Genome-Wide Identification and Expression Analysis of GATA Family Genes in Dimocarpus longan Lour
Abstract
1. Introduction
2. Results
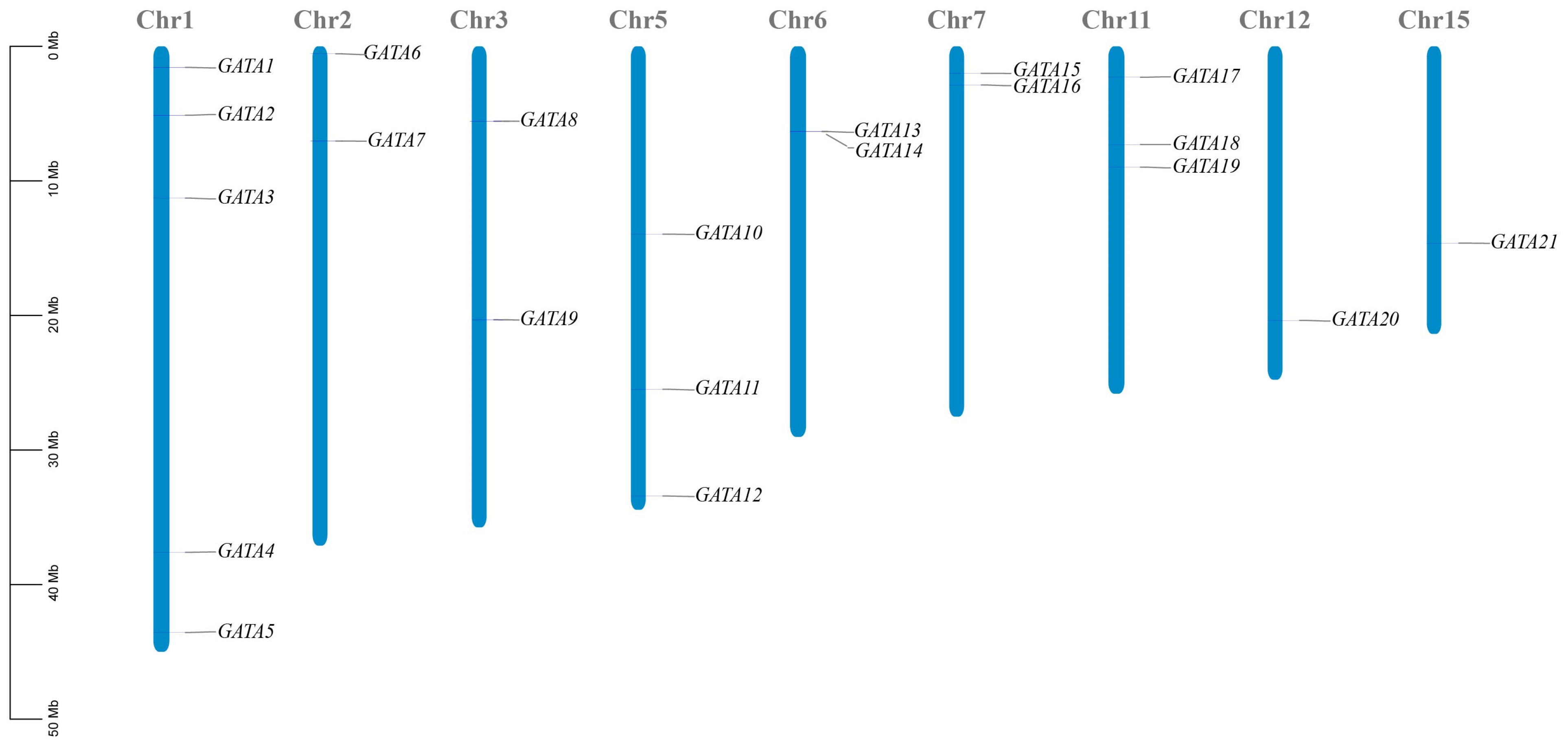
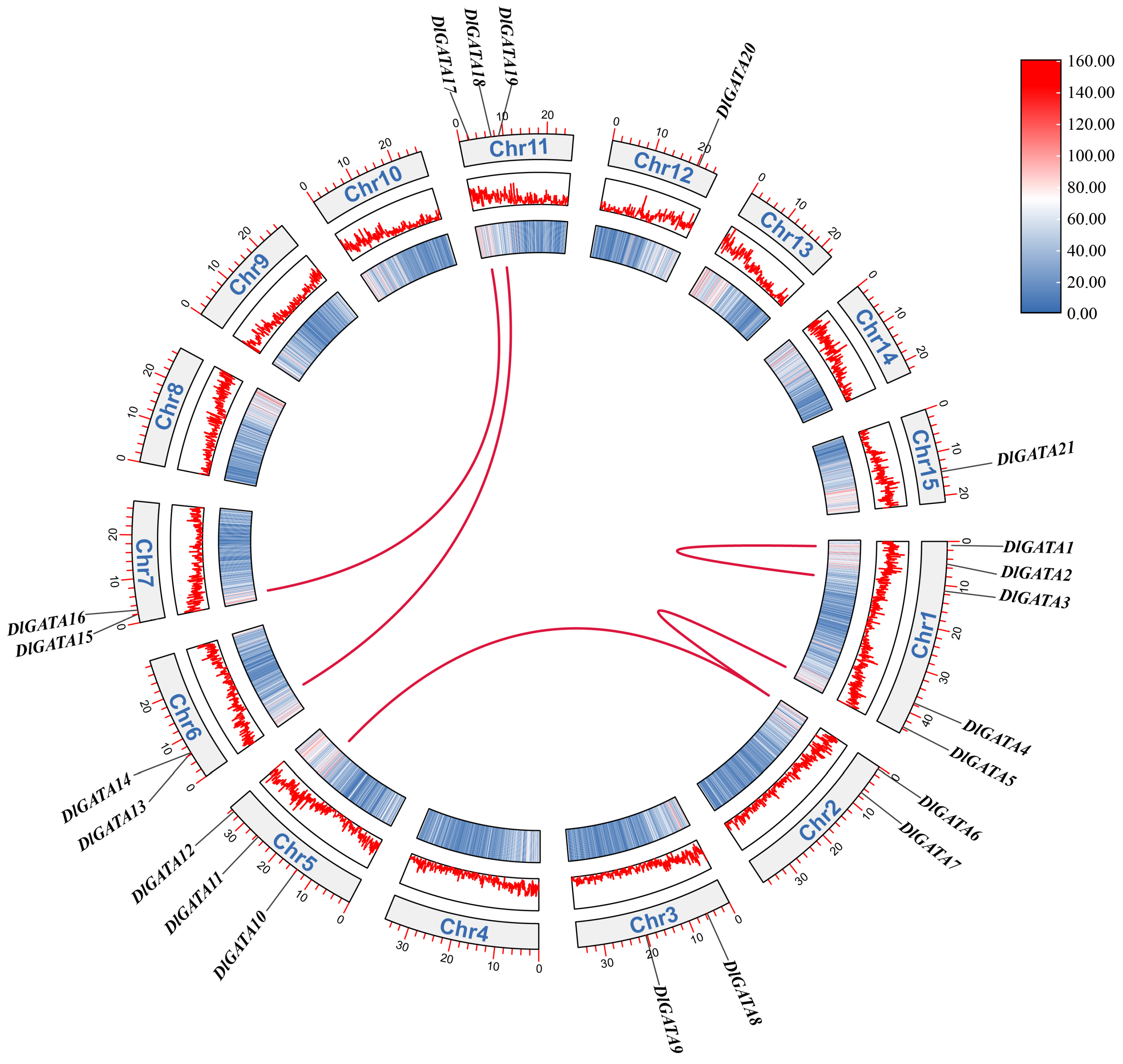
2.1. Characterization of the DlGATA Gene in Longan and Its Positional Distribution on Chromosomes
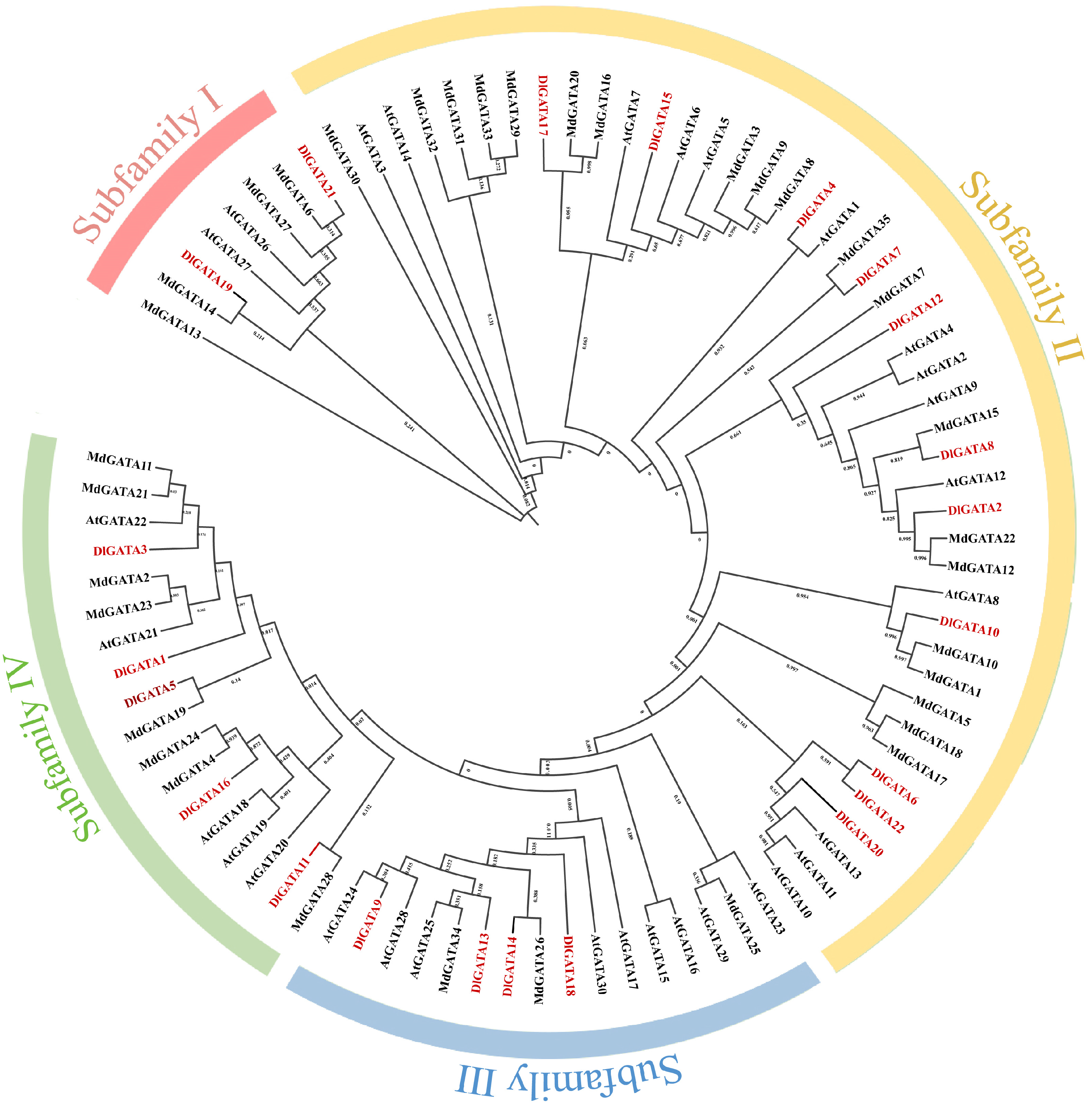
2.2. Phylogenetic Analysis between Longan, Apple, and Arabidopsis
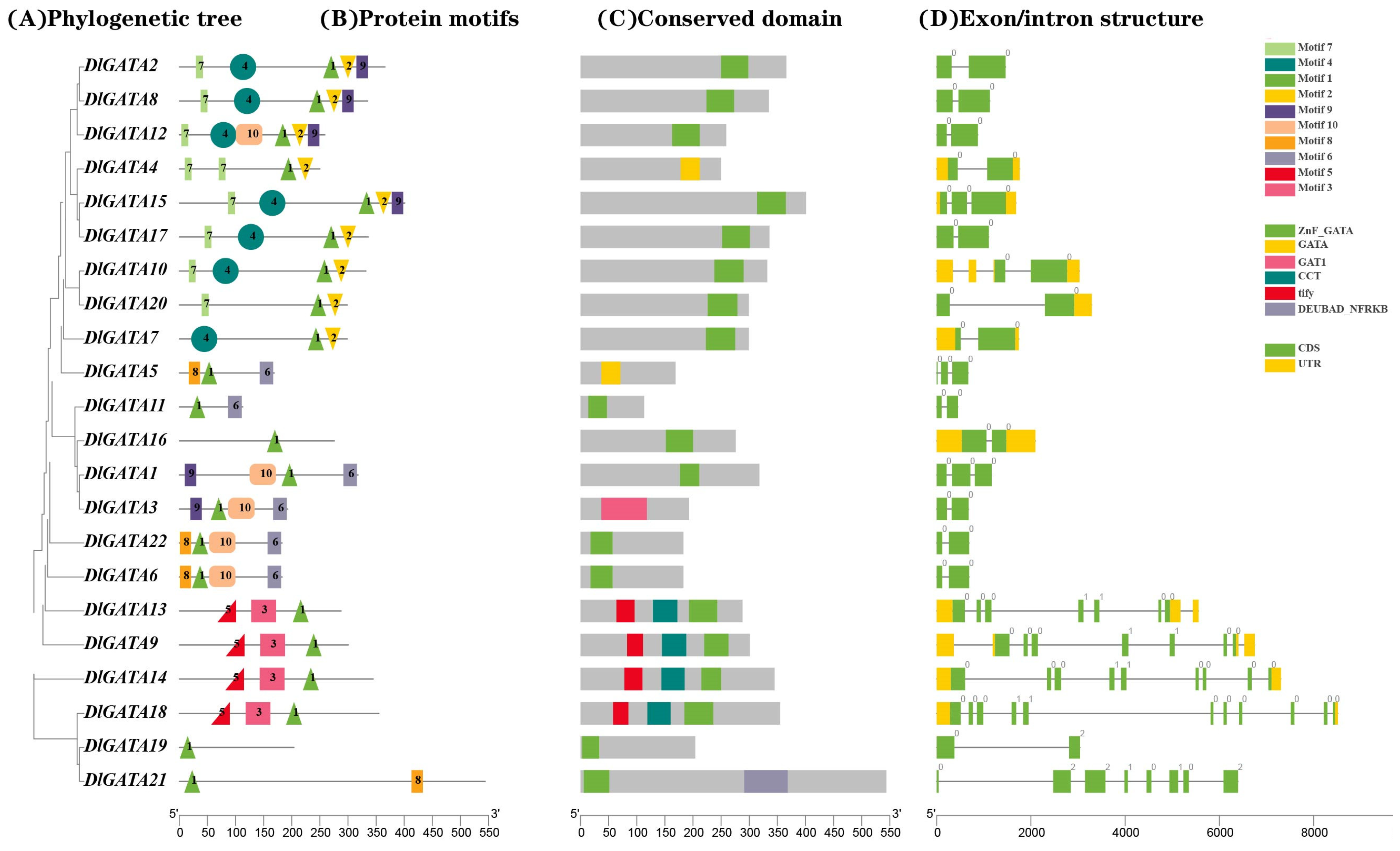
2.3. Analysis of the Gene Structure and Conserved Protein Sequence of DlGATA
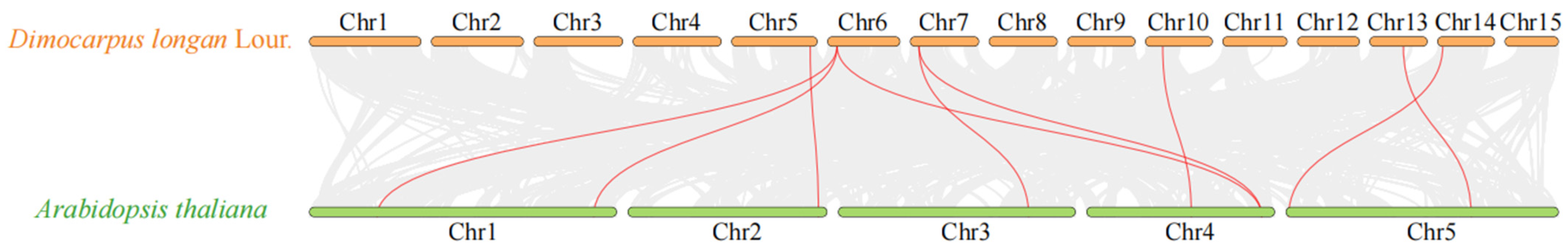
2.4. Synteny Analysis of Longan and Arabidopsis Genes and an Analysis of Replication Events within Gene Families
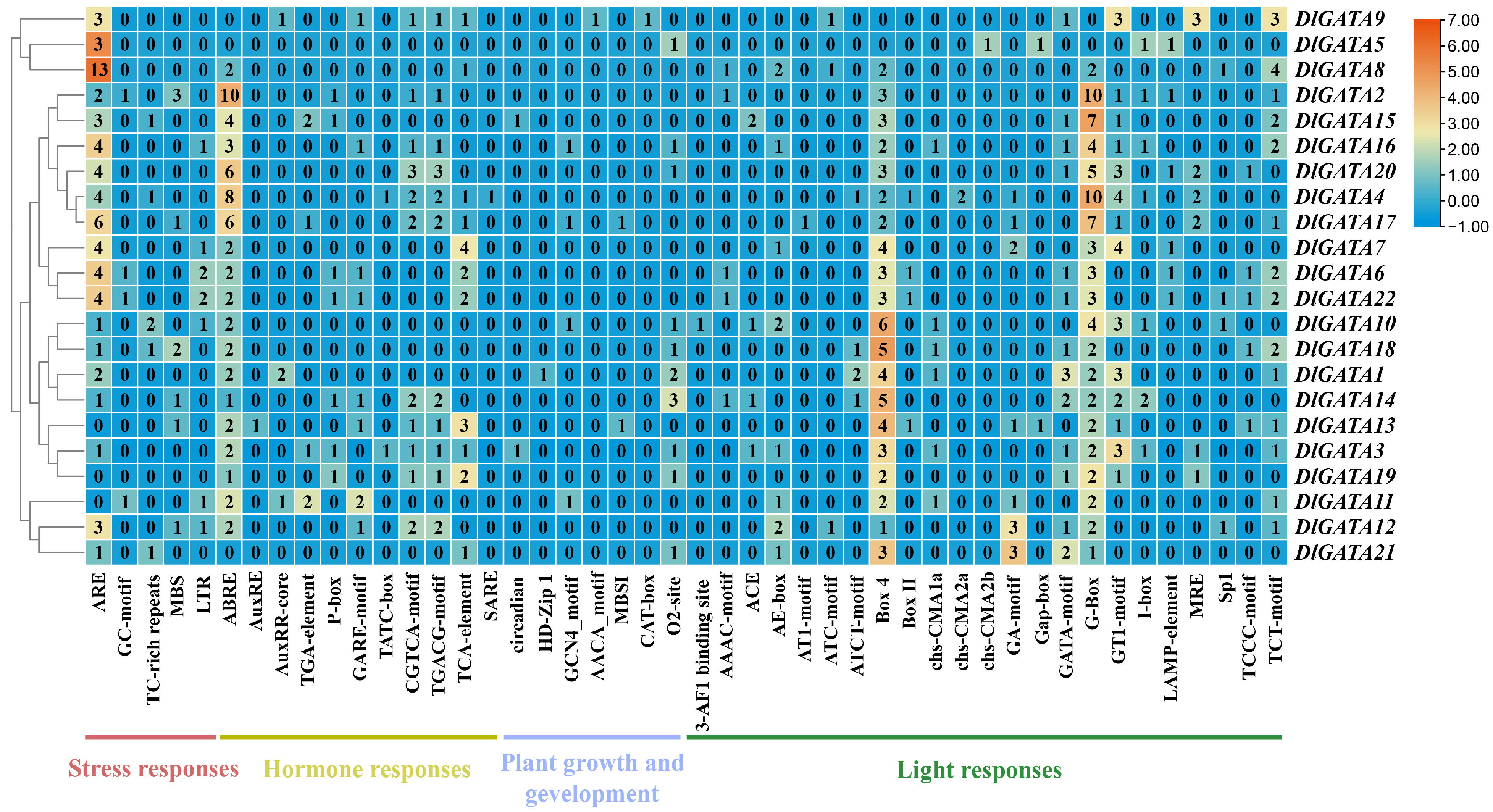
2.5. Analysis of the Cis-Acting Elements in the Promoter of the Longan GATA Genes
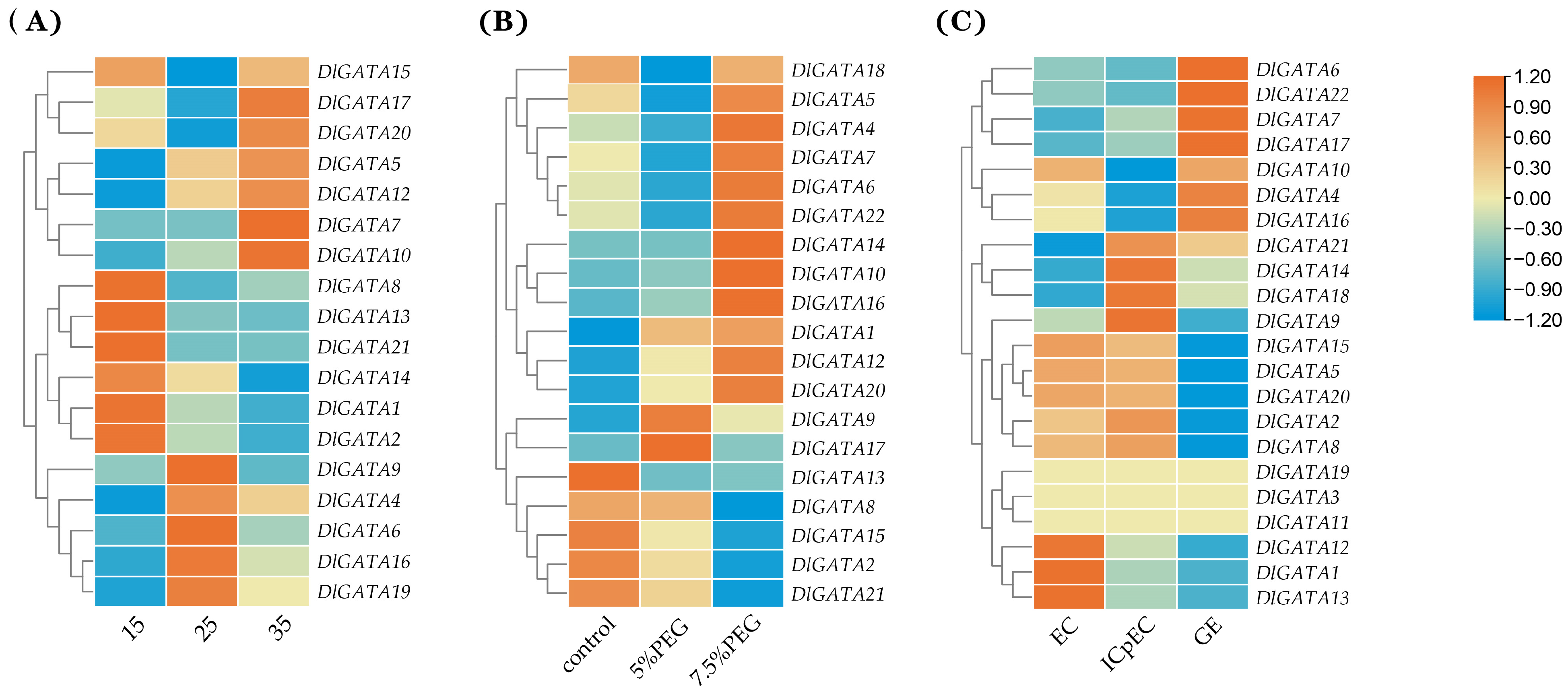
2.6. Expression Analysis of Longan at Different Temperatures, Concentration of PEG, and Early Developmental Stages of Somatic Embryos
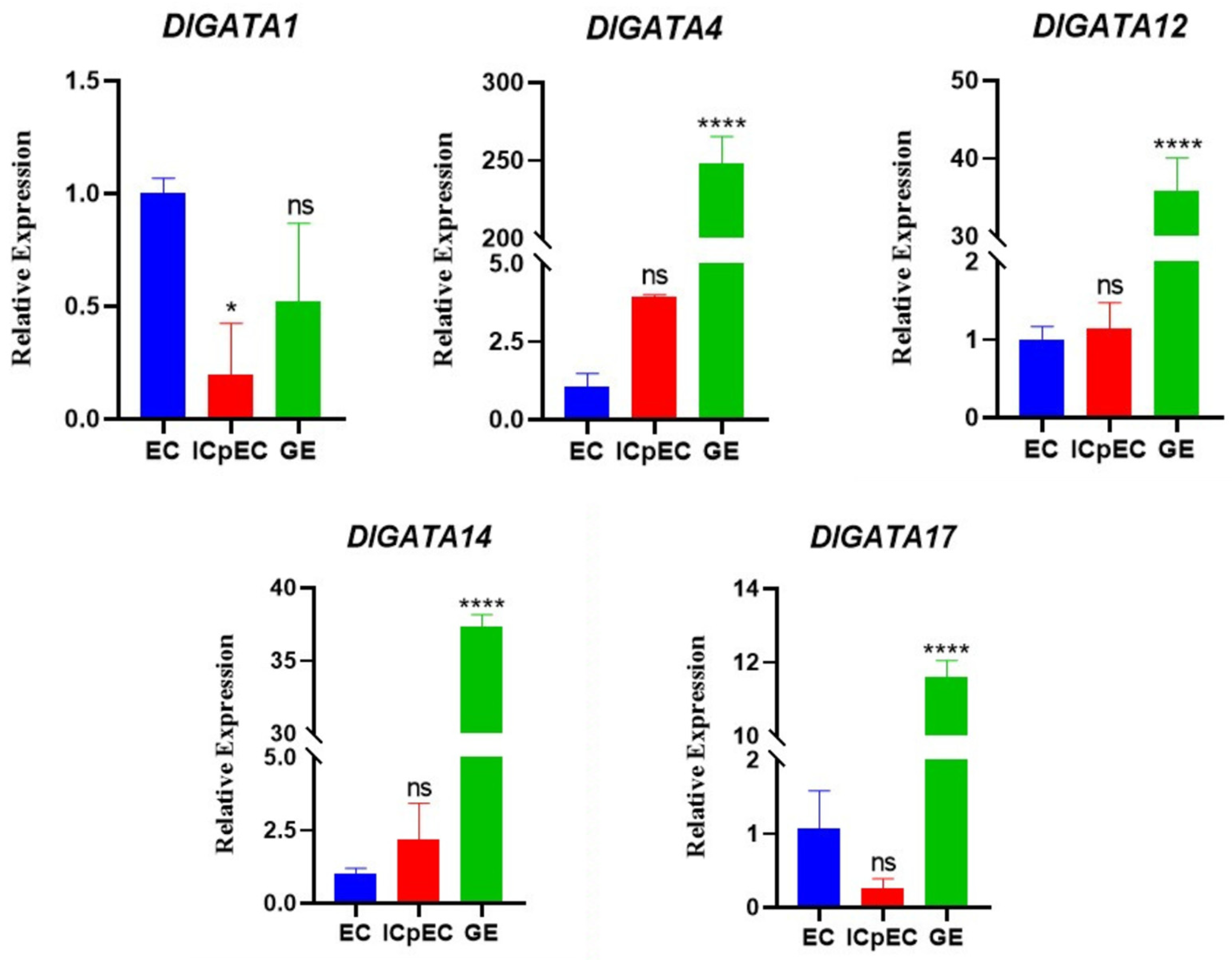
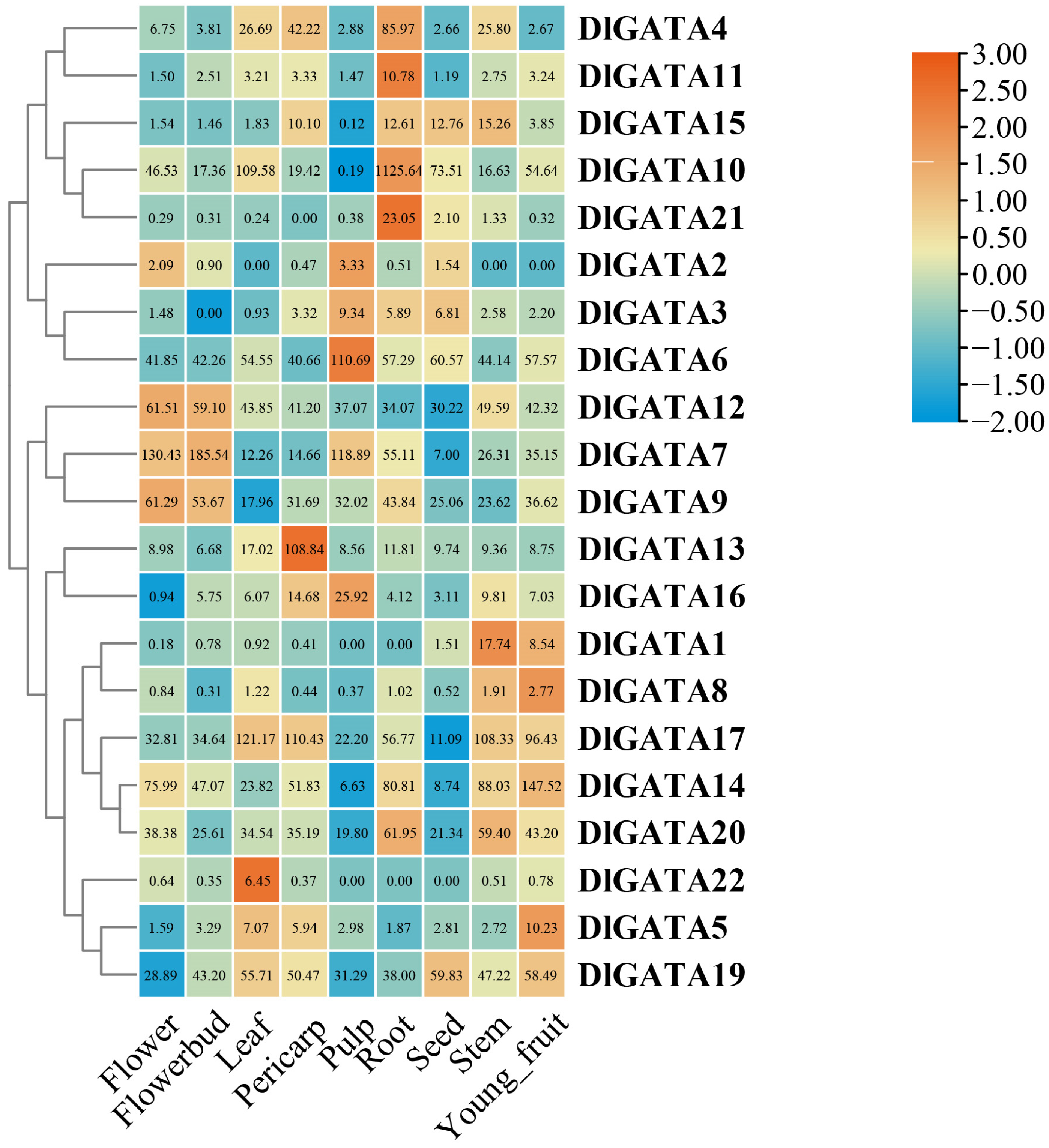
2.7. DlGATA Expression Profiles in Nine Different Tissues
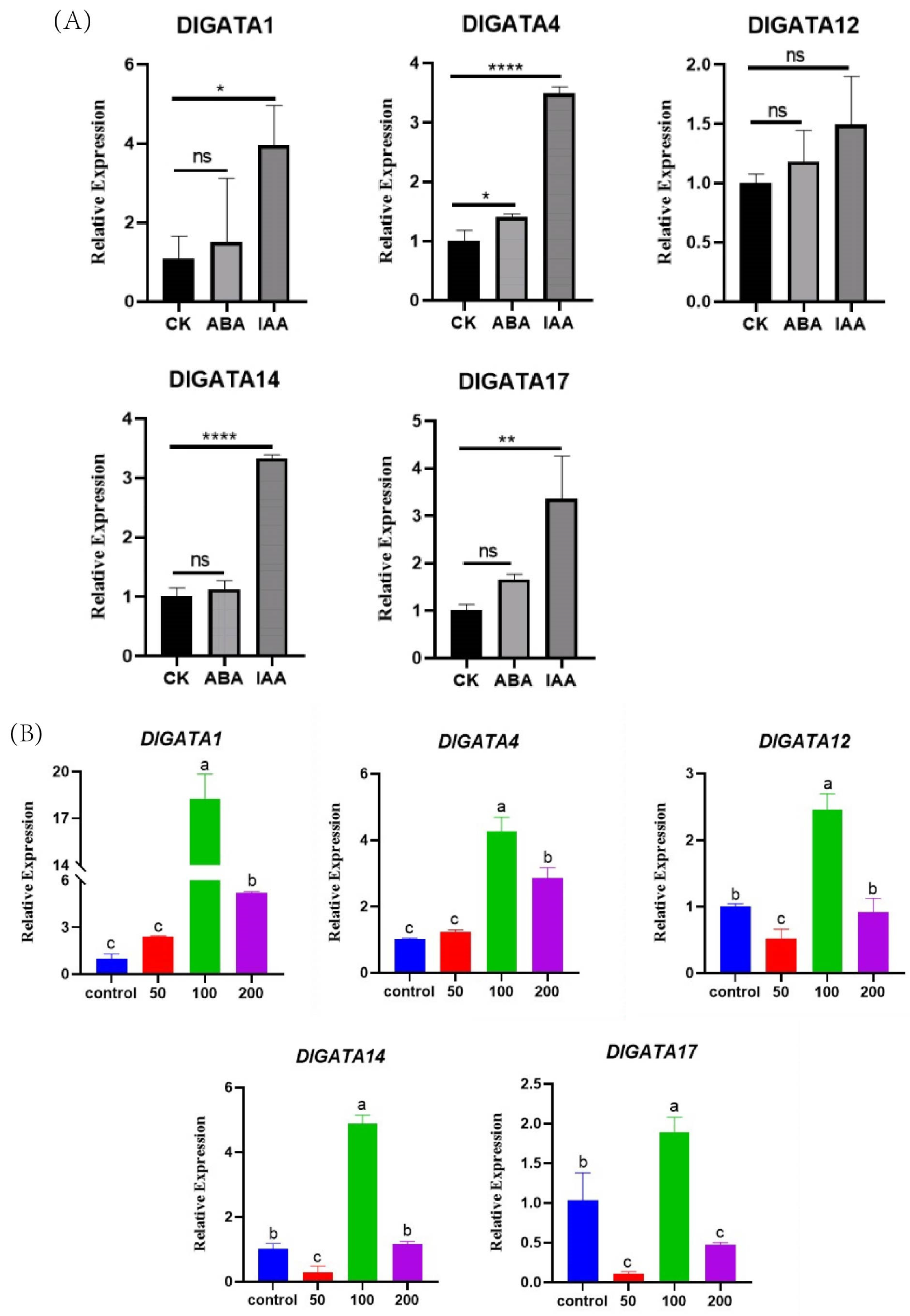
2.8. Expression of DlGATA Induced by Exogenous Hormone Treatments
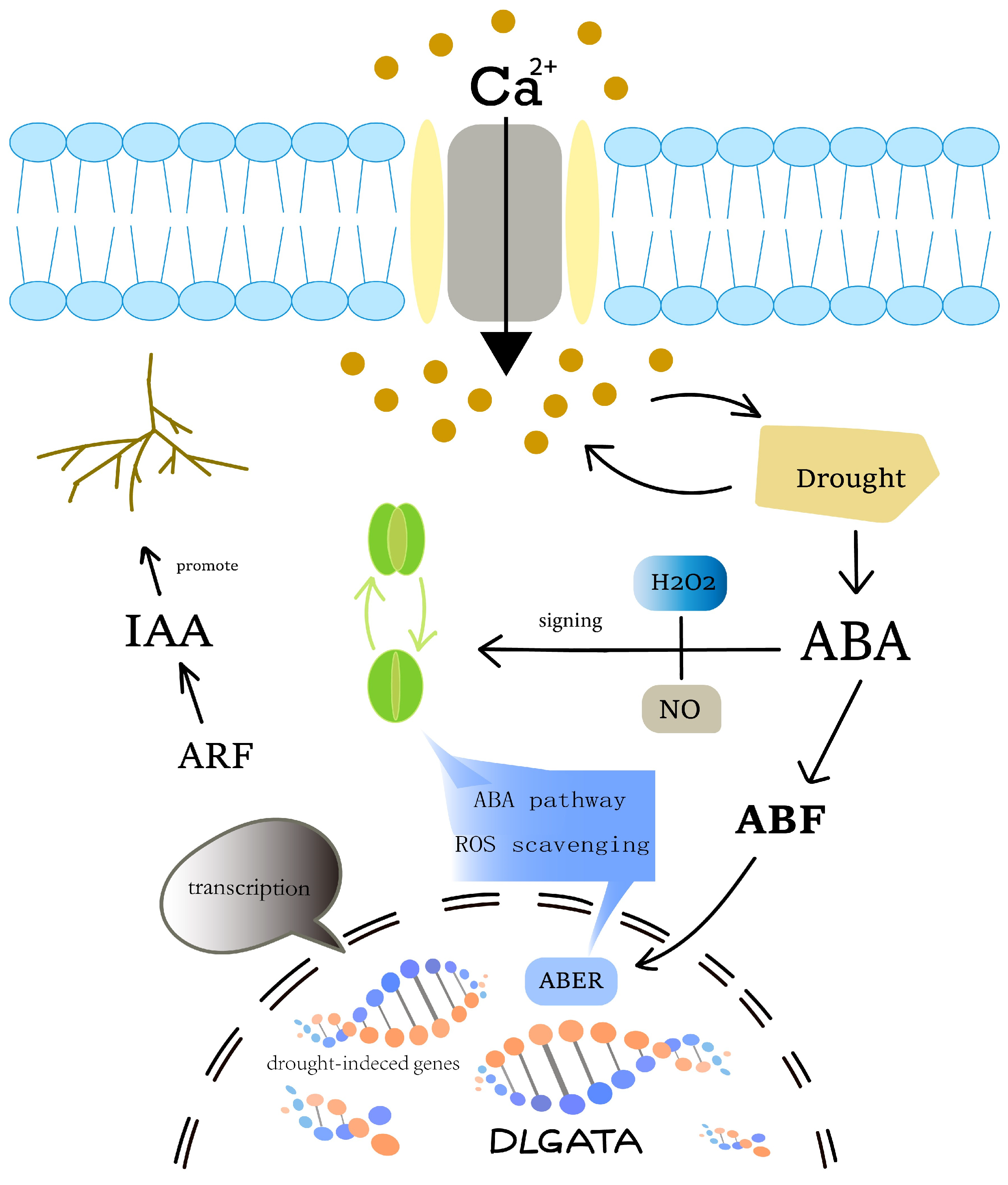
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of GATA Genes in Longan
4.3. Phylogenetic Analysis, Gene Structure, and Conserved Motif Analysis
4.4. Chromosomal Location, Cis-Acting Element Analyses, and Analysis of Collinearity with Other Species
4.5. Hormonal Processing of Longan EC, RNA Extraction, and qRT-PCR Analysis
4.6. Analysis of the Specific Expression of DlGATA Family Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitsuda, N.; Ohme-Takagi, M. Functional Analysis of Transcription Factors in Arabidopsis. Plant Cell Physiol. 2009, 50, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-N.; Cheong, Y.H.; Grant, J.J.; Pandey, G.K.; Luan, S. CIPK3, a Calcium Sensor–Associated Protein Kinase That Regulates Abscisic Acid and Cold Signal Transduction in Arabidopsis. Plant Cell 2003, 15, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Éva, C.; Moncsek, B.; Szőke-Pázsi, K.; Kunos, V.; Mészáros, K.; Makai, S.; Sági, L.; Juhász, A. bZIP transcription factors repress the expression of wheat (Triticum aestivum L.) high molecular weight glutenin subunit genes in vegetative tissues. Acta Physiol. 2023, 45, 29. [Google Scholar] [CrossRef]
- Abdullah-Zawawi, M.-R.; Ahmad-Nizammuddin, N.-F.; Govender, N.; Harun, S.; Mohd-Assaad, N.; Mohamed-Hussein, Z.-A. Comparative genome-wide analysis of WRKY, MADS-box and MYB transcription factor families in Arabidopsis and rice. Sci. Rep. 2021, 11, 19678. [Google Scholar] [CrossRef] [PubMed]
- Dudhate, A.; Shinde, H.; Yu, P.; Tsugama, D.; Gupta, S.K.; Liu, S.; Takano, T. Comprehensive analysis of NAC transcription factor family uncovers drought and salinity stress response in pearl millet (Pennisetum glaucum). BMC Genom. 2021, 22, 70. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Takasaki, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta—Gene Regul. Mech. 2012, 1819, 97–103. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef]
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Khan, I.; Asaf, S.; Jan, R.; Bilal, S.; Lubna; Khan, A.L.; Kim, K.-M.; Al-Harrasi, A. Genome-wide annotation and expression analysis of WRKY and bHLH transcriptional factor families reveal their involvement under cadmium stress in tomato (Solanum lycopersicum L.). Front. Plant Sci. 2023, 14, 1100895. [Google Scholar] [CrossRef]
- Khoudi, H. SHINE clade of ERF transcription factors: A significant player in abiotic and biotic stress tolerance in plants. Plant Physiol. Biochem. 2023, 195, 77–88. [Google Scholar] [CrossRef]
- Medina, J.; Bargues, M.; Terol, J.; Pérez-Alonso, M.; Salinas, J. The Arabidopsis CBF Gene Family Is Composed of Three Genes Encoding AP2 Domain-Containing Proteins Whose Expression Is Regulated by Low Temperature but Not by Abscisic Acid or Dehydration. Plant Physiol. 1999, 119, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Xia, Y.; Zhan, D.; Xu, T.; Lu, T.; Yang, J.; Kang, X. Genome-Wide Identification of the Eucalyptus urophylla GATA Gene Family and Its Diverse Roles in Chlorophyll Biosynthesis. Int. J. Mol. Sci. 2022, 23, 5251. [Google Scholar] [CrossRef] [PubMed]
- Gillis, W.Q.; St John, J.; Bowerman, B.; Schneider, S.Q. Whole genome duplications and expansion of the vertebrate GATA transcription factor gene family. Biology 2009, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Scazzocchio, C. The fungal GATA factors. Curr. Opin. Microbiol. 2000, 3, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA Family of Transcription Factors in Arabidopsis and Rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [PubMed]
- Lindemose, S.; O’Shea, C.; Jensen, M.; Skriver, K. Structure, Function and Networks of Transcription Factors Involved in Abiotic Stress Responses. Int. J. Mol. Sci. 2013, 14, 5842–5878. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic Stresses Cause Differential Regulation of Alternative Splice Forms of GATA Transcription Factor in Rice. Front. Plant Sci. 2017, 8, 1944. [Google Scholar] [CrossRef] [PubMed]
- Manzoor, M.A.; Sabir, I.A.; Shah, I.H.; Wang, H.; Yu, Z.; Rasool, F.; Mazhar, M.Z.; Younas, S.; Abdullah, M.; Cai, Y. Comprehensive Comparative Analysis of the GATA Transcription Factors in Four Rosaceae Species and Phytohormonal Response in Chinese Pear (Pyrus bretschneideri) Fruit. Int. J. Mol. Sci. 2021, 22, 12492. [Google Scholar] [CrossRef]
- Card, R.M. Functional Analysis of a Tobacco GATA Factor. 1999. Available online: https://www.researchgate.net/publication/263162054 (accessed on 24 November 2023).
- Nizamutdinova, I.T.; Kim, Y.M.; Il Chung, J.; Shin, S.C.; Jeong, Y.-K.; Seo, H.G.; Lee, J.H.; Chang, K.C.; Kim, H.J. Anthocyanins from Black Soybean Seed Coats Preferentially Inhibit TNF-α-Mediated Induction of VCAM-1 over ICAM-1 through the Regulation of GATAs and IRF-1. J. Agric. Food Chem. 2009, 57, 7324–7330. [Google Scholar] [CrossRef]
- Arens, P.; Odinot, P.; van Heusden, A.W.; Lindhout, P.; Vosman, B. GATA- and GACA-repeats are not evenly distributed throughout the tomato genome. Genome 1995, 38, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, S.; Wolf, A.; Picard, D. GATA4 is a regulator of astrocyte cell proliferation and apoptosis in the human and murine central nervous system. Oncogene 2009, 28, 3033–3046. [Google Scholar] [CrossRef] [PubMed]
- Reyes, R.; Haendel, M.; Grant, D.; Melancon, E.; Eisen, J.S. Slow degeneration of zebrafish Rohon-Beard neurons during programmed cell death. Dev. Dyn. 2003, 229, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Lowry, J.A.; Atchley, W.R. Molecular Evolution of the GATA Family of Transcription Factors: Conservation within the DNA-Binding Domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.S. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Tindall, H.D. Sapindaceous fruits: Botany and horticulture. Hortic. Rev. Am. Soc. Hortic. Sci. 1994, 16, 143–196. [Google Scholar]
- Zhang, X.; Guo, S.; Ho, C.-T.; Bai, N. Phytochemical constituents and biological activities of longan (Dimocarpus longan Lour.) fruit: A review. Food Sci. Hum. Wellness 2020, 9, 95–102. [Google Scholar] [CrossRef]
- Li, L.R.; Zhuang, Y.M. Longan Cultivation; China Agricultural Pres: Beijing, China, 1983; Volume 1, p. 83. [Google Scholar]
- Nickell, L.G. Plant Growth Regulators in Agriculture and Horticulture; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1994; Volume 557, pp. 1–14. [Google Scholar]
- Bandara, P.M.; Tanino, K.K. Paclobutrazol enhances minituber production in Norland potatoes. J. Plant Growth Regul. 1995, 14, 151–155. [Google Scholar] [CrossRef]
- Chai, M.N.; Cheng, H.; Yan, M.K.; Priyadarshani, S.; Zhang, M.; He, Q.; Huang, Y.M.; Chen, F.Q.; Liu, L.P.; Huang, X.Y.; et al. Identification and expression analysis of the DREB transcription factor family in pineapple (Ananas comosus (L.) Merr.). PeerJ 2020, 8, e9006. [Google Scholar] [CrossRef]
- Zhang, Z.; Zou, X.; Huang, Z.; Fan, S.; Qun, G.; Liu, A.; Gong, J.; Li, J.; Gong, W.; Shi, Y.; et al. Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene 2019, 680, 72–83. [Google Scholar] [CrossRef]
- Roy, S.W.; Penny, D. A Very High Fraction of Unique Intron Positions in the Intron-Rich Diatom Thalassiosira pseudonana Indicates Widespread Intron Gain. Mol. Biol. Evol. 2007, 24, 1447–1457. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants. Functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Xu, X.; Liu, Z.; Zhang, Z.; XuHan, X.; Lin, Y.; Lai, Z. Global scale transcriptome analysis reveals differentially expressed genes involve in early somatic embryogenesis in Dimocarpus longan Lour. BMC Genom. 2020, 21, 4. [Google Scholar] [CrossRef] [PubMed]
- Hellens, A.M.; Chabikwa, T.G.; Fichtner, F.; Brewer, P.B.; Beveridge, C.A. Identification of new potential downstream transcriptional targets of the strigolactone pathway including glucosinolate biosynthesis. Plant Direct 2023, 7, e486. [Google Scholar] [CrossRef] [PubMed]
- Anabel Lopez-Ruiz, B.; Thamara Juarez-Gonzalez, V.; Gomez-Felipe, A.; De Folter, S.; Dinkova, T.D. tasiR-ARFs Production and Target Regulation during In Vitro Maize Plant Regeneration. Plants 2020, 9, 849. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.-J.; Zheng, X.; da Silva, J.A.T.; Qin, P. Callus induction and plant regeneration from embryonic axes of Kosteletzkya virginica. Sci. Hortic. 2009, 120, 150–155. [Google Scholar] [CrossRef]
- Marimuthu, K.; Subbaraya, U.; Suthanthiram, B.; Marimuthu, S.S. Molecular analysis of somatic embryogenesis through proteomic approach and optimization of protocol in recalcitrant Musa spp. Physiol. Plant. 2019, 167, 282–301. [Google Scholar] [CrossRef]
- Lai, Z.; Lin, Y. Analysis of the global transcriptome of longan (Dimocarpus longan Lour.) embryogenic callus using Illumina paired-end sequencing. BMC Genom. 2013, 14, 561. [Google Scholar] [CrossRef]
- Otiende, M.A.; Fricke, K.; Nyabundi, J.O.; Ngamau, K.; Hajirezaei, M.R.; Druege, U. Involvement of the auxin–cytokinin homeostasis in adventitious root formation of rose cuttings as affected by their nodal position in the stock plant. Planta 2021, 254, 65. [Google Scholar] [CrossRef]
- Yang, L.; Zhu, S. The Interconnected Relationship between Auxin Concentration Gradient Changes in Chinese Fir Radial Stems and Dynamic Cambial Activity. Forests 2022, 13, 1698. [Google Scholar] [CrossRef]
- Yuan, R.; Kender, W.J.; Burns, J.K. Young Fruit and Auxin Transport Inhibitors Affect the Response of Mature ‘Valencia’ Oranges to Abscission Materials via Changing Endogenous Plant Hormones. J. Am. Soc. Hortic. 2003, 128, 302–308. [Google Scholar] [CrossRef]
- Hwarari, D.; Radani, Y.; Guan, Y.; Chen, J.; Liming, Y. Systematic Characterization of GATA Transcription Factors in Liriodendron chinense and Functional Validation in Abiotic Stresses. Plants 2023, 12, 2349. [Google Scholar] [CrossRef] [PubMed]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription Factors Interact with ABA through Gene Expression and Signaling Pathways to Mitigate Drought and Salinity Stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Lu, Y.; Sun, H.; Duan, W.; Hu, Y.; Yan, Y. Genome-Wide Analysis of Wheat GATA Transcription Factor Genes Reveals Their Molecular Evolutionary Characteristics and Involvement in Salt and Drought Tolerance. Int. J. Mol. Sci. 2022, 24, 27. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, T.S.; Andhare, R.; Cooper, T.G. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J. Biol. Chem. 2000, 275, 14408–14414. [Google Scholar] [CrossRef]
- Li, S.; Li, Q.; Tian, X.; Mu, L.; Ji, M.; Wang, X.; Li, N.; Liu, F.; Shu, J.; Crawford, N.M.; et al. PHB3 regulates lateral root primordia formation via NO-mediated degradation of AUXIN/INDOLE-3-ACETIC ACID proteins. J. Exp. Bot. 2022, 73, 4034–4045. [Google Scholar] [CrossRef]
- Shoaib, M.; Banerjee, B.P.; Hayden, M.; Kant, S. Roots’ Drought Adaptive Traits in Crop Improvement. Plants 2022, 11, 2256. [Google Scholar] [CrossRef]
- Steinemann, S.; Zeng, Z.H.; McKay, A.; Heuer, S.; Langridge, P.; Huang, C.Y. Dynamic root responses to drought and rewatering in two wheat (Triticum aestivum) genotypes. Plant Soil 2015, 391, 139–152. [Google Scholar] [CrossRef]
- Chen, Y.L.; Ghanem, M.E.; Siddique, K.H.M. Characterising root trait variability in chickpea (Cicer arietinum L.) germplasm. J. Exp. Bot. 2017, 68, 1987–1999. [Google Scholar]
- Manschadi, A.M.; Hammer, G.L.; Christopher, J.T.; deVoil, P. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 2008, 303, 115–129. [Google Scholar] [CrossRef]
- Andivia, E.; Zuccarini, P.; Grau, B.; de Herralde, F.; Villar-Salvador, P.; Savé, R. Rooting big and deep rapidly: The ecological roots of pine species distribution in southern Europe. Trees-Struct. Funct. 2019, 33, 293–303. [Google Scholar] [CrossRef]
- Bright, J.; Desikan, R.; Hancock, J.T.; Weir, I.S.; Neill, S.J. ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 2005, 45, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Guida, L.; Vigliarolo, T.; Passalacqua, M.; Begani, G.; Magnone, M.; Sturla, L.; Benzi, A.; Ameri, P.; Lazzarini, E.; et al. The ABA-LANCL1/2 Hormone-Receptors System Protects H9c2 Cardiomyocytes from Hypoxia-Induced Mitochondrial Injury via an AMPK- and NO-Mediated Mechanism. Cells 2022, 11, 2888. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Min, J.; Lai, R.; Wu, Z.; Chen, Y.; Yu, L.; Cheng, C.; Jin, Y.; Tian, Q.; Liu, Q.; et al. Genome-wide sequencing of longan (Dimocarpus longan Lour.) provides insights into molecular basis of its polyphenol-rich characteristics. GigaScience 2017, 6, gix023. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018, 289660. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, P.; Luo, J.; Jiang, Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 2003, 14, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lai, Z.X. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]
| Group | Gene Accession | Gene Id | Size/aa 1 | MW 2/Da | Theoretical pI 3 | Instability Index | Aliphatic Index | GRAVY 4 | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| I | Dlo023940 | DlGATA19 | 203 | 22,746.48 | 9.41 | 58.31 | 64.38 | −0.855 | Nuclear |
| Dlo031787 | DlGATA21 | 542 | 60,405.08 | 6.45 | 53.84 | 67.91 | −0.652 | Nuclear | |
| II | Dlo000544 | DlGATA2 | 365 | 40,147.31 | 5.75 | 50.23 | 56.41 | −0.715 | Nuclear |
| Dlo002694 | DlGATA4 | 249 | 27,832.37 | 8.81 | 48.32 | 59.08 | −0.642 | Nuclear | |
| Dlo003563 | DlGATA6 | 182 | 19,817.05 | 10.89 | 62.74 | 67.58 | −0.513 | Nuclear | |
| Dlo004225 | DlGATA7 | 298 | 32,967.26 | 8.07 | 56.55 | 67.01 | −0.606 | Nuclear | |
| Dol006515 | DlGATA8 | 334 | 36,529.19 | 6.26 | 45.76 | 56.11 | −0.685 | Nuclear | |
| Dlo011228 | DlGATA10 | 331 | 36,198.76 | 6.33 | 60.08 | 67.73 | −0.518 | Nuclear | |
| Dlo012990 | DlGATA12 | 285 | 28,487.45 | 8.25 | 47.23 | 56.78 | −0.728 | Nuclear | |
| Dlo015565 | DlGATA15 | 400 | 43,819.90 | 6.09 | 58.06 | 56.27 | −0.672 | Nuclear | |
| Dlo023262 | DlGATA17 | 335 | 37,210.04 | 4.74 | 70.62 | 58.78 | −0.671 | Nuclear | |
| Dlo026193 | DlGATA20 | 298 | 33,666.03 | 9.15 | 62.37 | 64.4 | −0.687 | Nuclear | |
| Dlo033619 | DlGATA22 | 182 | 19,817.05 | 10.89 | 62.74 | 67.58 | −0.513 | Nuclear | |
| III | Dlo007382 | DlGATA9 | 300 | 32,575.97 | 6.56 | 39.46 | 61.70 | −0.801 | Nuclear |
| Dlo013801 | DlGATA13 | 287 | 31,263.42 | 6.15 | 40.09 | 57.39 | −0.729 | Nuclear | |
| Dlo013802 | DlGATA14 | 344 | 37,626.45 | 4.62 | 48.36 | 66.02 | −0.697 | Nuclear | |
| Dlo023794 | DlGATA18 | 354 | 39,276.64 | 4.99 | 49.87 | 60.34 | −0.780 | Nuclear | |
| IV | Dlo000173 | DlGATA1 | 317 | 35,755.20 | 9.59 | 61.84 | 52.11 | −0.949 | Nuclear |
| Dlo001151 | DlGATA3 | 192 | 21,530.58 | 9.84 | 46.67 | 55.36 | −0.776 | Nuclear | |
| Dlo003321 | DlGATA5 | 168 | 18,519.80 | 9.58 | 58.8 | 67.38 | −0.943 | Nuclear | |
| Dlo012045 | DlGATA11 | 112 | 12,336.41 | 9.93 | 55.23 | 61.07 | −0.563 | Nuclear | |
| Dlo015654 | DlGATA16 | 275 | 30,480.31 | 8.32 | 66.27 | 34.11 | −0.946 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, K.; Lu, J.; He, X.; Lan, S.; Zhai, T.; Cao, S.; Lin, Y. Genome-Wide Identification and Expression Analysis of GATA Family Genes in Dimocarpus longan Lour. Int. J. Mol. Sci. 2024, 25, 731. https://doi.org/10.3390/ijms25020731
Zheng K, Lu J, He X, Lan S, Zhai T, Cao S, Lin Y. Genome-Wide Identification and Expression Analysis of GATA Family Genes in Dimocarpus longan Lour. International Journal of Molecular Sciences. 2024; 25(2):731. https://doi.org/10.3390/ijms25020731
Chicago/Turabian StyleZheng, Kehui, Jiayue Lu, Xinyu He, Shuoxian Lan, Tingkai Zhai, Shijiang Cao, and Yuling Lin. 2024. "Genome-Wide Identification and Expression Analysis of GATA Family Genes in Dimocarpus longan Lour" International Journal of Molecular Sciences 25, no. 2: 731. https://doi.org/10.3390/ijms25020731
APA StyleZheng, K., Lu, J., He, X., Lan, S., Zhai, T., Cao, S., & Lin, Y. (2024). Genome-Wide Identification and Expression Analysis of GATA Family Genes in Dimocarpus longan Lour. International Journal of Molecular Sciences, 25(2), 731. https://doi.org/10.3390/ijms25020731








