Genome-Wide and Expression Pattern Analysis of the DVL Gene Family Reveals GhM_A05G1032 Is Involved in Fuzz Development in G. hirsutum
Abstract
1. Introduction
2. Results
2.1. Genomic Identification of DVL Gene
2.2. Phylogenetic Analysis of DVL Gene
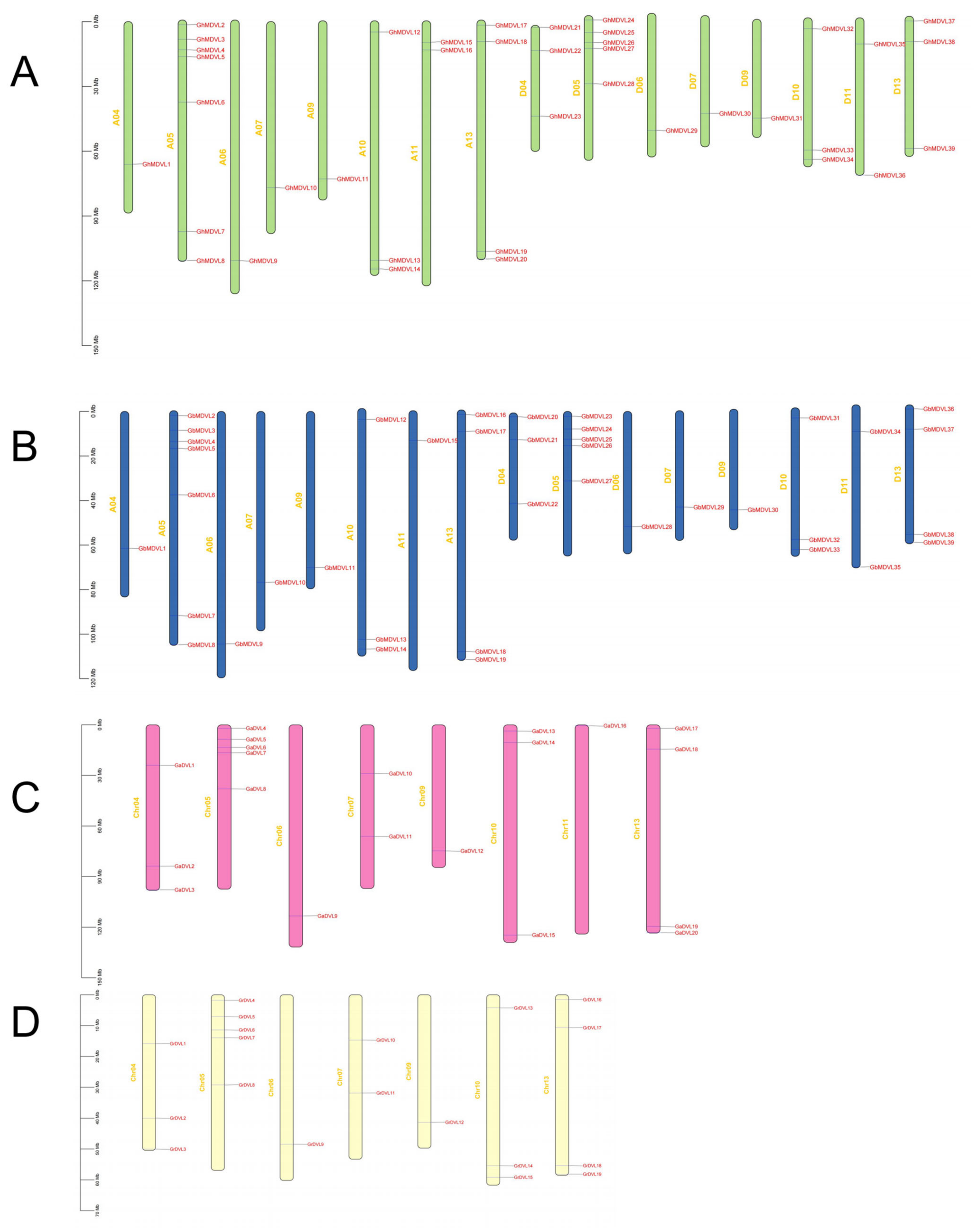
2.3. The Chromosomal Mapping of DVL Genes in Four Cotton Species
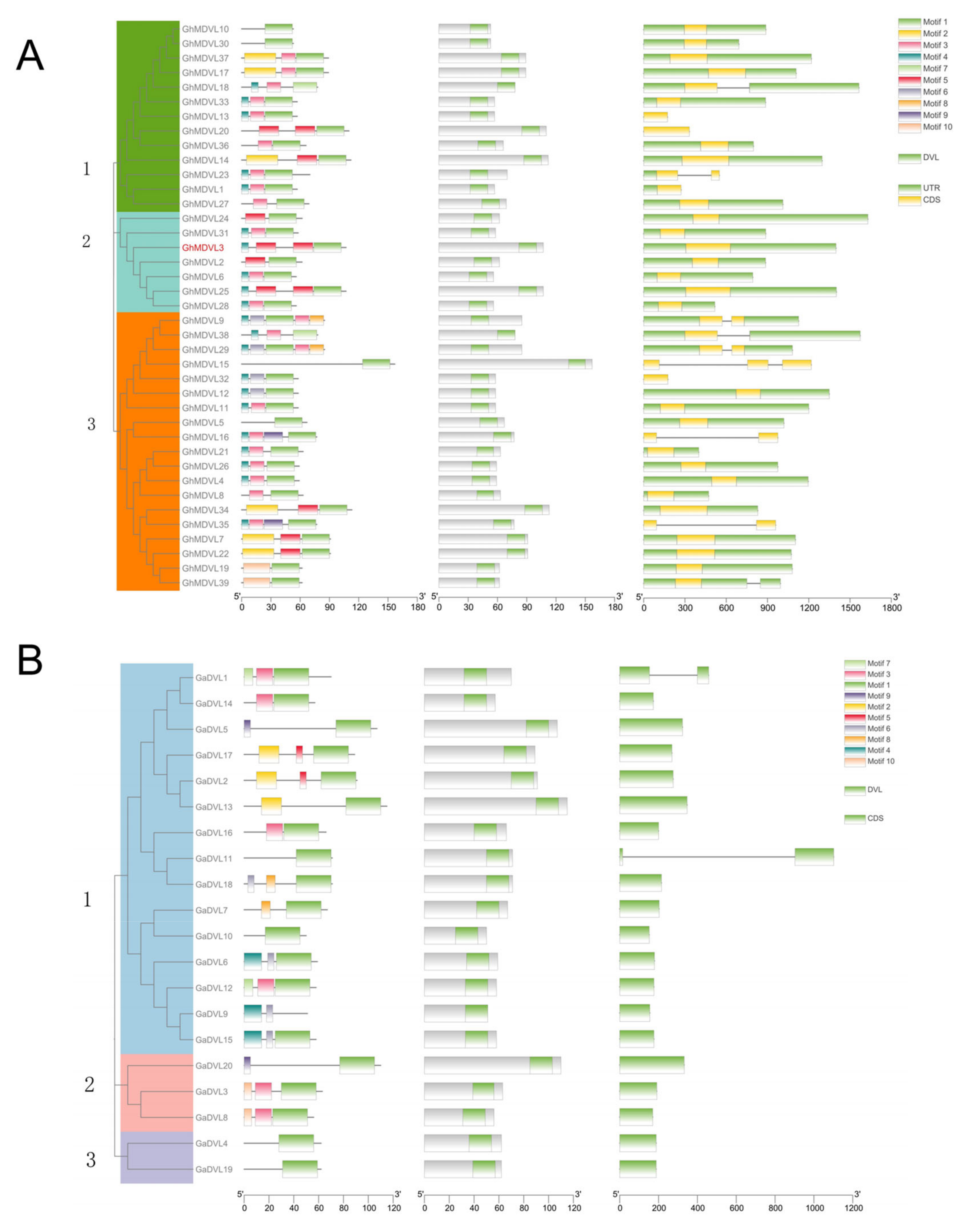
2.4. Gene Structure and Protein Motif Analysis
2.5. Cis-Regulatory Element Analysis of DVL Genes
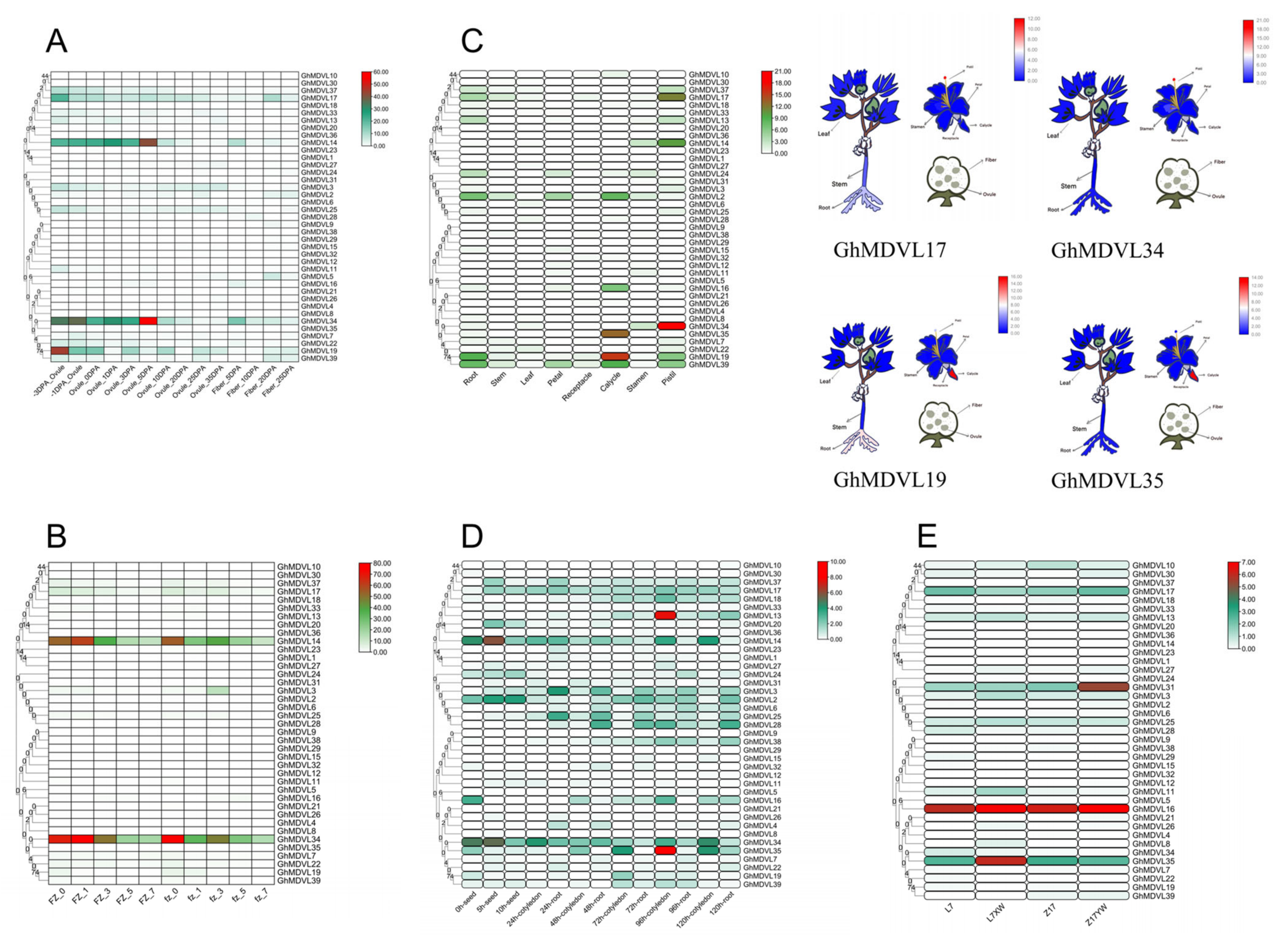
2.6. Tissue Expression Patterns of DVL Genes in G. hirsutum
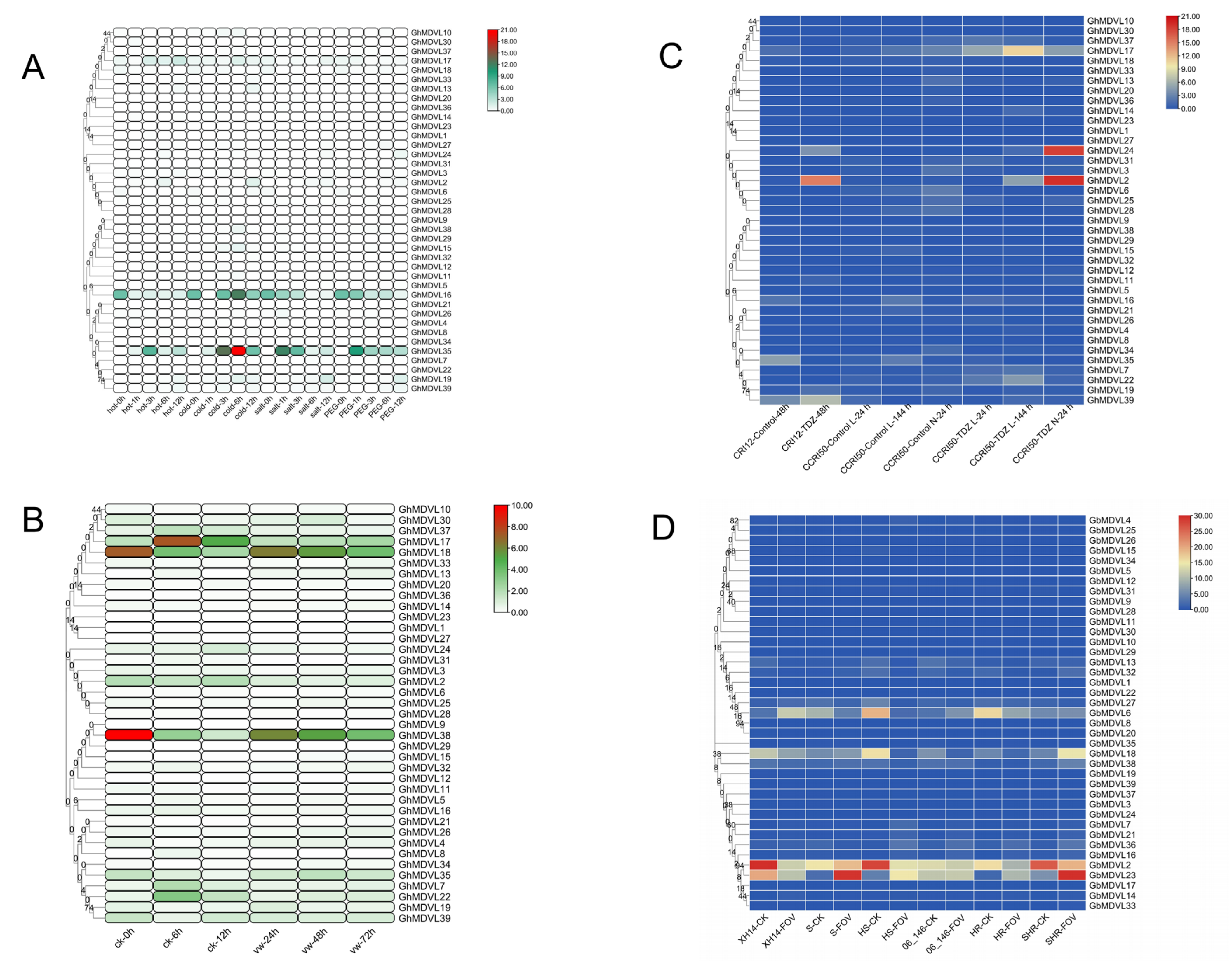
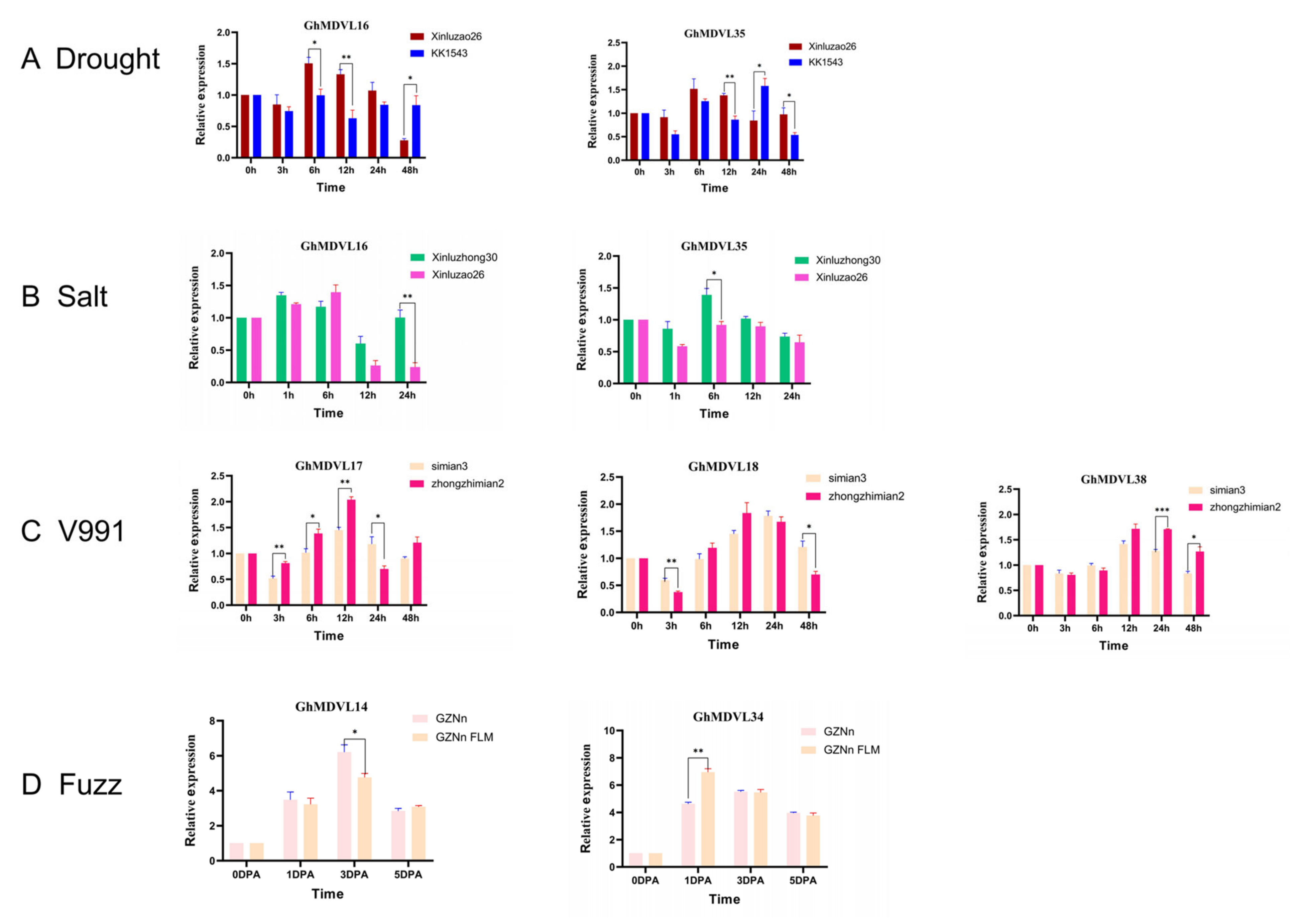
2.7. Expression Patterns of DVL Genes in G. hirsutum under Stress Conditions
2.8. Expression Analysis of the DVL Genes in G. hirsutum in Response to Verticillium Wilt Stress
2.9. Expression Analysis of the DVL Genes in G. hirsutum in Response to TDZ Treatment
2.10. Expression Analysis of the DVL Genes in G. barbadense in Response to FOV Stress
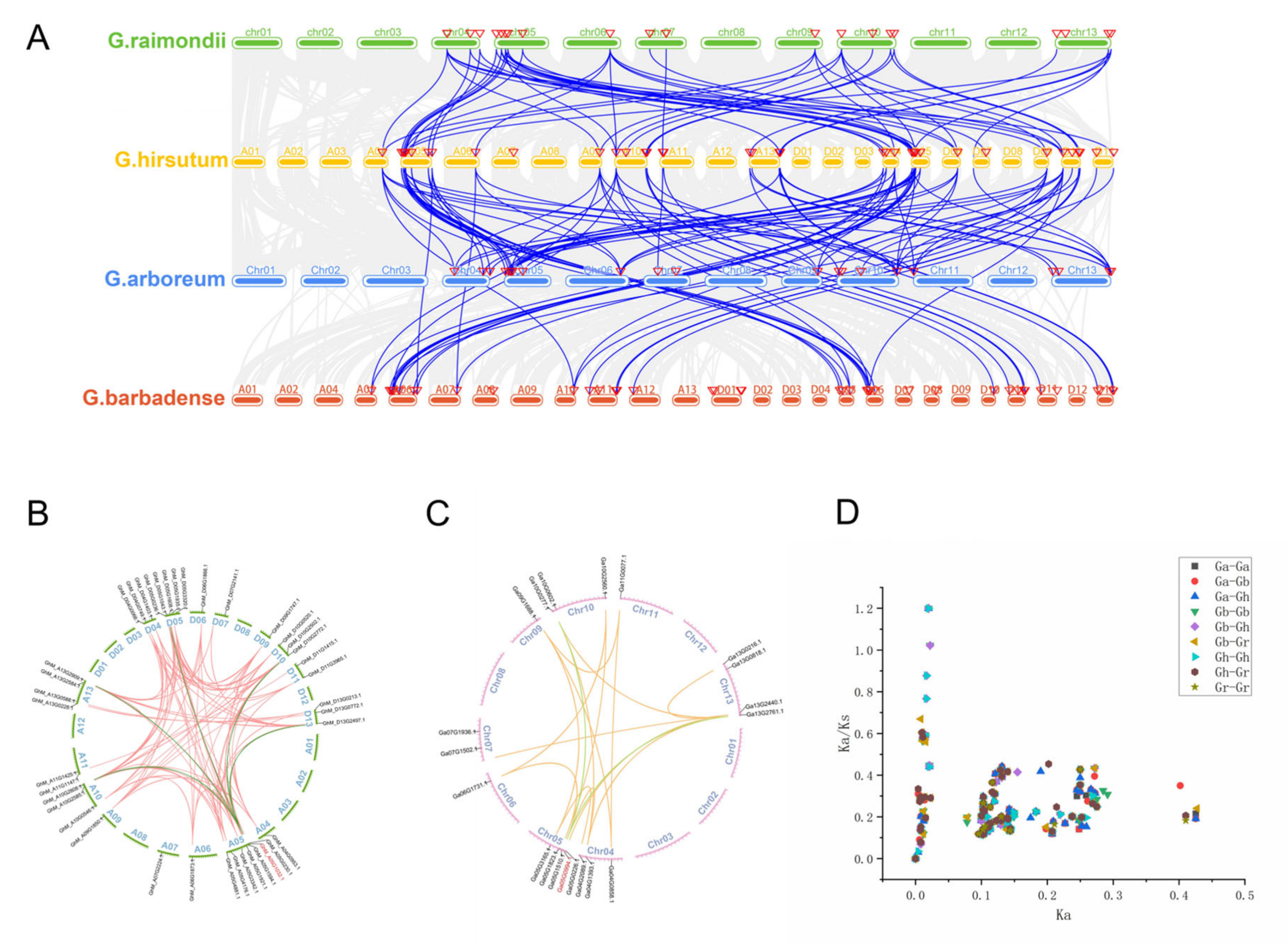
2.11. Gene Replication and Collinearity Analysis
2.12. Calculation of Selection Pressure
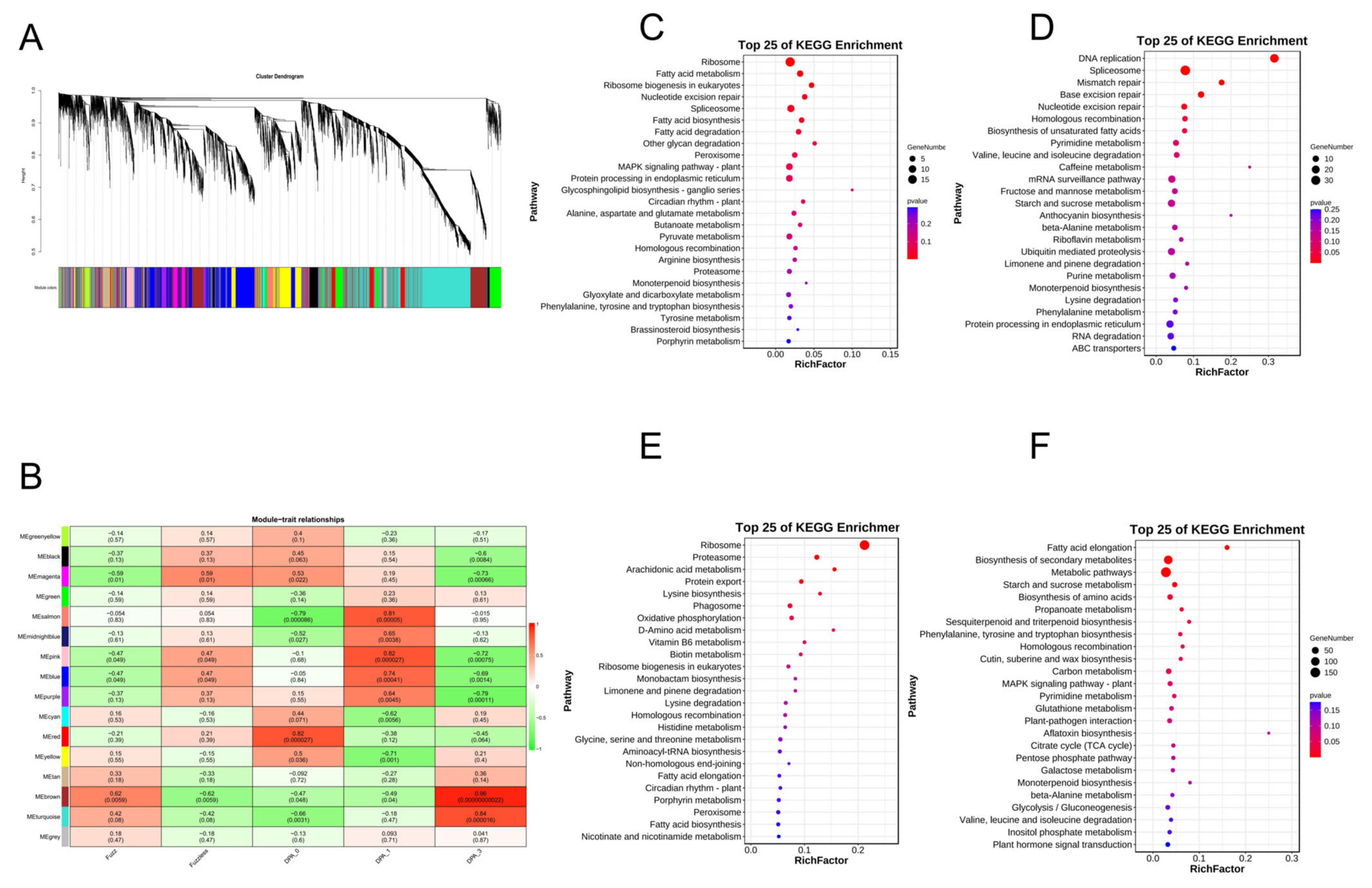
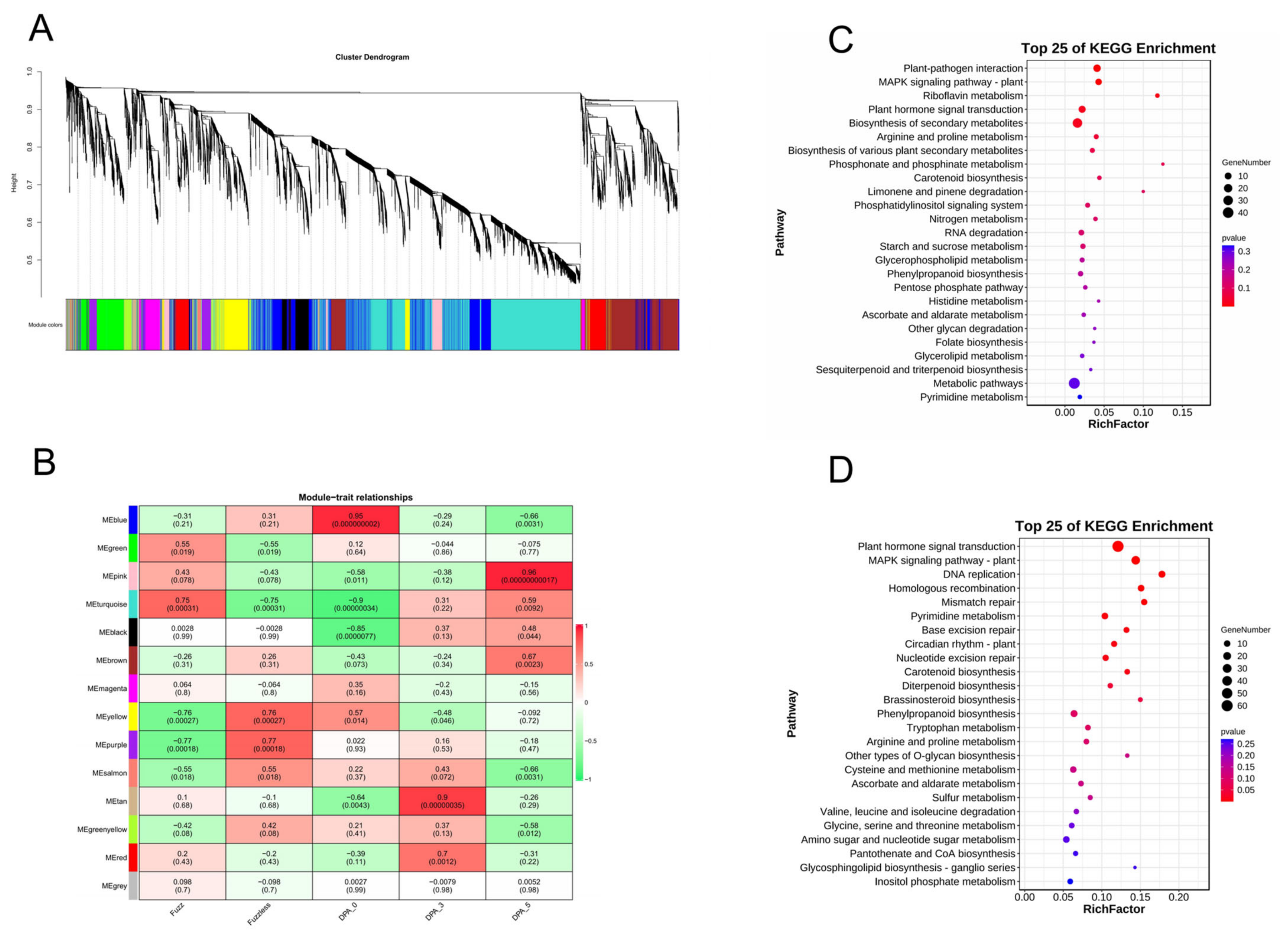
2.13. Transcription Analysis DVL Members
2.14. Functional Validation and Interaction Network Construction for GhM_A05G1032
2.15. qRT-PCR Analysis of DVL Gene in G. hirsutum
3. Discussion
3.1. Basic Analysis of DVL Family Genes
3.2. Gene Expression and Regulation Patterns of DVL Family
4. Materials and Methods
4.1. Identification of Members of the Cotton DVL Gene Family
4.2. Localization of DVL Genes on Chromosomes and Analysis of Gene Duplication in Cotton
4.3. The Construction of a Phylogenetic Tree for the DVL Family Proteins
4.4. Analysis of Gene Structure and Conserved Protein Motifs in the DVL Gene Family
4.5. Analysis of Expression Patterns and Cis-Elements in DVL Family Genes
4.6. Collinearity Analysis of DVL Family Genes
4.7. Selective Pressure Calculation
4.8. An Investigation into Gene Expression Patterns and the Construction of Weighted Gene Co-Expression Networks
4.9. KEGG Enrichment Analysis and Construction of Interaction Networks
4.10. Cotton Material and qRT-PCR Analysis
4.11. VIGS Silencing of Gene GhMDVL3
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fang, L.; Gong, H.; Hu, Y.; Liu, C.; Zhou, B.; Huang, T.; Wang, Y.; Chen, S.; Fang, D.D.; Du, X.; et al. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 2017, 18, 33. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Wang, B.; Chee, P. Recent advances in cotton genomics. Int. J. Plant Genom. 2008, 2008, 742304. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; He, X.; Tu, L.; Zhu, L.; Zhu, S.; Ge, Z.; Zhang, X. GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 2016, 88, 921–935. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhu, Y.; Hu, W.; Zhang, X.; Cai, C.; Guo, W. Comparative Transcriptomics Reveals Jasmonic Acid-Associated Metabolism Related to Cotton Fiber Initiation. Public Libr. Sci. One 2015, 10, e0129854. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.; Pierce, G.J.; Waghmare, V.N.; Rogers, C.J.; Desai, A.; Chee, P.W.; May, O.L.; Gannaway, J.R.; Wendel, J.F.; Wilkins, T.A.; et al. Genetic mapping and comparative analysis of seven mutants related to seed fiber development in cotton. Theor. Appl. Genet. 2005, 111, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hinchliffe, D.J.; Triplett, B.A.; Chen, Z.J.; Stelly, D.M.; Yeater, K.M.; Moon, H.S.; Gilbert, M.K.; Thyssen, G.N.; Turley, R.B.; et al. Phytohormonal networks promote differentiation of fiber initials on pre-anthesis cotton ovules grown in vitro and in planta. Public Libr. Sci. One 2015, 10, e0125046. [Google Scholar] [CrossRef]
- Liang, W.; Fang, L.; Xiang, D.; Hu, Y.; Feng, H.; Chang, L.; Zhang, T. Transcriptome Analysis of Short Fiber Mutant Ligon lintless-1 (Li1) Reveals Critical Genes and Key Pathways in Cotton Fiber Elongation and Leaf Development. Public Libr. Sci. One 2015, 10, e0143503. [Google Scholar] [CrossRef]
- Lee, J.; Woodward, A.; Chen, Z. Gene expression changes and early events in cotton fibre development. Ann. Bot. 2007, 100, 1391–1401. [Google Scholar] [CrossRef]
- Wan, Q.; Guan, X.; Yang, N.; Wu, H.; Pan, M.; Liu, B.; Fang, L.; Yang, S.; Hu, Y.; Ye, W.; et al. Small interfering RNAs from bidirectional transcripts of GhMML3_A12 regulate cotton fiber development. New Phytol. 2016, 210, 1298–1310. [Google Scholar] [CrossRef]
- Feng, X.; Cheng, H.; Zuo, D.; Zhang, Y.; Wang, Q.; Liu, K.; Ashraf, J.; Yang, Q.; Li, S.; Chen, X.; et al. Fine mapping and identification of the fuzzless gene GaFzl in DPL972 (Gossypium arboreum). Theor. Appl. Genet. 2019, 132, 2169–2179. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, E.; Chevalier, D.; Sampedro, J.; Taylor, I.; Niederhuth, C.; Walker, J. DVL genes play a role in the coordination of socket cell recruitment and differentiation. J. Exp. Bot. 2012, 63, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Zhang, H.; Ye, W.; Wu, H.; Zhang, T. Genome-wide transcriptome profiling revealed cotton fuzz fiber development having a similar molecular model as Arabidopsis trichome. Public Libr. Sci. One 2014, 9, e97313. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef]
- Jonathan, F.; Wendel, R. Cronn.Polyploidy and the evolutionary history of cotton. Adv. Agron. 2002, 87, 139–186. [Google Scholar]
- Hu, Y.; Chen, J.; Fang, L.; Zhang, Z.; Ma, W.; Niu, Y.; Ju, L.; Deng, J.; Zhao, T.; Lian, J.; et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 2019, 51, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Long, Y.; Xu, K.; Zhao, F.; Zhao, J.; Li, S.; Geng, S.; Gao, W.; Sun, P.; Deng, X.; et al. Weighted Gene Co-Expression Network Analysis Reveals Hub Genes for Fuzz Development in Gossypium hirsutum. Genes 2023, 14, 208. [Google Scholar] [CrossRef]
- Dai, Y.; Liu, S.; Zuo, D.; Wang, Q.; Lv, L.; Zhang, Y.; Cheng, H.; Yu, J.; Song, G. Identification of MYB gene family and functional analysis of GhMYB4 in cotton (Gossypium spp.). Mol. Genet. Genom. 2023, 298, 755–766. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhao, M.; Li, T.; Wang, L.; Liao, C.; Liu, D.; Zhang, H.; Zhao, Y.; Liu, L.; Ge, X.; et al. Interactions between Verticillium dahliae and cotton: Pathogenic mechanism and cotton resistance mechanism to Verticillium wilt. Front. Plant Sci. 2023, 14, 1174281. [Google Scholar] [CrossRef]
- Shu, H.; Sun, S.; Wang, X.; Yang, C.; Zhang, G.; Meng, Y.; Wang, Y.; Hu, W.; Liu, R. Low Temperature Inhibits the Defoliation Efficiency of Thidiazuron in Cotton by Regulating Plant Hormone Synthesis and the Signaling Pathway. Int. J. Mol. Sci. 2022, 23, 14208. [Google Scholar] [CrossRef]
- Yao, Z.; Chen, Q.; Chen, D.; Zhan, L.; Zeng, K.; Gu, A.; Zhou, J.; Zhang, Y.; Zhu, Y.; Gao, W.; et al. The susceptibility of sea-island cotton recombinant inbred lines to Fusarium oxysporum f. sp. vasinfectum infection is characterized by altered expression of long noncoding RNAs. Sci. Rep. 2019, 9, 2894. [Google Scholar] [CrossRef]
- Wang, X.; Miao, Y.; Cai, Y.; Sun, G.; Jia, Y.; Song, S.; Pan, Z.; Zhang, Y.; Wang, L.; Fu, G.; et al. Large-fragment insertion activates gene GaFZ (Ga08G0121) and is associated with the fuzz and trichome reduction in cotton (Gossypium arboreum). Plant Biotechnol. J. 2021, 19, 1110–1124. [Google Scholar] [CrossRef]
- Feng, X.; Cheng, H.; Zuo, D.; Zhang, Y.; Wang, Q.; Lv, L.; Li, S.; Yu, J.; Song, G. Genome-wide identification and expression analysis of GL2-interacting-repressor (GIR) genes during cotton fiber and fuzz development. Planta 2021, 255, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shi, Y.; Xiao, X.; Song, J.; Li, P.; Gong, J.; Zhang, H.; Gong, W.; Liu, A.; Peng, R.; et al. Genome-wide characterization of trichome birefringence-like genes provides insights into fiber yield improvement. Front. Plant Sci. 2023, 14, 1127760. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Jiao, Y.; Jiang, Z.; Liu, P.; Chen, Q.; Qu, Y.; Deng, X. GBSOT4 Enhances the Resistance of Gossypium barbadense to Fusarium oxysporum f. sp. vasinfectum (FOV) by Regulating the Content of Flavonoid. Plants 2023, 12, 3529. [Google Scholar] [CrossRef]
- Shuya, M.; Le, L.; Huiyun, S.; Yu, G.; Yujun, L.; Qanmber, G. Genomic identification of cotton SAC genes branded ovule and stress-related key genes in Gossypium hirsutum. Front. Plant Sci. 2023, 14, 1123745. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, Y.; Li, P.; Gong, W.; Wang, X.; Yan, H.; Ge, Q.; Liu, A.; Shi, Y.; Shang, H.; et al. Genome-Wide Analysis and Functional Characterization of LACS Gene Family Associated with Lipid Synthesis in Cotton (Gossypium spp.). Int. J. Mol. Sci. 2023, 24, 8530. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Pan, A.; Lei, M.; Song, Z.; Chen, Y.; Wang, X.; Gao, Y.; Zhang, J.; Wang, S.; Zhao, Y.; et al. Genome-Wide Characterization and Functional Analysis of ABCG Subfamily Reveal Its Role in Cutin Formation in Cotton. Int. J. Mol. Sci. 2023, 24, 2379. [Google Scholar] [CrossRef]
- Li, J.; Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Zheng, L.; Liu, Z.; Wu, H.; Liu, X.; Chen, Q.; et al. GhKLCR1, a kinesin light chain-related gene, induces drought-stress sensitivity in Arabidopsis. Sci. China-Life Sci. 2019, 62, 63–75. [Google Scholar] [CrossRef]
- Qanmber, G.; Yu, D.; Li, J.; Wang, L.; Ma, S.; Lu, L.; Yang, Z.; Li, F. Genome-wide identification and expression analysis of Gossypium RING-H2 finger E3 ligase genes revealed their roles in fiber development, and phytohormone and abiotic stress responses. J. Cotton Res. 2018, 1, 1. [Google Scholar] [CrossRef]
- Wang, L.; Yang, Z.; Zhang, B.; Yu, D.; Liu, J.; Gong, Q.; Qanmber, G.; Li, Y.; Lu, L.; Lin, Y.; et al. Genome-wide characterization and phylogenetic analysis of GSK gene family in three species of cotton: Evidence for a role of some GSKs in fiber development and responses to stress. BMC Plant Biol. 2018, 18, 330. [Google Scholar] [CrossRef]
- Yu, D.; Qanmber, G.; Lu, L.; Wang, L.; Li, J.; Yang, Z.; Liu, Z.; Li, Y.; Chen, Q.; Mendu, V.; et al. Genome-wide analysis of cotton GH3 subfamily II reveals functional divergence in fiber development, hormone response and plant architecture. BMC Plant Biol. 2018, 18, 350. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.; Mitra, A.; Baumgarten, A.; Young, N.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S. Evolution of Gene Duplication in Plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef]
- Zhu, T.; Liang, C.; Meng, Z.; Sun, G.; Meng, Z.; Guo, S.; Zhang, R. CottonFGD: An integrated functional genomics database for cotton. BMC Plant Biol. 2017, 17, 101. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.; Coggill, P.; Eberhardt, R.; Eddy, S.; Mistry, J.; Mitchell, A.; Potter, S.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.; Gasteiger, E.; Bairoch, A.; Sanchez, J.; Williams, K.; Appel, R.; Hochstrasser, D. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiao, Y.; Zhao, F.; Geng, S.; Li, S.; Su, Z.; Chen, Q.; Yu, Y.; Qu, Y. Genome-Wide and Expression Pattern Analysis of the DVL Gene Family Reveals GhM_A05G1032 Is Involved in Fuzz Development in G. hirsutum. Int. J. Mol. Sci. 2024, 25, 1346. https://doi.org/10.3390/ijms25021346
Jiao Y, Zhao F, Geng S, Li S, Su Z, Chen Q, Yu Y, Qu Y. Genome-Wide and Expression Pattern Analysis of the DVL Gene Family Reveals GhM_A05G1032 Is Involved in Fuzz Development in G. hirsutum. International Journal of Molecular Sciences. 2024; 25(2):1346. https://doi.org/10.3390/ijms25021346
Chicago/Turabian StyleJiao, Yang, Fuxiang Zhao, Shiwei Geng, Shengmei Li, Zhanlian Su, Quanjia Chen, Yu Yu, and Yanying Qu. 2024. "Genome-Wide and Expression Pattern Analysis of the DVL Gene Family Reveals GhM_A05G1032 Is Involved in Fuzz Development in G. hirsutum" International Journal of Molecular Sciences 25, no. 2: 1346. https://doi.org/10.3390/ijms25021346
APA StyleJiao, Y., Zhao, F., Geng, S., Li, S., Su, Z., Chen, Q., Yu, Y., & Qu, Y. (2024). Genome-Wide and Expression Pattern Analysis of the DVL Gene Family Reveals GhM_A05G1032 Is Involved in Fuzz Development in G. hirsutum. International Journal of Molecular Sciences, 25(2), 1346. https://doi.org/10.3390/ijms25021346






