Biology of Healthy Aging: Biological Hallmarks of Stress Resistance Related and Unrelated to Longevity in Humans
Abstract
1. Introduction
2. Results
2.1. Stress Resistance
2.1.1. Databases and Overview of Stress Resistance
2.1.2. Stress Resistance: Cell-Cycle-Related Pathways
2.1.3. Stress Resistance: Signal Transduction–Gene Expression Pathways
2.1.4. Stress Resistance: Metabolic Pathways
2.1.5. Stress Resistance: Metabolism of Proteins, Metabolism of RNA, and Others
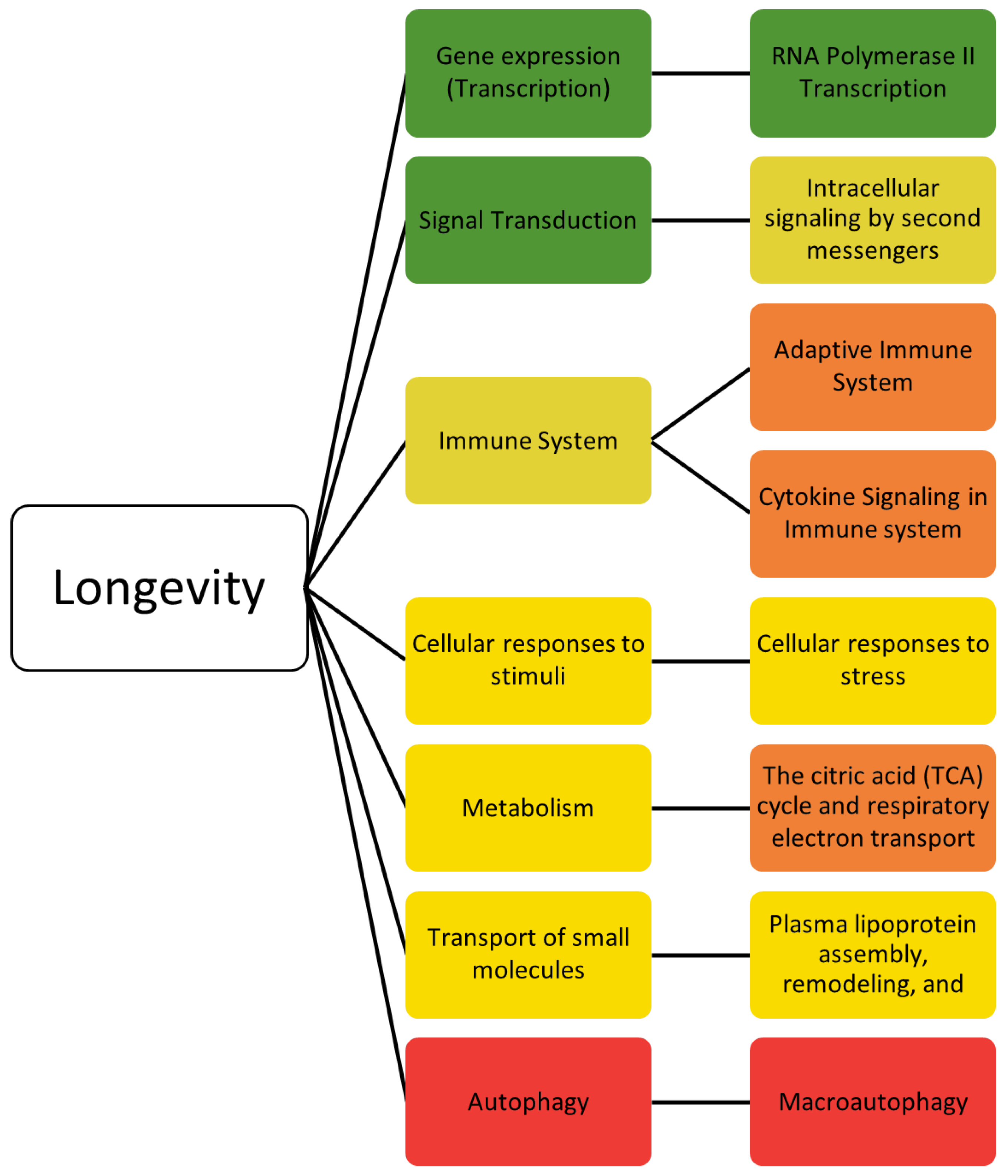
2.2. Longevity
2.3. Overlap for Stress Resistance and Longevity
3. Discussion
3.1. Similarity among Stress Resistance and Longevity Pathways
3.2. Differences among Stress Resistance and Longevity Pathways
3.3. Translation of the Results into Potential Therapeutic Targets or Interventions
3.3.1. Overlapped Pathway Categories between Stress Resistance and Longevity
3.3.2. Non-Overlapped Pathway Categories between Stress Resistance and Longevity
3.4. Advantages and Limitations of the Method
3.5. Nuclear Transport and Integrity Emphasized Stress Resistance and Aging but Not Longevity
4. Materials and Methods
4.1. Nomenclatures
4.2. Datasets
4.3. Gene Ontology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harman, D. Aging: A theory based on free radical and radiation chemistry. J. Gerontol. 1956, 11, 298–300. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.L.; Lustgarten, M.S.; Jang, Y.; Richardson, A.; Van Remmen, H. Trends in oxidative aging theories. Free Radic. Biol. Med. 2007, 43, 477–503. [Google Scholar] [CrossRef] [PubMed]
- Doonan, R.; McElwee, J.J.; Matthijssens, F.; Walker, G.A.; Houthoofd, K.; Back, P.; Matscheski, A.; Vanfleteren, J.R.; Gems, D. Against the oxidative damage theory of aging: Superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 2008, 22, 3236–3241. [Google Scholar] [CrossRef] [PubMed]
- Pérez, V.I.; Bokov, A.; Van Remmen, H.; Mele, J.; Ran, Q.; Ikeno, Y.; Richardson, A. Is the oxidative stress theory of aging dead? Biochim. Biophys. Acta 2009, 1790, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Dues, D.J.; Andrews, E.K.; Schaar, C.E.; Bergsma, A.L.; Senchuk, M.M.; Van Raamsdonk, J.M. Aging causes decreased resistance to multiple stresses and a failure to activate specific stress response pathways. Aging 2016, 8, 777–795. [Google Scholar] [CrossRef]
- Salmon, A.B.; Richardson, A.; Pérez, V.I. Update on the oxidative stress theory of aging: Does oxidative stress play a role in aging or healthy aging? Free Radic. Biol. Med. 2010, 48, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Ziada, A.S.; Smith, M.R.; Côté, H.C.F. Updating the Free Radical Theory of Aging. Front. Cell Dev. Biol. 2020, 8, 575645. [Google Scholar] [CrossRef]
- Douglas, P.M.; Baird, N.A.; Simic, M.S.; Uhlein, S.; McCormick, M.A.; Wolff, S.C.; Kennedy, B.K.; Dillin, A. Heterotypic Signals from Neural HSF-1 Separate Thermotolerance from Longevity. Cell Rep. 2015, 12, 1196–1204. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, J.; Robida-Stubbs, S.; Tullet, J.M.; Rual, J.F.; Vidal, M.; Blackwell, T.K. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. 2010, 6, e1001048. [Google Scholar] [CrossRef]
- Zhang, P.; Zhai, Y.; Cregg, J.; Ang, K.K.; Arkin, M.; Kenyon, C. Stress Resistance Screen in a Human Primary Cell Line Identifies Small Molecules That Affect Aging Pathways and Extend Caenorhabditis elegans’ Lifespan. G3 Genes Genomes Genet. 2020, 10, 849–862. [Google Scholar] [CrossRef]
- Derisbourg, M.J.; Wester, L.E.; Baddi, R.; Denzel, M.S. Mutagenesis screen uncovers lifespan extension through integrated stress response inhibition without reduced mRNA translation. Nat. Commun. 2021, 12, 1678. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.M.; Salmon, A.B.; Chang, Y.; Bonkowski, M.; Bartke, A.; Miller, R.A. Stress resistance and aging: Influence of genes and nutrition. Mech. Ageing Dev. 2006, 127, 687–694. [Google Scholar] [CrossRef]
- Murakami, S. Stress resistance in long-lived mouse models. Exp. Gerontol. 2006, 41, 1014–1019. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.I.; Pincus, Z.; Slack, F.J. Longevity and stress in Caenorhabditis elegans. Aging 2011, 3, 733–753. [Google Scholar] [CrossRef]
- Soo, S.K.; Rudich, Z.D.; Ko, B.; Moldakozhayev, A.; AlOkda, A.; Van Raamsdonk, J.M. Biological resilience and aging: Activation of stress response pathways contributes to lifespan extension. Ageing Res. Rev. 2023, 88, 101941. [Google Scholar] [CrossRef]
- Murakami, S.; Johnson, T.E. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 1996, 143, 1207–1218. [Google Scholar] [CrossRef]
- Murakami, S.; Salmon, A.; Miller, R.A. Multiplex stress resistance in cells from long-lived dwarf mice. FASEB J. 2003, 17, 1565–1566. [Google Scholar] [CrossRef] [PubMed]
- Salmon, A.B.; Murakami, S.; Bartke, A.; Kopchick, J.; Yasumura, K.; Miller, R.A. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am. J. Physiol. Endocrinol. Metab. 2005, 289, E23–E29. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Murakami, S. Serotonin receptors antagonistically modulate Caenorhabditis elegans longevity. Aging Cell 2007, 6, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Bessinger, K.; Hellmann, J.; Murakami, S. Manipulation of serotonin signal suppresses early phase of behavioral aging in Caenorhabditis elegans. Neurobiol. Aging 2008, 29, 1093–1100. [Google Scholar] [CrossRef] [PubMed]
- Murakami, S.; Cabana, K.; Anderson, D. Current Advances in the Studies of Oxidative Stress and Age-Related Memory Impairment in C. elegans. In Oxidative Stress in Vertebrates and Invertebrates; Farooqui, T., Farooqui, A.A., Eds.; John Wiley & Sons, Inc.: New York, NY, USA, 2011. [Google Scholar] [CrossRef]
- Murakami, S. Age-dependent modulation of learning and memory in Caenorhabditis elegans, in Invertebrate learning and memory. In Handbook of Behavioral Neuroscience; Menzel, R., Benjamin, P.R., Eds.; Elsevier/Academic Press: Cambridge, MA, USA, 2013; pp. 140–150. [Google Scholar] [CrossRef]
- Finch, C.E. Longevity, Senescence, and the Genome; University of Chicago Press: Chicago, IL, USA, 1994. [Google Scholar]
- Miller, J.K. Escaping senescence: Demographic data from the three-toed box turtle (Terrapene carolina triunguis). Exp. Gerontol. 2001, 36, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Stenvinkel, P.; Shiels, P.G. Long-lived animals with negligible senescence: Clues for ageing research. Biochem. Soc. Trans. 2019, 47, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Møller, A.P. An explanation for negligible senescence in animals. Ecol. Evol. 2022, 12, e8970. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ruby, J.G.; Smith, M.; Buffenstein, R. Naked Mole-Rat mortality rates defy gompertzian laws by not increasing with age. eLife 2018, 7, e31157. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oka, K.; Yamakawa, M.; Kawamura, Y.; Kutsukake, N.; Miura, K. The Naked Mole-Rat as a Model for Healthy Aging. Annu. Rev. Anim. Biosci. 2023, 11, 207–226. [Google Scholar] [CrossRef] [PubMed]
- Rando, T.A.; Chang, H.Y. Aging, rejuvenation, and epigenetic reprogramming: Resetting the aging clock. Cell 2012, 148, 46–57. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ocampo, A.; Reddy, P.; Belmonte, J.C.I. Anti-Aging Strategies Based on Cellular Reprogramming. Trends Mol. Med. 2016, 22, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.J.; Olova, N.N.; Chandra, T. Cellular reprogramming and epigenetic rejuvenation. Clin. Epigenet. 2021, 13, 170. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Alle, Q.; Le Borgne, E.; Milhavet, O.; Lemaitre, J.M. Reprogramming: Emerging Strategies to Rejuvenate Aging Cells and Tissues. Int. J. Mol. Sci. 2021, 22, 3990. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yousefzadeh, M.; Henpita, C.; Vyas, R.; Soto-Palma, C.; Robbins, P.; Niedernhofer, L. DNA damage-how and why we age? eLife 2021, 10, e62852. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, J.-H.; Hayano, M.; Griffin, P.T.; Amorim, J.A.; Bonkowski, M.S.; Apostolides, J.K.; Salfati, E.L.; Blanchette, M.; Munding, E.M.; Bhakta, M.; et al. Loss of epigenetic information as a cause of mammalian aging. Cell 2023, 186, 305–326.e27, Erratum in Cell 2024, 187, 1312–1313. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Manukyan, M.; Singh, P.B. Epigenome rejuvenation: HP1β mobility as a measure of pluripotent and senescent chromatin ground states. Sci. Rep. 2014, 4, 4789. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sarkar, T.J.; Quarta, M.; Mukherjee, S.; Colville, A.; Paine, P.; Doan, L.; Tran, C.M.; Chu, C.R.; Horvath, S.; Qi, L.S.; et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 2020, 11, 1545. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yücel, A.D.; Gladyshev, V.N. The long and winding road of reprogramming-induced rejuvenation. Nat. Commun. 2024, 15, 1941. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mitchell, W.; Goeminne, L.J.E.; Tyshkovskiy, A.; Zhang, S.; Chen, J.Y.; Paulo, J.A.; Pierce, K.A.; Choy, A.H.; Clish, C.B.; Gygi, S.P.; et al. Multi-omics characterization of partial chemical reprogramming reveals evidence of cell rejuvenation. eLife 2024, 12, RP90579. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Balmorez, T.; Sakazaki, A.; Murakami, S. Genetic Networks of Alzheimer’s Disease, Aging, and Longevity in Humans. Int. J. Mol. Sci. 2023, 24, 5178. [Google Scholar] [CrossRef]
- Murakami, S.; Lacayo, P. Biological and disease hallmarks of Alzheimer’s disease defined by Alzheimer’s disease genes. Front. Aging Neurosci. 2022, 14, 996030. [Google Scholar] [CrossRef] [PubMed]
- Stünkel, W.; Campbell, R.M. Sirtuin 1 (SIRT1): The misunderstood HDAC. J. Biomol. Screen. 2011, 16, 1153–1169. [Google Scholar] [CrossRef] [PubMed]
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Crimmins, E.M. A Genetic Network Associated with Stress Resistance, Longevity, and Cancer in Humans. J. Gerontol. A Biol. Sci. Med. Sci. 2016, 71, 703–712. [Google Scholar] [CrossRef][Green Version]
- Machino, K.; Link, C.D.; Wang, S.; Murakami, H.; Murakami, S. A semi-automated motion-tracking analysis of locomotion speed in the C. elegans transgenics overexpressing beta-amyloid in neurons. Front. Genet. 2014, 5, 202. [Google Scholar] [CrossRef] [PubMed]
- Tauffenberger, A.; Fiumelli, H.; Almustafa, S.; Magistretti, P.J. Lactate and pyruvate promote oxidative stress resistance through hormetic ROS signaling. Cell Death Dis. 2019, 10, 653. [Google Scholar] [CrossRef] [PubMed]
- Dues, D.J.; Andrews, E.K.; Senchuk, M.M.; Van Raamsdonk, J.M. Resistance to Stress Can Be Experimentally Dissociated from Longevity. J. Gerontol. A Biol. Sci. Med. Sci. 2019, 74, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Petrucci, G.; Rizzi, A.; Hatem, D.; Tosti, G.; Rocca, B.; Pitocco, D. Role of Oxidative Stress in the Pathogenesis of Atherothrombotic Diseases. Antioxidants 2022, 11, 1408. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lyons, T.J. Glycation and oxidation: A role in the pathogenesis of atherosclerosis. Am. J. Cardiol. 1993, 71, 26B–31B. [Google Scholar] [CrossRef] [PubMed]
- WHO. Global Health Estimates: Life Expectancy and Leading Causes of Death and Disability. 2020. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 13 July 2023).
- CDC. Mortality in the United States, 2022. In Deaths and Mortality, National Center for Health Statistics. Available online: https://www.cdc.gov/nchs/fastats/deaths.htm (accessed on 13 July 2023).
- Gonzalez-Freire, M.; Diaz-Ruiz, A.; Hauser, D.; Martinez-Romero, J.; Ferrucci, L.; Bernier, M.; de Cabo, R. The road ahead for health and lifespan interventions. Ageing Res. Rev. 2020, 59, 101037. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Le Couteur, D.G.; Barzilai, N. New horizons in life extension, healthspan extension and exceptional longevity. Age Ageing 2022, 51, afac156. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- de Cabo, R.; Le Couteur, D. Biology of Aging. In Harrison’s Principles of Internal Medicine, 21st ed.; Loscalzo, J., Fauci, A., Kasper, D., Hauser, S., Longo, D., Jameson, J., Eds.; McGraw-Hill Education: New York, NY, USA, 2022; Available online: https://accessmedicine.mhmedical.com/content.aspx?bookid=3095§ionid=265470089 (accessed on 17 September 2024).
- Mackay, T.F. Epistasis and quantitative traits: Using model organisms to study gene-gene interactions. Nat. Rev. Genet. 2014, 15, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Mackay, T.F.; Moore, J.H. Why epistasis is important for tackling complex human disease genetics. Genome Med. 2014, 6, 124, Erratum in Genome Med. 2015, 7, 85. [Google Scholar] [CrossRef]
- Vahdati Nia, B.; Kang, C.; Tran, M.G.; Lee, D.; Murakami, S. Meta Analysis of Human AlzGene Database: Benefits and Limitations of Using, C. elegans for the Study of Alzheimer’s Disease and Co-morbid Conditions. Front. Genet. 2017, 8, 55. [Google Scholar] [CrossRef]
- Le, D.; Brown, L.; Malik, K.; Murakami, S. Two Opposing Functions of Angiotensin-Converting Enzyme (ACE) That Links Hypertension, Dementia, and Aging. Int. J. Mol. Sci. 2021, 22, 13178. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, M.A.; Hetzer, M.W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008, 18, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.; Sousa, J.; Pereira, C.D.; da Cruz ESilva, O.A.B.; Rebelo, S. Nuclear envelope dysfunction and its contribution to the aging process. Aging Cell 2020, 19, e13143. [Google Scholar] [CrossRef]
- Eriksson, M.; Brown, W.T.; Gordon, L.B.; Glynn, M.W.; Singer, J.; Scott, L.; Erdos, M.R.; Robbins, C.M.; Moses, T.Y.; Berglund, P.; et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 2003, 423, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Cenni, V.; Capanni, C.; Mattioli, E.; Schena, E.; Squarzoni, S.; Bacalini, M.G.; Garagnani, P.; Salvioli, S.; Franceschi, C.; Lattanzi, G. Lamin A involvement in ageing processes. Ageing Res. Rev. 2020, 62, 101073. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari Fuchs, S.; Lieder, I.; Stelzer, G.; Mazor, Y.; Buzhor, E.; Kaplan, S.; Bogoch, Y.; Plaschkes, I.; Shitrit, A.; Rappaport, N.; et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. OMICS 2016, 20, 139–151. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer: Singapore, 2021. [Google Scholar] [CrossRef]
- von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Friedrich, K.; Lee, L.; Leistritz, D.F.; Nürnberg, G.; Saha, B.; Hisama, F.M.; Eyman, D.K.; Lessel, D.; Nürnberg, P.; Li, C.; et al. WRN mutations in Werner syndrome patients: Genomic rearrangements, unusual intronic mutations and ethnic-specific alterations. Hum. Genet. 2010, 128, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.M.; Hisama, F.M.; Oshima, J. Review of How Genetic Research on Segmental Progeroid Syndromes Has Documented Genomic Instability as a Hallmark of Aging but Let Us Now Pursue Antigeroid Syndromes! J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, 253–259. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]



| Functional Enrichment Dimensions | Number of Hits |
|---|---|
| Biological Process (Gene Ontology) | 1359 GO-terms |
| Molecular Function (Gene Ontology) | 162 GO-terms |
| Cellular Component (Gene Ontology) | 163 GO-terms |
| Reference publications (PubMed) | 10,000 publications |
| Local network cluster (STRING) | 116 clusters |
| KEGG Pathways | 125 pathways |
| Reactome Pathways | 468 pathways |
| GO (gene ontology) |
| Pathways | No. of Pathways |
|---|---|
| Total Number of Pathways Identified | 172 |
| Total Number of Pathway Categories Grouped * | 14 |
| Breakdowns of 172 Pathway Categories | No. of pathways |
| 1 Cell Cycle | 40 |
| 2 DNA Repair | 37 |
| 3 Gene Expression (Transcription) | 19 |
| 4 Metabolism of Proteins | 17 |
| 5 Metabolism of RNA | 14 |
| 6 Signal Transduction | 14 |
| 7 Cellular Responses to Stimuli | 11 |
| 8 Immune System | 6 |
| 9 Metabolism | 4 |
| 10 Autophagy | 3 |
| 11 Chromatin Organization | 2 |
| 12 Developmental Biology | 2 |
| 13 Organelle Biogenesis | 2 |
| 14 DNA Replication | 1 |
| Total | 172 |
| Selected Pathways for Stress-Resistance Pathway Enrichment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reactome Pathway | Ratio of Protein in Pathway | Number of Protein in Pathway | Protein from Gene Set | p-Value | FDR | Hit Genes | General | Specific | More Specific |
| Cell Cycle, Mitotic | 0.0478 | 530 | 86 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,SMC3,SMC4,SMC2,MYC,AKT1,NUP210,PRKCA,NUP93,PSME1,NUP98,NUP205,SEH1L,NUP85,RAD21,NUP88,PLK1,TUBA1B,TUBA1A,EMD,NDC1,SIRT2,TUBB2A,TUBB4B,PSMC6,PSMC3,NCAPD2,ANAPC7,LEMD2,KPNB1,ANAPC1,HSP90AB1,MCM7,FOXM1,TNPO1,RFC5,RFC3,HSP90AA1,RFC4,RFC1,RFC2,RANGAP1,SMC1A,MCM6,MCM2,HDAC1,RBBP4,H2AZ2,CDC27,RCC1,H4C1,SKP2,TMPO,TP53,WAPL,TOP2A,CETN2,SEC13,TUBB,SFI1,PCNA,LMNA,RAE1,H2BC3,RPA1,RPA2,RPA3,NUP188,CCND1,BANF1,PSMD11,PDS5A,NUP160,UBE2I,NUP155,LMNB1,NUP62,NUP133,NUP58,POLD3,POLD2,NUP43,BUB3,NUP35,RAN,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Cell Cycle, Mitotic |
| Mitotic Prophase | 0.0101 | 112 | 33 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,SMC4,SMC2,NUP210,PRKCA,NUP93,NUP98,NUP205,SEH1L,NUP85,NUP88,PLK1,EMD,NDC1,LEMD2,H2AZ2,H4C1,TMPO,SEC13,LMNA,RAE1,H2BC3,NUP188,BANF1,NUP160,NUP155,LMNB1,NUP62,NUP133,NUP58,NUP43,NUP35,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Mitotic Prophase |
| Mitotic Anaphase | 0.0212 | 235 | 48 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,SMC3,NUP93,PSME1,NUP98,NUP205,SEH1L,NUP85,RAD21,PLK1,TUBA1B,TUBA1A,EMD,NDC1,SIRT2,TUBB2A,TUBB4B,PSMC6,PSMC3,ANAPC7,LEMD2,KPNB1,ANAPC1,TNPO1,RANGAP1,SMC1A,CDC27,RCC1,TMPO,WAPL,SEC13,LMNA,NUP188,BANF1,PSMD11,PDS5A,NUP160,UBE2I,NUP155,LMNB1,NUP62,NUP133,NUP58,NUP43,BUB3,NUP35,RAN,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Mitotic Anaphase |
| Nuclear Envelope Breakdown | 0.0048 | 53 | 28 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,NUP210,PRKCA,NUP93,NUP98,NUP205,SEH1L,NUP85,NUP88,PLK1,EMD,NDC1,LEMD2,TMPO,SEC13,LMNA,RAE1,NUP188,BANF1,NUP160,NUP155,LMNB1,NUP62,NUP133,NUP58,NUP43,NUP35,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Nuclear Envelope Breakdown |
| M Phase | 0.0348 | 386 | 62 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,SMC3,SMC4,SMC2,NUP210,PRKCA,NUP93,PSME1,NUP98,NUP205,SEH1L,NUP85,RAD21,NUP88,PLK1,TUBA1B,TUBA1A,EMD,NDC1,SIRT2,TUBB2A,TUBB4B,PSMC6,PSMC3,NCAPD2,ANAPC7,LEMD2,KPNB1,ANAPC1,TNPO1,HSP90AA1,RANGAP1,SMC1A,H2AZ2,CDC27,RCC1,H4C1,TMPO,WAPL,CETN2,SEC13,TUBB,SFI1,LMNA,RAE1,H2BC3,NUP188,BANF1,PSMD11,PDS5A,NUP160,UBE2I,NUP155,LMNB1,NUP62,NUP133,NUP58,NUP43,BUB3,NUP35,RAN,NUP37 | Cell Cycle | Cell Cycle, Mitotic | M Phase |
| Nuclear Pore Complex (NPC) Disassembly | 0.0032 | 36 | 20 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,NUP210,NUP93,NUP98,NUP205,SEH1L,NUP85,NUP88,NDC1,SEC13,RAE1,NUP188,NUP160,NUP155,NUP62,NUP133,NUP58,NUP43,NUP35,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Nuclear Pore Complex (NPC) Disassembly |
| Mitotic Metaphase and Anaphase | 0.0213 | 236 | 48 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,SMC3,NUP93,PSME1,NUP98,NUP205,SEH1L,NUP85,RAD21,PLK1,TUBA1B,TUBA1A,EMD,NDC1,SIRT2,TUBB2A,TUBB4B,PSMC6,PSMC3,ANAPC7,LEMD2,KPNB1,ANAPC1,TNPO1,RANGAP1,SMC1A,CDC27,RCC1,TMPO,WAPL,SEC13,LMNA,NUP188,BANF1,PSMD11,PDS5A,NUP160,UBE2I,NUP155,LMNB1,NUP62,NUP133,NUP58,NUP43,BUB3,NUP35,RAN,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Mitotic Metaphase and Anaphase |
| Postmitotic Nuclear Pore Complex (NPC) Reformation | 0.0024 | 27 | 23 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,NUP93,NUP98,NUP205,SEH1L,NUP85,NDC1,KPNB1,TNPO1,RANGAP1,RCC1,SEC13,NUP188,NUP160,UBE2I,NUP155,NUP62,NUP133,NUP58,NUP43,NUP35,RAN,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Postmitotic Nuclear Pore Complex (NPC) Reformation |
| Nuclear Envelope (NE) Reassembly | 0.0068 | 76 | 34 | 1.11 × 10−16 | 4.44 × 10−15 | NUP107,NUP93,NUP98,NUP205,SEH1L,NUP85,TUBA1B,TUBA1A,EMD,NDC1,SIRT2,TUBB2A,TUBB4B,LEMD2,KPNB1,TNPO1,RANGAP1,RCC1,TMPO,SEC13,LMNA,NUP188,BANF1,NUP160,UBE2I,NUP155,LMNB1,NUP62,NUP133,NUP58,NUP43,NUP35,RAN,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Nuclear Envelope (NE) Reassembly |
| S Phase | 0.0147 | 163 | 31 | 2.68 × 10−12 | 6.43 × 10−11 | SMC3,MYC,AKT1,PSME1,RAD21,PSMC6,PSMC3,ANAPC7,ANAPC1,MCM7,RFC5,RFC3,RFC4,RFC1,RFC2,SMC1A,MCM6,MCM2,RBBP4,CDC27,SKP2,WAPL,PCNA,RPA1,RPA2,RPA3,CCND1,PSMD11,PDS5A,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | S Phase |
| DNA Strand Elongation | 0.0029 | 32 | 14 | 1.08 × 10−10 | 2.38 × 10−9 | MCM7,RFC5,RFC3,RFC4,RFC1,RFC2,MCM6,MCM2,PCNA,RPA1,RPA2,RPA3,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | DNA Strand Elongation |
| Lagging Strand Synthesis | 0.0018 | 20 | 11 | 1.05 × 10−9 | 1.99 × 10−8 | RFC5,RFC3,RFC4,RFC1,RFC2,PCNA,RPA1,RPA2,RPA3,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | Lagging Strand Synthesis |
| Synthesis of DNA | 0.0109 | 121 | 22 | 9.19 × 10−9 | 1.38 × 10−7 | PSME1,PSMC6,PSMC3,ANAPC7,ANAPC1,MCM7,RFC5,RFC3,RFC4,RFC1,RFC2,MCM6,MCM2,CDC27,SKP2,PCNA,RPA1,RPA2,RPA3,PSMD11,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | Synthesis of DNA |
| Mitotic Prometaphase | 0.0169 | 188 | 26 | 9.57 × 10−8 | 1.15 × 10−6 | NUP107,SMC3,SMC4,SMC2,NUP98,SEH1L,NUP85,RAD21,PLK1,TUBA1A,TUBB4B,NCAPD2,HSP90AA1,RANGAP1,SMC1A,WAPL,CETN2,SEC13,TUBB,SFI1,PDS5A,NUP160,NUP133,NUP43,BUB3,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Mitotic Prometaphase |
| Polymerase Switching | 0.0013 | 14 | 8 | 1.51 × 10−7 | 1.81 × 10−6 | RFC5,RFC3,RFC4,RFC1,RFC2,PCNA,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | Polymerase switching |
| Leading Strand Synthesis | 0.0013 | 14 | 8 | 1.51 × 10−7 | 1.81 × 10−6 | RFC5,RFC3,RFC4,RFC1,RFC2,PCNA,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | Leading Strand Synthesis |
| Separation of Sister Chromatids | 0.0155 | 172 | 24 | 2.55 × 10−7 | 2.81 × 10−6 | NUP107,SMC3,PSME1,NUP98,SEH1L,NUP85,RAD21,PLK1,PSMC6,PSMC3,ANAPC7,ANAPC1,RANGAP1,SMC1A,CDC27,WAPL,SEC13,PSMD11,PDS5A,NUP160,NUP133,NUP43,BUB3,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Separation of Sister Chromatids |
| Initiation of Nuclear Envelope (NE) Reformation | 0.0017 | 19 | 8 | 1.46 × 10−6 | 1.39 × 10−5 | EMD,SIRT2,LEMD2,KPNB1,TMPO,LMNA,BANF1,LMNB1 | Cell Cycle | Cell Cycle, Mitotic | Initiation of Nuclear Envelope (NE) Reformation |
| Resolution of Sister Chromatid Cohesion | 0.0096 | 107 | 17 | 2.70 × 10−6 | 2.43 × 10−5 | NUP107,SMC3,NUP98,SEH1L,NUP85,RAD21,PLK1,RANGAP1,SMC1A,WAPL,SEC13,PDS5A,NUP160,NUP133,NUP43,BUB3,NUP37 | Cell Cycle | Cell Cycle, Mitotic | Resolution of Sister Chromatid Cohesion |
| Mitotic Telophase/Cytokinesis | 0.0012 | 13 | 6 | 1.83 × 10−5 | 1.46 × 10−4 | SMC3,RAD21,PLK1,SMC1A,WAPL,PDS5A | Cell Cycle | Cell Cycle, Mitotic | Mitotic Telophase/Cytokinesis |
| Removal of the Flap Intermediate | 0.0013 | 14 | 6 | 2.75 × 10−5 | 1.93 × 10−4 | PCNA,RPA1,RPA2,RPA3,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | Removal of the Flap Intermediate |
| G1/S Transition | 0.0118 | 131 | 17 | 3.50 × 10−5 | 2.45 × 10−4 | MYC,AKT1,PSME1,PSMC6,PSMC3,MCM7,MCM6,MCM2,HDAC1,RBBP4,SKP2,PCNA,RPA1,RPA2,RPA3,CCND1,PSMD11 | Cell Cycle | Cell Cycle, Mitotic | G1/S Transition |
| Processive Synthesis on the Lagging Strand | 0.0014 | 15 | 6 | 4.03 × 10−5 | 2.82 × 10−4 | PCNA,RPA1,RPA2,RPA3,POLD3,POLD2 | Cell Cycle | Cell Cycle, Mitotic | Processive Synthesis on the Lagging Strand |
| Depolymerization of the Nuclear Lamina | 0.0014 | 15 | 6 | 4.03 × 10−5 | 2.82 × 10−4 | PRKCA,EMD,LEMD2,TMPO,LMNA,LMNB1 | Cell Cycle | Cell Cycle, Mitotic | Depolymerization of the Nuclear Lamina |
| Mitotic G1 phase and G1/S transition | 0.0134 | 149 | 18 | 5.17 × 10−5 | 3.29 × 10−4 | MYC,AKT1,PSME1,PSMC6,PSMC3,MCM7,MCM6,MCM2,HDAC1,RBBP4,SKP2,TOP2A,PCNA,RPA1,RPA2,RPA3,CCND1,PSMD11 | Cell Cycle | Cell Cycle, Mitotic | Mitotic G1 Phase and G1/S Transition |
| Cohesin Loading Onto Chromatin | 0.0009 | 10 | 5 | 6.34 × 10−5 | 3.81 × 10−4 | SMC3,RAD21,SMC1A,WAPL,PDS5A | Cell Cycle | Cell Cycle, Mitotic | Cohesin Loading onto Chromatin |
| Establishment of Sister Chromatid Cohesion | 0.001 | 11 | 5 | 9.88 × 10−5 | 5.93 × 10−4 | SMC3,RAD21,SMC1A,WAPL,PDS5A | Cell Cycle | Cell Cycle, Mitotic | Establishment of Sister Chromatid Cohesion |
| Selected Pathways for Longevity Pathway Enrichment | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Reactome Pathway | Ratio of Protein in Pathway | Number of Proteins in Pathway | Protein from Gene Set | p-Value | FDR | Hit Genes | General | Specific | More Specific |
| MTOR signaling | 0.0037 | 41 | 19 | 1.11 × 10−16 | 5.34 × 10−14 | AKT1,RRAGA,RRAGC,RRAGB,RRAGD,RPTOR,LAMTOR2,LAMTOR3,RPS6,TSC2,TSC1,EIF4EBP1,RHEB,MLST8,AKT1S1,EIF4E,EIF4B,MTOR,RPS6KB1 | Signal transduction | MTOR signaling | |
| mTORC1-mediated signaling | 0.0022 | 24 | 16 | 1.11 × 10−16 | 5.34 × 10−14 | RRAGA,RRAGC,RRAGB,RRAGD,RPTOR,LAMTOR2,LAMTOR3,RPS6,EIF4EBP1,RHEB,MLST8,AKT1S1,EIF4E,EIF4B,MTOR,RPS6KB1 | Signal transduction | MTOR signaling | mTORC1-mediated signaling |
| Intracellular signaling by second messengers | 0.0294 | 326 | 34 | 1.29 × 10−14 | 4.12 × 10−12 | AKT1,PRKCA,RRAGA,RRAGC,RRAGB,RRAGD,TP53,MAPKAP1,TGFA,INSR,CDKN1A,RPTOR,PPARG,EGFR,RICTOR,CAMK4,LAMTOR2,LAMTOR3,IRS2,TSC2,FOXO4,FOXO3,FOXO1,KL,ESR1,NBEA,RHEB,FGFR1,PRR5,MLST8,PIK3CA,AKT1S1,INS,MTOR | Signal transduction | Intracellular signaling by second messengers | |
| PIP3 activates AKT signaling | 0.0258 | 286 | 31 | 7.83 × 10−14 | 1.88 × 10−11 | AKT1,RRAGA,RRAGC,RRAGB,RRAGD,TP53,MAPKAP1,TGFA,INSR,CDKN1A,RPTOR,PPARG,EGFR,RICTOR,LAMTOR2,LAMTOR3,IRS2,TSC2,FOXO4,FOXO3,FOXO1,KL,ESR1,RHEB,FGFR1,PRR5,MLST8,PIK3CA,AKT1S1,INS,MTOR | Signal transduction | Intracellular signaling by second messengers | PIP3 activates AKT signaling |
| Generic transcription pathway | 0.1113 | 1235 | 66 | 2.29 × 10−13 | 4.40 × 10−11 | SERPINE1,AKT1,RUNX3,RRAGA,RRAGC,RRAGB,RRAGD,TP53,MAPKAP1,TGFA,GATA4,APOE,ATRIP,PPARGC1A,ATM,CDKN1A,MSTN,RPTOR,SREBF1,SIRT1,SIRT3,RAD51D,RARB,PPARG,SGK1,EGFR,WRN,RICTOR,WWOX,CDKN2B,GSR,MLH1,VEGFA,CAMK4,FAS,LAMTOR2,LAMTOR3,NR3C1,YY1,KCTD1,TSC2,TSC1,TBL1XR1,CSF1R,FOXO4,FOXO3,FOXO1,PLXNA4,TGFB1,ESRRG,ESR1,NFKB1,IL6,CDK6,RHEB,PRR5,EXO1,MLST8,YWHAG,H2AFX,IFNG,INS,TXNRD1,SOD2,MTOR,ERCC2 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Energy-dependent regulation of mTOR by LKB1-AMPK | 0.0026 | 29 | 12 | 1.95 × 10−12 | 3.12 × 10−10 | RRAGA,RRAGC,RRAGB,RRAGD,RPTOR,LAMTOR2,LAMTOR3,TSC2,TSC1,RHEB,MLST8,MTOR | Signal transduction | MTOR signaling | Energy-dependent regulation of mTOR by LKB1-AMPK |
| RNA polymerase II transcription | 0.1229 | 1364 | 67 | 6.72 × 10−12 | 9.20 × 10−10 | SERPINE1,AKT1,RUNX3,RRAGA,RRAGC,RRAGB,RRAGD,TP53,MAPKAP1,TGFA,GATA4,APOE,ATRIP,PPARGC1A,ATM,CDKN1A,MSTN,RPTOR,SREBF1,SIRT1,SIRT3,RAD51D,RARB,PPARG,SGK1,EGFR,WRN,RICTOR,WWOX,CDKN2B,GSR,MLH1,VEGFA,CAMK4,FAS,LAMTOR2,LAMTOR3,NR3C1,YY1,KCTD1,TSC2,TSC1,TBL1XR1,CSF1R,FOXO4,FOXO3,FOXO1,PLXNA4,TGFB1,ESRRG,ESR1,NFKB1,IL6,CDK6,RHEB,PRR5,EXO1,MLST8,YWHAG,H2AFX,IFNG,POLDIP3,INS,TXNRD1,SOD2,MTOR,ERCC2 | Gene expression (transcription) | RNA polymerase II transcription | |
| TP53 regulates metabolic genes | 0.008 | 89 | 17 | 9.43 × 10−12 | 1.13 × 10−9 | AKT1,RRAGA,RRAGC,RRAGB,RRAGD,TP53,RPTOR,GSR,LAMTOR2,LAMTOR3,TSC2,TSC1,RHEB,MLST8,YWHAG,TXNRD1,MTOR | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| FOXO-mediated transcription | 0.006 | 67 | 15 | 1.84 × 10−11 | 1.95 × 10−9 | AKT1,PPARGC1A,CDKN1A,MSTN,SREBF1,SIRT1,SIRT3,NR3C1,FOXO4,FOXO3,FOXO1,PLXNA4,YWHAG,INS,SOD2 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Transcriptional regulation by TP53 | 0.0332 | 368 | 30 | 2.09 × 10−10 | 1.82 × 10−8 | AKT1,RRAGA,RRAGC,RRAGB,RRAGD,TP53,MAPKAP1,ATRIP,ATM,CDKN1A,RPTOR,RAD51D,SGK1,WRN,RICTOR,GSR,MLH1,FAS,LAMTOR2,LAMTOR3,TSC2,TSC1,RHEB,PRR5,EXO1,MLST8,YWHAG,TXNRD1,MTOR,ERCC2 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| FOXO-mediated transcription of oxidative stress and metabolic and neuronal genes | 0.0028 | 31 | 10 | 1.49 × 10−9 | 1.19 × 10−7 | PPARGC1A,SREBF1,SIRT3,NR3C1,FOXO4,FOXO3,FOXO1,PLXNA4,INS,SOD2 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Regulation of PTEN gene transcription | 0.0055 | 61 | 12 | 8.21 × 10−9 | 5.58 × 10−7 | RRAGA,RRAGC,RRAGB,RRAGD,TP53,RPTOR,PPARG,LAMTOR2,LAMTOR3,RHEB,MLST8,MTOR | Signal transduction | Intracellular signaling by second messengers | PIP3 activates AKT signaling |
| Transcriptional regulation by the AP-2 (TFAP2) family of transcription factors | 0.0032 | 36 | 9 | 8.53 × 10−8 | 4.78 × 10−6 | TGFA,APOE,CDKN1A,EGFR,WWOX,VEGFA,YY1,KCTD1,ESR1 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Regulation of TP53 degradation | 0.0032 | 36 | 9 | 8.53 × 10−8 | 4.78 × 10−6 | AKT1,TP53,MAPKAP1,ATM,SGK1,RICTOR,PRR5,MLST8,MTOR | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Regulation of TP53 expression and degradation | 0.0033 | 37 | 9 | 1.07 × 10−7 | 5.69 × 10−6 | AKT1,TP53,MAPKAP1,ATM,SGK1,RICTOR,PRR5,MLST8,MTOR | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| VEGFA-VEGFR2 pathway | 0.0086 | 95 | 12 | 9.06 × 10−7 | 3.35 × 10−5 | AKT1,PRKCA,MAPKAP1,HRAS,RHOA,RICTOR,VEGFA,PRR5,ITGB3,MLST8,PIK3CA,MTOR | Signal transduction | Signaling by receptor tyrosine kinases | Signaling by VEGF |
| Signaling by VEGF | 0.0094 | 104 | 12 | 2.28 × 10−6 | 7.99 × 10−5 | AKT1,PRKCA,MAPKAP1,HRAS,RHOA,RICTOR,VEGFA,PRR5,ITGB3,MLST8,PIK3CA,MTOR | Signal transduction | Signaling by receptor tyrosine kinases | Signaling by VEGF |
| Regulation of FOXO transcriptional activity by acetylation | 0.0009 | 10 | 5 | 2.52 × 10−6 | 8.33 × 10−5 | SIRT1,SIRT3,FOXO4,FOXO3,FOXO1 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Regulation of localization of FOXO transcription factors | 0.0011 | 12 | 5 | 6.07 × 10−6 | 1.76 × 10−4 | AKT1,FOXO4,FOXO3,FOXO1,YWHAG | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Signaling by receptor tyrosine kinases | 0.0458 | 508 | 27 | 7.81 × 10−6 | 2.19 × 10−4 | AKT1,PRKCA,MAPKAP1,TGFA,HIF1A,APOE,LYN,INSR,IGF2,HRAS,RHOA,SGK1,EGFR,RICTOR,WWOX,VEGFA,IRS2,IGF1R,KL,ESR1,FGFR1,PRR5,ITGB3,MLST8,PIK3CA,INS,MTOR | Signal transduction | Signaling by receptor tyrosine kinases | |
| TFAP2 (AP-2) family regulates transcription of growth factors and their receptors | 0.0012 | 13 | 5 | 8.91 × 10−6 | 2.22 × 10−4 | TGFA,EGFR,VEGFA,YY1,ESR1 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| RUNX3 regulates CDKN1A transcription | 0.0006 | 7 | 4 | 1.58 × 10−5 | 3.63 × 10−4 | RUNX3,TP53,CDKN1A,TGFB1 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| VEGFR2 mediated vascular permeability | 0.0024 | 27 | 6 | 2.45 × 10−5 | 5.39 × 10−4 | AKT1,MAPKAP1,RICTOR,PRR5,MLST8,MTOR | Signal transduction | Signaling by receptor tyrosine kinases | Signaling by VEGF |
| PTEN regulation | 0.0142 | 158 | 13 | 3.09 × 10−5 | 6.69 × 10−4 | AKT1,RRAGA,RRAGC,RRAGB,RRAGD,TP53,RPTOR,PPARG,LAMTOR2,LAMTOR3,RHEB,MLST8,MTOR | Signal transduction | Intracellular signaling by second messengers | PIP3 activates AKT signaling |
| FOXO-mediated transcription of cell cycle genes | 0.0015 | 17 | 5 | 3.19 × 10−5 | 6.69 × 10−4 | CDKN1A,MSTN,FOXO4,FOXO3,FOXO1 | Gene expression (transcription) | RNA polymerase II transcription | Generic transcription pathway |
| Signaling by nuclear receptors | 0.0245 | 272 | 17 | 6.09 × 10−5 | 1.22 × 10−3 | AKT1,CETP,TGFA,APOE,HRAS,SREBF1,RARB,DLD,EGFR,IGF1R,YY1,TBL1XR1,FOXO3,ESR1,H2AFX,PIK3CA,APOC1 | Signal transduction | Signaling by nuclear receptors | |
| AKT phosphorylates targets in the nucleus | 0.0009 | 10 | 4 | 6.26 × 10−5 | 1.22 × 10−3 | AKT1,FOXO4,FOXO3,FOXO1 | Signal transduction | Intracellular signaling by second messengers | PIP3 activates AKT signaling |
| PI5P, PP2A, and IER3 regulate PI3K/AKT signaling | 0.0096 | 106 | 10 | 8.53 × 10−5 | 1.62 × 10−3 | AKT1,TGFA,INSR,EGFR,IRS2,KL,ESR1,FGFR1,PIK3CA,INS | Signal transduction | Intracellular signaling by second messengers | PIP3 activates AKT signaling |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badial, K.; Lacayo, P.; Murakami, S. Biology of Healthy Aging: Biological Hallmarks of Stress Resistance Related and Unrelated to Longevity in Humans. Int. J. Mol. Sci. 2024, 25, 10493. https://doi.org/10.3390/ijms251910493
Badial K, Lacayo P, Murakami S. Biology of Healthy Aging: Biological Hallmarks of Stress Resistance Related and Unrelated to Longevity in Humans. International Journal of Molecular Sciences. 2024; 25(19):10493. https://doi.org/10.3390/ijms251910493
Chicago/Turabian StyleBadial, Komalpreet, Patricia Lacayo, and Shin Murakami. 2024. "Biology of Healthy Aging: Biological Hallmarks of Stress Resistance Related and Unrelated to Longevity in Humans" International Journal of Molecular Sciences 25, no. 19: 10493. https://doi.org/10.3390/ijms251910493
APA StyleBadial, K., Lacayo, P., & Murakami, S. (2024). Biology of Healthy Aging: Biological Hallmarks of Stress Resistance Related and Unrelated to Longevity in Humans. International Journal of Molecular Sciences, 25(19), 10493. https://doi.org/10.3390/ijms251910493






