Cas9 system [15]. Four different sets of guide RNAs (gRNAs, Supplementary Table S2) were designed and co-injected with Cas9 into 1-cell embryos. The gRNAs were designed prior to the construction of the Genome-Scale Guide Set using the CHOPCHOP website (https://chopchop.rc.fas.harvard.edu (accessed on 11 June 2021)) [29]. gRNAs were generated by PCR with a forward primer containing a T7 promoter, the guide sequence, a scaffold sequence, and a reverse primer encoding the standard chimeric gRNA scaffold “tracer rev” [30]. The efficacy of gRNA was verified by extracting crude genomic DNA from whole zebrafish embryos, followed by PCR amplification with the indicated primers (Supplementary Table S2) and sequencing. A morpholino targeting upf3a was also injected at the dosage of 4 pg per embryo to inhibit the compensatory effects of DNA damage [31]. An amer1-targeting morpholino (5′-CGATCTCCATAATGACACAGGTGAC-3′), synthetized by Gene Tools, LLC, was applied to knockdown amer1 aiming to replicate and to confirm the phenotypic changes associated with amer1 knockdown. Homozygotic embryos were bred, whereas unless stated otherwise, F0 amer1 knockdown embryos were used in most of the experiments.
4.6. Cartilage Staining and Immunofluorescence Staining
Embryos were fixed in 4% paraformaldehyde (PFA) at 4 °C overnight, washed in phosphate-buffered saline containing 0.1% Tween-20 (PBST) for 8 h, and subjected to Alcian blue staining, as described in [32]. Embryos were also fixed and stained with wheat germ agglutinin (WGA; Invitrogen, Carlsbad, CA, USA) and 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, St. Louis, MO, USA) [33].
Tg (sox10:EGFP) zebrafish embryos were fixed with 4% phosphate-buffered paraformaldehyde, washed with PBST for 20 min, and immunostained with antibody to phosphohistone H (PHH3; 1:400; sc-374669, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays were performed using the In Situ Cell Death Detection Kit, TMR red (12156792910, Roche, Basel, Basel state, Switzerland), according to the manufacturer’s instructions. Nuclei were visualized by staining with DAPI; proliferation by staining with anti-PHH3; and apoptosis by TUNEL assays. To ensure that only cranial neural crest cells (CNCCs) were assessed, only cells that were doubly positive for anti-PHH3/TUNEL and EGFP were counted manually.
In the immunofluorescence experiments, embryos were sectioned at the plane of ceratohyal cartilage. Cells were stained with monoclonal mouse anti-β-catenin antibody (1:500, C7207, Sigma-Aldrich, St. Louis, MO, USA), followed by staining with Alexa Fluor 647-conjugated goat anti-mouse IgG H&L (1:500, ab150115, Abcam, Cambridge, UK) or Alexa Fluor 594 conjugated WGA (5 μg/mL, W11262, ThermoFisher, Waltham, MA, USA).
4.7. Statistical Analysis
In all phenotypic observational experiments, at least 20 embryos were observed in each condition. Our experiments showed congruent results, affecting >85% of embryos observed, and all selected images are representative. All statistical analyses were carried out using 3 randomly picked embryos in each group and statistical significance was assessed by unpaired t-tests using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, USA), with p < 0.05 considered statistically significant.
5. Conclusions
In conclusion, the present study identified a gene in zebrafish that was homologous to human AMER1, a candidate gene associated with the pathogenesis of microtia-atresia. The expression profile of amer1 in zebrafish embryos was determined by RT-PCR and in situ hybridization. Genetic manipulation of amer1 in zebrafish using the CRISPR/Cas9 technique resulted in severe pharyngeal malformation and cartilage deformity, with a stable inbred fish line established after excluding potential compensation effects. Chondrocyte differentiation, proliferation, and apoptosis were all affected by amer1 variation. Variations in amer1 upregulated the expression of multiple proteins of the Wnt/β-catenin signaling pathway, as well as altering the intracellular distribution of β-catenin. Treatment of zebrafish embryos with an inhibitor of the Wnt/β-catenin signaling pathway successfully rescued the malformation of the mutant embryos. Taken together, these results support the hypothesis that amer1 regulates zebrafish craniofacial development by interacting with the Wnt/β-catenin pathway.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25020734/s1.
Author Contributions
L.S.: Conceptualization, data curation, formal analysis, investigation, writing—original draft. L.P.: Conceptualization, data curation, formal analysis, investigation, writing—original draft, writing—review and editing. X.F.: Formal analysis, investigation. Y.F.: Data curation, investigation. B.Z.: Conceptualization, project administration, resources, supervision. X.C.: Conceptualization, funding acquisition, project administration, resources, supervision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grant to Xiaowei Chen from: 1. the General Programs of National Natural Science Foundation of China (82271186); 2. the General Programs of National Natural Science Foundation of China (81974143); 3. National High Level Hospital Clinical Research Funding (2022-PUMCH-C-029).
Institutional Review Board Statement
All zebrafish experiments were approved by the Animal Ethics Committee of Peking University (protocol code: LSC-ZhangB-3, date of approval: 1 September 2019) and conformed to institutional, local, and national rules and guidelines.
Informed Consent Statement
Not applicable.
Data Availability Statement
All figures are associated with raw data. Raw images can be provided upon request. Sequences used for CRISPR/Cas9 and for in situ hybridization are included in the Supplementary Materials.
Acknowledgments
We thank all colleagues from Bo Zhang’s laboratory for their generous help and instruction to our experiments. We also thank all colleagues and friends for their critical discussions and valuable support to our work.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Deng, K.; Dai, L.; Yi, L.; Deng, C.; Li, X.; Zhu, J. Epidemiologic characteristics and time trend in the prevalence of anotia and microtia in China. Birth Defects Res. A Clin. Mol. Teratol. 2016, 106, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Cabrejo, R.; Persing, J.; Alperovich, M. Epidemiologic Assessment of Microtia in Over 23 Million Consecutive United States Births. J. Craniofacial Surg. 2019, 30, 342–346. [Google Scholar] [CrossRef] [PubMed]
- Bartel-Friedrich, S. Congenital Auricular Malformations: Description of Anomalies and Syndromes. Facial Plast. Surg. 2015, 31, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Ping, L.; Sun, H.; Chen, Y.; Wang, P.; Liu, T.; Jiang, R.; Zhang, X.; Chen, X. Whole-Exome Sequencing of Discordant Monozygotic Twin Families for Identification of Candidate Genes for Microtia-Atresia. Front. Genet. 2020, 11, 568052. [Google Scholar] [CrossRef] [PubMed]
- Arora, V.; Bijarnia-Mahay, S.; Saxena, K.K.; Suman, P.; Kukreja, S. Osteopathia Striata with Cranial Sclerosis: A Face-to-Radiograph-to-Gene Diagnosis. J. Pediatr. Genet. 2022, 11, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, Z.A.; van Kogelenberg, M.; Morgan, T.; Jeffs, A.; Fukuzawa, R.; Pearl, E.; Thaller, C.; Hing, A.V.; Porteous, M.E.; Garcia-Miñaur, S.; et al. Germline mutations in WTX cause a sclerosing skeletal dysplasia but do not predispose to tumorigenesis. Nat. Genet. 2009, 41, 95–100. [Google Scholar] [CrossRef]
- Li, J.; Rodriguez, G.; Han, X.; Janečková, E.; Kahng, S.; Song, B.; Chai, Y. Regulatory Mechanisms of Soft Palate Development and Malformations. J. Dent. Res. 2019, 98, 959–967. [Google Scholar] [CrossRef]
- Yin, X.; Li, J.; Salmon, B.; Huang, L.; Lim, W.H.; Liu, B.; Hunter, D.J.; Ransom, R.C.; Singh, G.; Gillette, M.; et al. Wnt Signaling and Its Contribution to Craniofacial Tissue Homeostasis. J. Dent. Res. 2015, 94, 1487–1494. [Google Scholar] [CrossRef]
- Sisson, B.E.; Dale, R.M.; Mui, S.R.; Topczewska, J.M.; Topczewski, J. A role of glypican4 and wnt5b in chondrocyte stacking underlying craniofacial cartilage morphogenesis. Mech. Dev. 2015, 138 Pt 3, 279–290. [Google Scholar] [CrossRef]
- Boutet, A.; Comai, G.; Schedl, A. The WTX/AMER1 gene family: Evolution, signature and function. BMC Evol. Biol. 2010, 10, 280. [Google Scholar] [CrossRef]
- Regimbald-Dumas, Y.; He, X. Wnt signalling: What The X@# is WTX? EMBO J. 2011, 30, 1415–1417. [Google Scholar] [PubMed]
- Fukuzawa, R.; Anaka, M.R.; Weeks, R.J.; Morison, I.M.; Reeve, A.E. Canonical WNT signalling determines lineage specificity in Wilms tumour. Oncogene 2009, 28, 1063–1075. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, L.; Liu, L.; Wang, S.; Yang, Y.; Ding, Y.; Jeevan, D.; Li, X.; Ding, Y. LMP1 antagonizes WNT/β-catenin signalling through inhibition of WTX and promotes nasopharyngeal dysplasia but not tumourigenesis in LMP1(B95-8) transgenic mice. J. Pathol. 2011, 223, 574–583. [Google Scholar]
- Große, A.; Perner, B.; Naumann, U.; Englert, C. Zebrafish Wtx is a negative regulator of Wnt signaling but is dispensable for embryonic development and organ homeostasis. Dev. Dyn. 2019, 248, 866–881. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.S.; Lam, I.I.; Clay, H.; Duong, D.N.; Deo, R.C.; Coughlin, S.R. A Rapid Method for Directed Gene Knockout for Screening in G0 Zebrafish. Dev. Cell 2018, 46, 112–125.e4. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Moisan, A.; Rivera, M.N.; Lotinun, S.; Akhavanfard, S.; Coffman, E.J.; Cook, E.B.; Stoykova, S.; Mukherjee, S.; Schoonmaker, J.A.; Burger, A.; et al. The WTX tumor suppressor regulates mesenchymal progenitor cell fate specification. Dev. Cell 2011, 20, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Major, M.B.; Camp, N.D.; Berndt, J.D.; Yi, X.; Goldenberg, S.J.; Hubbert, C.; Biechele, T.L.; Gingras, A.C.; Zheng, N.; Maccoss, M.J.; et al. Wilms tumor suppressor WTX negatively regulates WNT/beta-catenin signaling. Science 2007, 316, 1043–1046. [Google Scholar] [CrossRef]
- Chen, Q.; Zhao, Y.; Shen, G.; Dai, J. Etiology and Pathogenesis of Hemifacial Microsomia. J. Dent. Res. 2018, 97, 1297–1305. [Google Scholar] [CrossRef]
- Lefebvre, V.; de Crombrugghe, B. Toward understanding SOX9 function in chondrocyte differentiation. Matrix Biol. 1998, 16, 529–540. [Google Scholar] [CrossRef]
- Zhao, Q.; Eberspaecher, H.; Lefebvre, V.; De Crombrugghe, B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev. Dyn. 1997, 209, 377–386. [Google Scholar] [CrossRef]
- Tanneberger, K.; Pfister, A.S.; Kriz, V.; Bryja, V.; Schambony, A.; Behrens, J. Structural and functional characterization of the Wnt inhibitor APC membrane recruitment 1 (Amer1). J. Biol. Chem. 2011, 286, 19204–19214. [Google Scholar] [CrossRef] [PubMed]
- Tanneberger, K.; Pfister, A.S.; Brauburger, K.; Schneikert, J.; Hadjihannas, M.V.; Kriz, V.; Schulte, G.; Bryja, V.; Behrens, J. Amer1/WTX couples Wnt-induced formation of PtdIns(4,5)P2 to LRP6 phosphorylation. EMBO J. 2011, 30, 1433–1443. [Google Scholar] [CrossRef] [PubMed]
- Hans, S.; Westerfield, M. Changes in retinoic acid signaling alter otic patterning. Development 2007, 134, 2449–2458. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Mao, A.; Wang, P.; Ning, G.; Cao, Y.; Wang, Q. Endodermal pouch-expressed dmrt2b is important for pharyngeal cartilage formation. Biol. Open 2018, 7, bio035444. [Google Scholar]
- Liu, C.; Li, R.; Li, Y.; Lin, X.; Zhao, K.; Liu, Q.; Wang, S.; Yang, X.; Shi, X.; Ma, Y.; et al. Spatiotemporal mapping of gene expression landscapes and developmental trajectories during zebrafish embryogenesis. Dev. Cell 2022, 57, 1284–1298.e5. [Google Scholar] [CrossRef]
- Mm, W. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio) Rerio; University of Oregon Press: Eugene, Oregon, 1994. [Google Scholar]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef]
- Chang, N.; Sun, C.; Gao, L.; Zhu, D.; Xu, X.; Zhu, X.; Xiong, J.W.; Xi, J.J. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013, 23, 465–472. [Google Scholar] [CrossRef]
- Shum, E.Y.; Jones, S.H.; Shao, A.; Dumdie, J.; Krause, M.D.; Chan, W.K.; Lou, C.H.; Espinoza, J.L.; Song, H.W.; Phan, M.H.; et al. The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell 2016, 165, 382–395. [Google Scholar] [CrossRef]
- Wang, Y.; Ping, L.; Luan, X.; Chen, Y.; Fan, X.; Li, L.; Liu, Y.; Wang, P.; Zhang, S.; Zhang, B.; et al. A Mutation in VWA1, Encoding von Willebrand Factor A Domain-Containing Protein 1, Is Associated With Hemifacial Microsomia. Front. Cell Dev. Biol. 2020, 8, 571004. [Google Scholar] [CrossRef] [PubMed]
- Dale, R.M.; Topczewski, J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev. Biol. 2011, 357, 518–531. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).