Rli51 Attenuates Transcription of the Listeria Pathogenicity Island 1 Gene mpl and Functions as a Trans-Acting sRNA in Intracellular Bacteria
Abstract
1. Introduction
2. Results
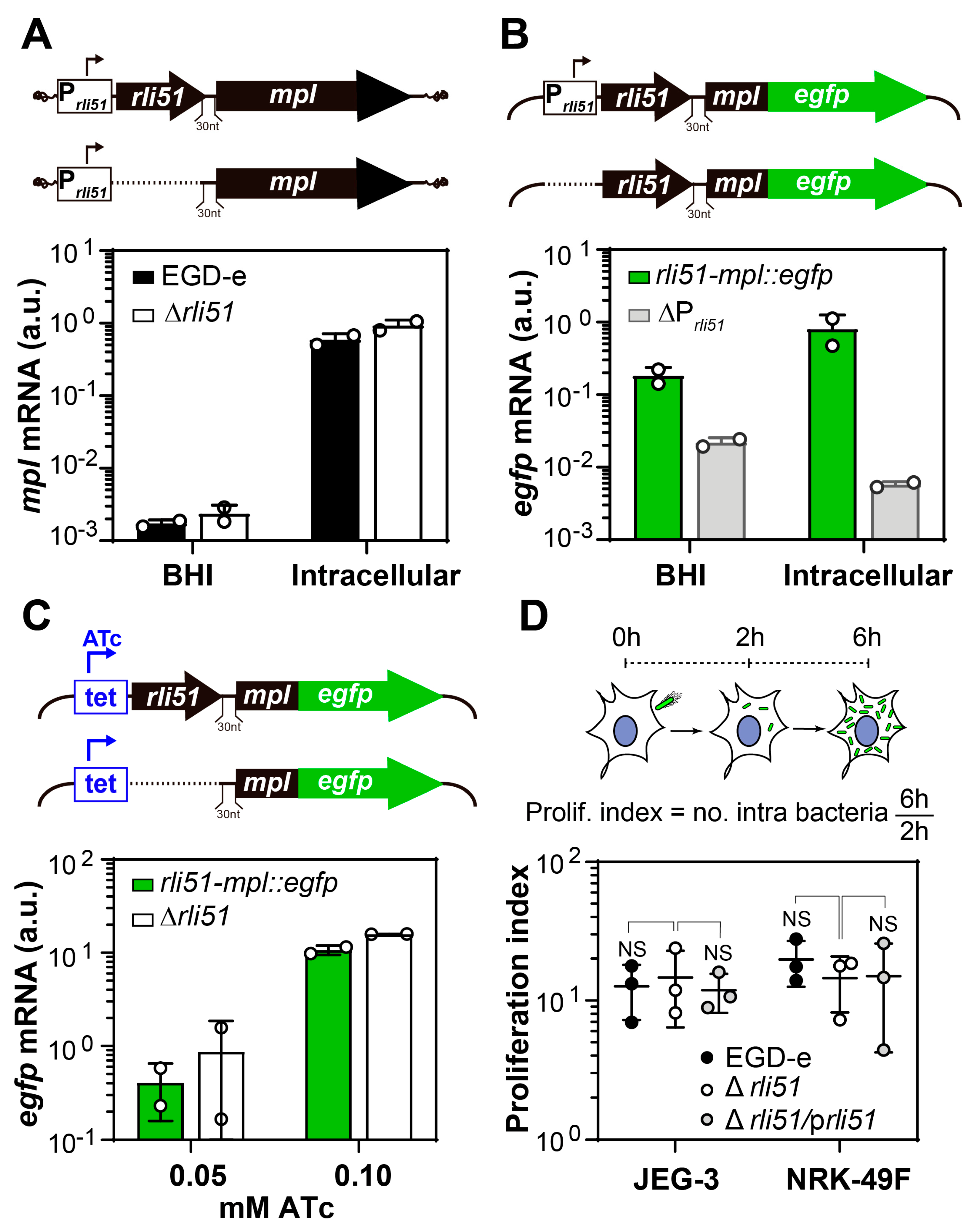
2.1. Rli51 Does Not Play a Relevant Role in the Upregulation of mpl during Infection
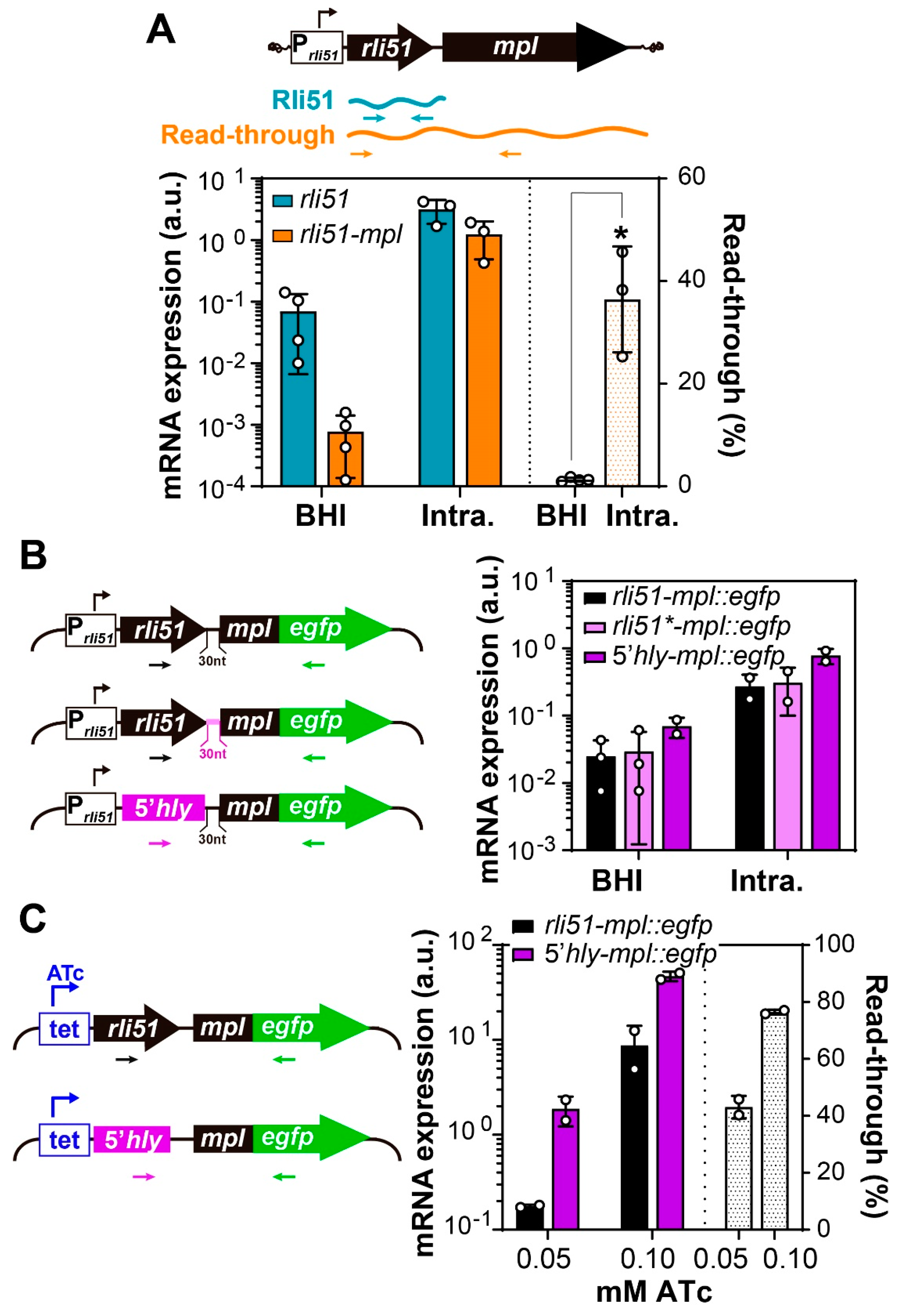
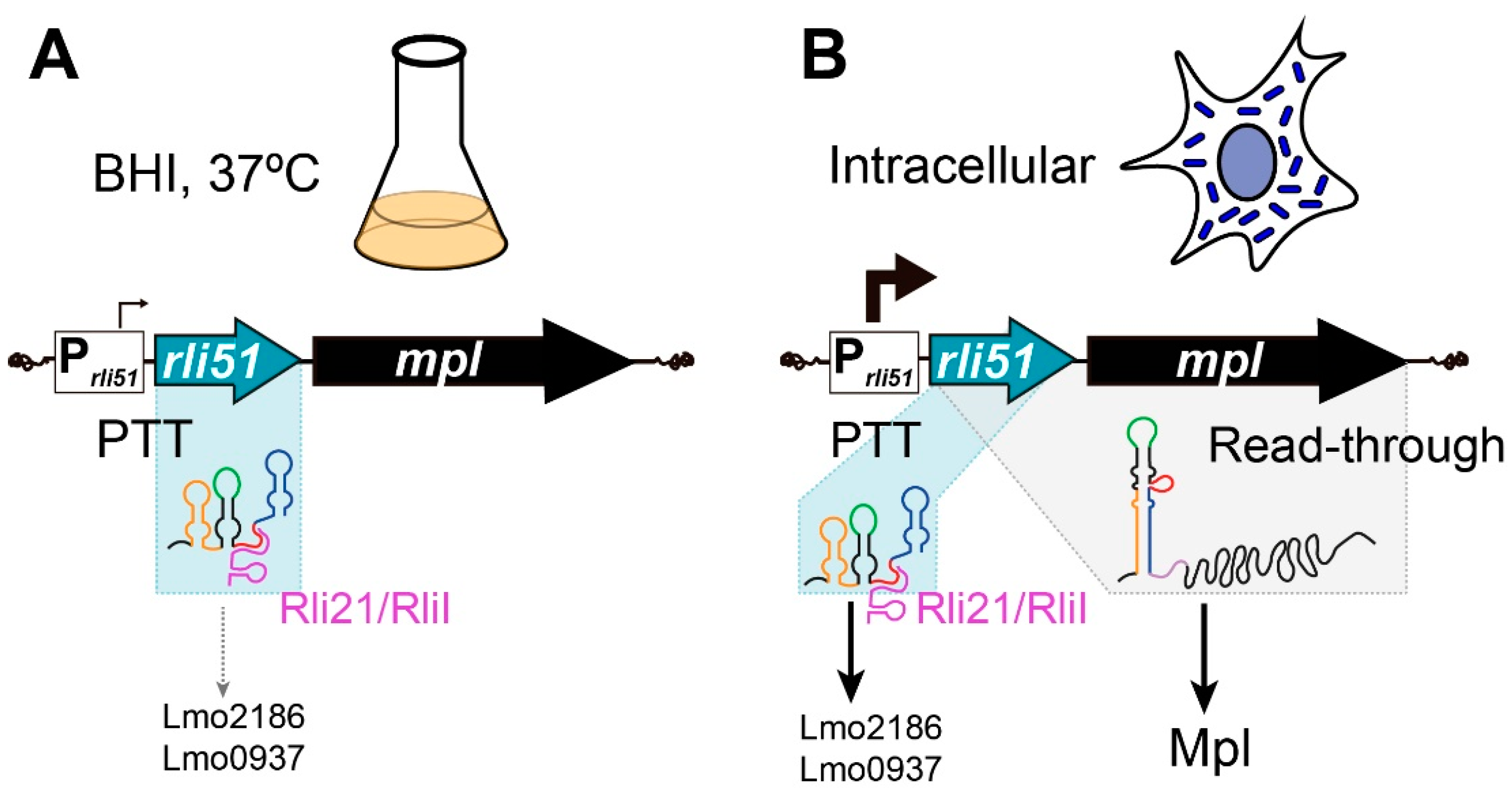
2.2. Condition-Specific Premature Termination of Rli51-mpl Transcription
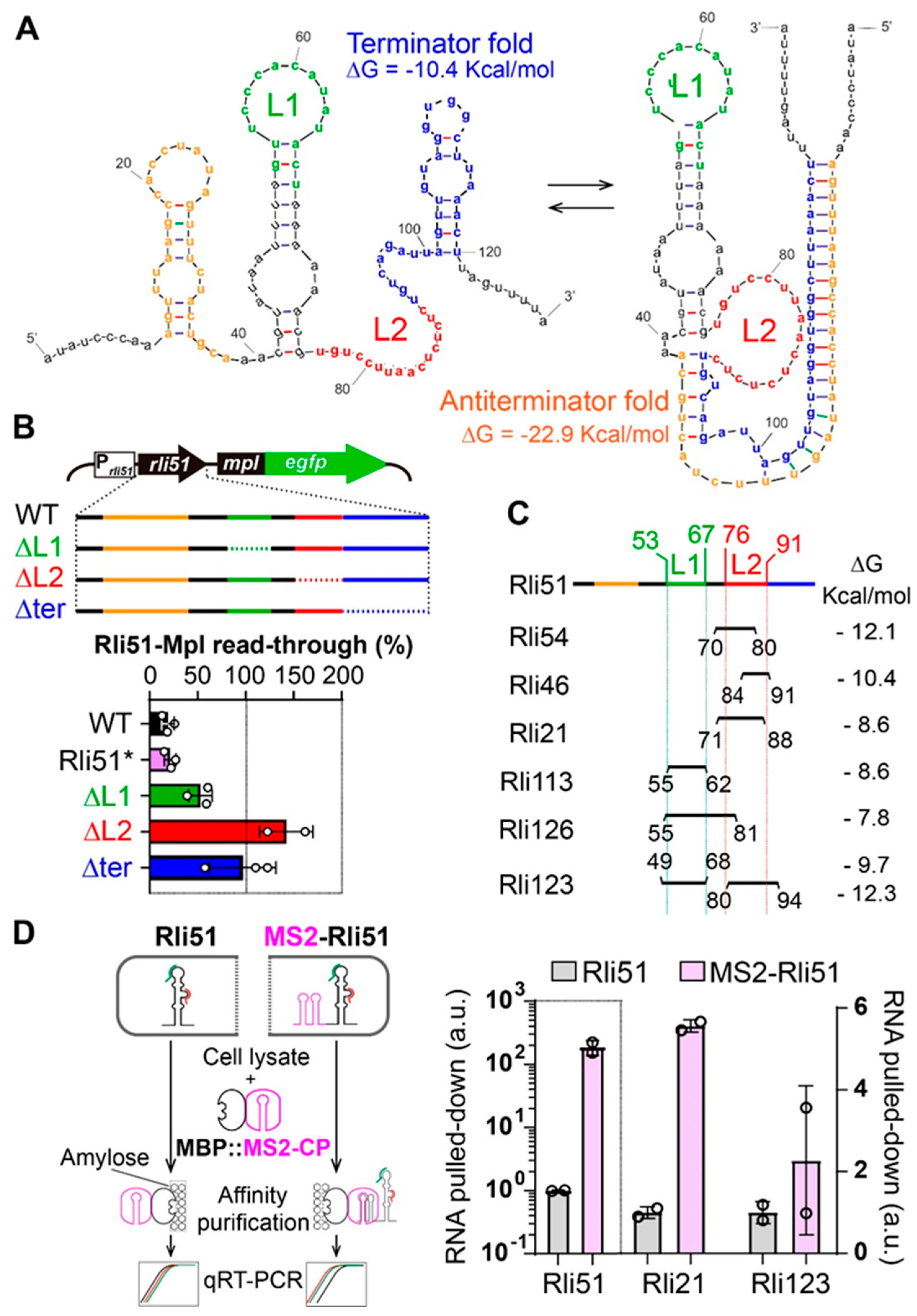
2.3. Trans-Acting Regulation of Rli51-Mediated Transcriptional Termination
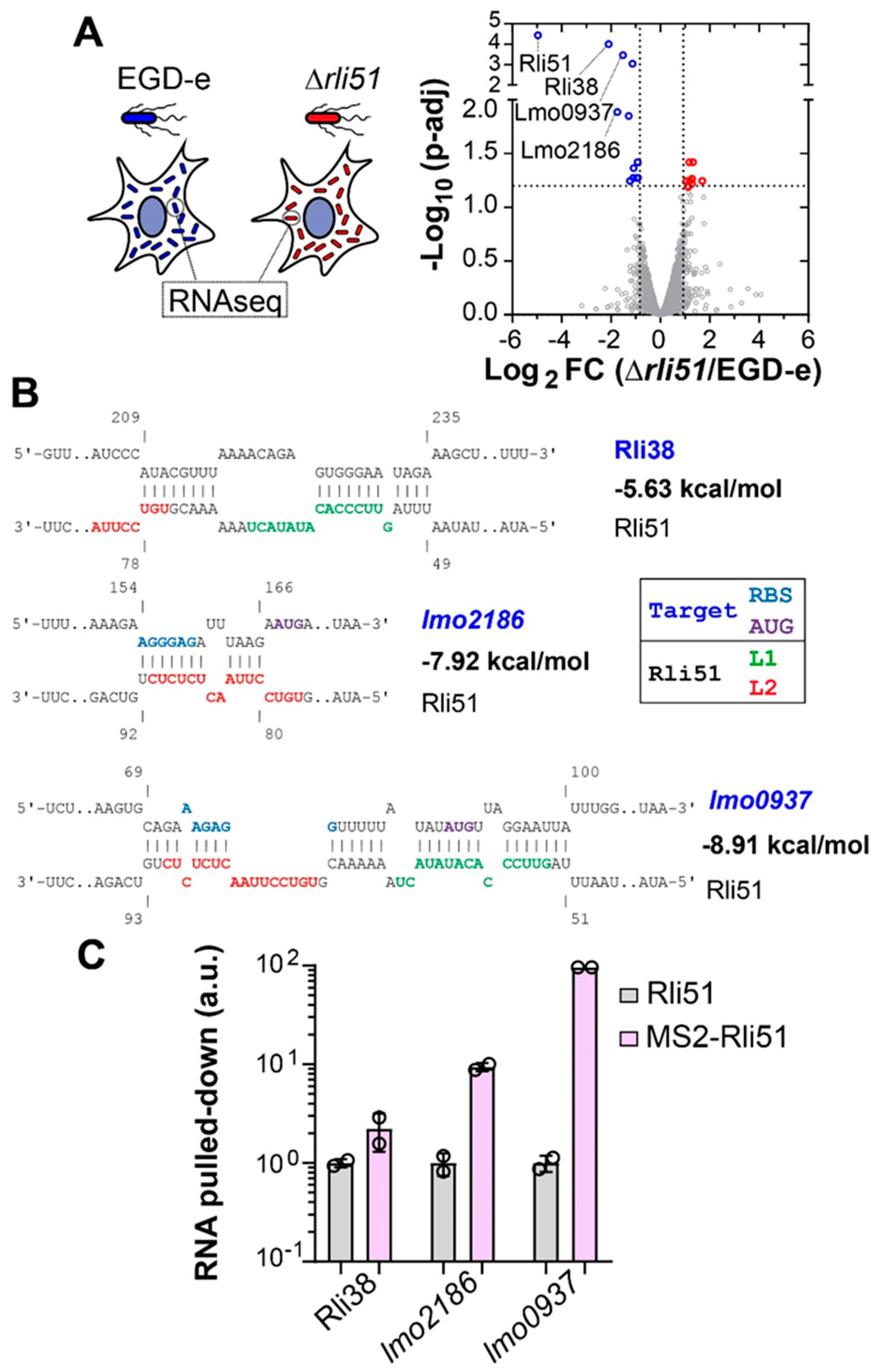
2.4. Rli51 Is a Trans-Acting sRNA in Bacteria Infecting Epithelial Cells
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Cloning Procedures
4.3. Determination of L. monocytogenes Intracellular Proliferation (Gentamicin-Resistance Assay)
4.4. RNA Isolation from Cultured and Intracellular Bacteria
4.5. Reverse Transcription and Real-Time Quantitative PCR (qPCR)
4.6. Production of Recombinant His-MBP-CP
4.7. MS2-Affinity Purification of Rli51-Binding RNAs (RNA Pull Down)
4.8. PASIFIC Structural Analysis of Rli51
4.9. sRNA-sRNA Interaction Prediction
4.10. RNA Sequencing: Library Construction and Next-Generation DNA Sequencing
4.11. RNA Sequencing Data Analysis
5. Conclusions
- Rli51 is an sRNA located in the 5′UTR of the LIPI-1 virulence factor mpl that functions both as a cis- and trans-acting sRNA.
- Under nutrient-rich laboratory conditions, the transcription of rli51-mpl is prematurely terminated, producing a 121-nucleotide-long sRNA at very low levels.
- The sRNA Rli21/RliI binds to a single-stranded RNA loop in Rli51, which is crucial for premature transcription termination.
- Under intracellular infection conditions, rli51 is induced, leading to a higher abundance of the short Rli51 sRNA and allowing for transcriptional read-through into mpl.
- Transcriptional read-through allows Mpl virulence factor expression in intracellular bacteria and the short Rli51 sRNA regulates in trans transcripts encoding iron scavenging and cell surface proteins.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dutta, T.; Srivastava, S. Small RNA-Mediated Regulation in Bacteria: A Growing Palette of Diverse Mechanisms. Gene 2018, 656, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Felden, B.; Augagneur, Y. Diversity and Versatility in Small RNA-Mediated Regulation in Bacterial Pathogens. Front. Microbiol. 2021, 12, 719977. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Cossart, P. Regulating Bacterial Virulence with RNA. Annu. Rev. Microbiol. 2017, 71, 263–280. [Google Scholar] [CrossRef]
- Ortega, A.D.; Quereda, J.J.; Pucciarelli, M.G.; García-del Portillo, F. Non-Coding RNA Regulation in Pathogenic Bacteria Located inside Eukaryotic Cells. Front. Cell. Infect. Microbiol. 2014, 4, 162. [Google Scholar] [CrossRef]
- Padalon-Brauch, G.; Hershberg, R.; Elgrably-Weiss, M.; Baruch, K.; Rosenshine, I.; Margalit, H.; Altuvia, S. Small RNAs Encoded within Genetic Islands of Salmonella typhimurium Show Host-Induced Expression and Role in Virulence. Nucleic Acids Res. 2008, 36, 1913–1927. [Google Scholar] [CrossRef]
- Wurtzel, O.; Sesto, N.; Mellin, J.R.; Karunker, I.; Edelheit, S.; Bécavin, C.; Archambaud, C.; Cossart, P.; Sorek, R. Comparative Transcriptomics of Pathogenic and Non-pathogenic Listeria Species. Mol. Syst. Biol. 2012, 8, 583. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; et al. The Listeria Transcriptional Landscape from Saprophytism to Virulence. Nature 2009, 459, 950–956. [Google Scholar] [CrossRef]
- Mraheil, M.A.; Billion, A.; Mohamed, W.; Mukherjee, K.; Kuenne, C.; Pischimarov, J.; Krawitz, C.; Retey, J.; Hartsch, T.; Chakraborty, T.; et al. The Intracellular SRNA Transcriptome of Listeria monocytogenes during Growth in Macrophages. Nucleic Acids Res. 2011, 39, 4235–4248. [Google Scholar] [CrossRef]
- Ortega, A.D.; Gonzalo-Asensio, J.; García-del Portillo, F. Dynamics of Salmonella Small RNA Expression in Non-Growing Bacteria Located inside Eukaryotic Cells. RNA Biol. 2012, 9, 469–488. [Google Scholar] [CrossRef]
- Srikumar, S.; Kröger, C.; Hébrard, M.; Colgan, A.; Owen, S.V.; Sivasankaran, S.K.; Cameron, A.D.S.; Hokamp, K.; Hinton, J.C.D. RNA-Seq Brings New Insights to the Intra-Macrophage Transcriptome of Salmonella typhimurium. PLoS Pathog. 2015, 11, e1005262. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ji, C.; Xia, X.; Cai, X.; Meng, Q.; Qiao, J. A Regulatory SRNA Rli43 Is Involved in the Modulation of Biofilm Formation and Virulence in Listeria monocytogenes. Pathogens 2022, 11, 1137. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-Del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and Virulence of Listeria monocytogenes: A Trip from Environmental to Medical Microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and Molecular Virulence Determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef]
- Wong, K.K.Y.; Bouwer, H.G.A.; Freitag, N.E. Evidence Implicating the 5′ Untranslated Region of Listeria monocytogenes ActA in the Regulation of Bacterial Actin-Based Motility. Cell Microbiol. 2004, 6, 155–166. [Google Scholar] [CrossRef]
- Domann, E.; Leimeister-Wächter, M.; Goebel, W.; Chakraborty, T. Molecular Cloning, Sequencing, and Identification of a Metalloprotease Gene from Listeria monocytogenes That Is Species Specific and Physically Linked to the Listeriolysin Gene. Infect. Immun. 1991, 59, 65–72. [Google Scholar] [CrossRef]
- Loh, E.; Gripenland, J.; Johansson, J. Control of Listeria monocytogenes Virulence by 5′-Untranslated RNA. Trends Microbiol. 2006, 14, 294–298. [Google Scholar] [CrossRef]
- Dar, D.; Shamir, M.; Mellin, J.R.; Koutero, M.; Stern-Ginossar, N.; Cossart, P.; Sorek, R. Term-Seq Reveals Abundant Ribo-Regulation of Antibiotics Resistance in Bacteria. Science 2016, 352, aad9822. [Google Scholar] [CrossRef]
- Ignatov, D.; Vaitkevicius, K.; Durand, S.; Cahoon, L.; Sandberg, S.S.; Liu, X.; Kallipolitis, B.H.; Rydén, P.; Freitag, N.; Condon, C.; et al. An MRNA-MRNA Interaction Couples Expression of a Virulence Factor and Its Chaperone in Listeria monocytogenes. Cell Rep. 2020, 30, 4027–4040.e7. [Google Scholar] [CrossRef]
- Shen, A.; Higgins, D.E. The 5′ Untranslated Region-Mediated Enhancement of Intracellular Listeriolysin O Production Is Required for Listeria monocytogenes Pathogenicity. Mol. Microbiol. 2005, 57, 1460–1473. [Google Scholar] [CrossRef]
- Bohne, J.; Sokolovic, Z.; Goebel, W. Transcriptional Regulation of PrfA and PrfA-Regulated Virulence Genes in Listeria monocytogenes. Mol. Microbiol. 1994, 11, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Marquis, H.; Goldfine, H.; Portnoy, D.A. Proteolytic Pathways of Activation and Degradation of a Bacterial Phospholipase C during Intracellular Infection by Listeria monocytogenes. J. Cell Biol. 1997, 137, 1381–1392. [Google Scholar] [CrossRef]
- Alvarez, D.E.; Agaisse, H. The Metalloprotease Mpl Supports Listeria monocytogenes Dissemination through Resolution of Membrane Protrusions into Vacuoles. Infect. Immun. 2016, 84, 1806–1814. [Google Scholar] [CrossRef]
- Santangelo, T.J.; Artsimovitch, I. Termination and Antitermination: RNA Polymerase Runs a Stop Sign. Nat. Rev. Microbiol. 2011, 9, 319–329. [Google Scholar] [CrossRef]
- Millman, A.; Dar, D.; Shamir, M.; Sorek, R. Computational Prediction of Regulatory, Premature Transcription Termination in Bacteria. Nucleic Acids Res. 2017, 45, 886–893. [Google Scholar] [CrossRef] [PubMed]
- Loh, E.; Dussurget, O.; Gripenland, J.; Vaitkevicius, K.; Tiensuu, T.; Mandin, P.; Repoila, F.; Buchrieser, C.; Cossart, P.; Johansson, J. A Trans-Acting Riboswitch Controls Expression of the Virulence Regulator PrfA in Listeria monocytogenes. Cell 2009, 139, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Salvail, H.; Breaker, R.R. Riboswitches. Curr. Biol. 2023, 33, R343–R348. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and Customizable Prediction of RNA–RNA Interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef] [PubMed]
- Bécavin, C.; Koutero, M.; Tchitchek, N.; Cerutti, F.; Lechat, P.; Maillet, N.; Hoede, C.; Chiapello, H.; Gaspin, C.; Cossart, P. Listeriomics: An Interactive Web Platform for Systems Biology of Listeria. mSystems 2017, 2, e00186-16. [Google Scholar] [CrossRef]
- Bardwell, V.J.; Wickens, M. Purification or RNA and RNA—Protein Complexes by an R17 Coat Protein Affinity Method. Nucleic Acids Res. 1991, 19, 1980. [Google Scholar] [CrossRef][Green Version]
- Lalaouna, D.; Prévost, K.; Eyraud, A.; Massé, E. Identification of Unknown RNA Partners Using MAPS. Methods 2017, 117, 28–34. [Google Scholar] [CrossRef]
- Poyart, C.; Abachin, E.; Razafimanantsoa, I.; Berche, P. The Zinc Metalloprotease of Listeria monocytogenes Is Required for Maturation of Phosphatidylcholine Phospholipase C: Direct Evidence Obtained by Gene Complementation. Infect. Immun. 1993, 61, 1576–1580. [Google Scholar] [CrossRef] [PubMed]
- Raveneau, J.; Geoffroy, C.; Beretti, J.L.; Gaillard, J.L.; Alouf, J.E.; Berche, P. Reduced Virulence of a Listeria monocytogenes Phospholipase-Deficient Mutant Obtained by Transposon Insertion into the Zinc Metalloprotease Gene. Infect. Immun. 1992, 60, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, S.; Massé, E. RNA-Dependent Regulation of Virulence in Pathogenic Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 337. [Google Scholar] [CrossRef]
- Krawczyk-Balska, A.; Ładziak, M.; Burmistrz, M.; Ścibek, K.; Kallipolitis, B.H. RNA-Mediated Control in Listeria monocytogenes: Insights Into Regulatory Mechanisms and Roles in Metabolism and Virulence. Front. Microbiol. 2021, 12, 622829. [Google Scholar] [CrossRef] [PubMed]
- Stritzker, J.; Schoen, C.; Goebel, W. Enhanced Synthesis of Internalin A in Aro Mutants of Listeria monocytogenes Indicates Posttranscriptional Control of the InlAB MRNA. J. Bacteriol. 2005, 187, 2836–2845. [Google Scholar] [CrossRef][Green Version]
- Johansson, J.; Mandin, P.; Renzoni, A.; Chiaruttini, C.; Springer, M.; Cossart, P. An RNA Thermosensor Controls Expression of Virulence Genes in Listeria monocytogenes. Cell 2002, 110, 551–561. [Google Scholar] [CrossRef]
- Turnbough, C.L. Regulation of Bacterial Gene Expression by Transcription Attenuation. Microbiol. Mol. Biol. Rev. 2019, 83, e00019. [Google Scholar] [CrossRef]
- Sedlyarova, N.; Shamovsky, I.; Bharati, B.K.; Epshtein, V.; Chen, J.; Gottesman, S.; Schroeder, R.; Nudler, E. SRNA-Mediated Control of Transcription Termination in E. coli. Cell 2016, 167, 111–121.e13. [Google Scholar] [CrossRef]
- Malmirchegini, G.R.; Sjodt, M.; Shnitkind, S.; Sawaya, M.R.; Rosinski, J.; Newton, S.M.; Klebba, P.E.; Clubb, R.T. Novel Mechanism of Hemin Capture by Hbp2, the Hemoglobin-Binding Hemophore from Listeria monocytogenes. J. Biol. Chem. 2014, 289, 34886–34899. [Google Scholar] [CrossRef]
- Wu, R.; Skaar, E.P.; Zhang, R.; Joachimiak, G.; Gornicki, P.; Schneewind, O.; Joachimiak, A. Staphylococcus aureus IsdG and IsdI, Heme-Degrading Enzymes with Structural Similarity to Monooxygenases. J. Biol. Chem. 2005, 280, 2840–2846. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, P.T.; Menendez-Gil, P.; Sabharwal, D.; Christensen, J.H.; Brunhede, M.Z.; Lillebæk, E.M.S.; Kallipolitis, B.H. The Small Regulatory RNAs LhrC1–5 Contribute to the Response of Listeria monocytogenes to Heme Toxicity. Front. Microbiol. 2018, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, O.; Livny, J.; Bhattacharyya, R.; Amster-Choder, O. Wisdom of the Crowds: A Suggested Polygenic Plan for Small-RNA-Mediated Regulation in Bacteria. iScience 2021, 24, 103096. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Wagner, E.G.H. Target Identification of Small Noncoding RNAs in Bacteria. Curr. Opin. Microbiol. 2007, 10, 262–270. [Google Scholar] [CrossRef]
- Acebo, P.; Herranz, C.; Espenberger, L.B.; Gómez-sanz, A.; Terrón, M.C.; Luque, D.; Amblar, M. A Small Non-coding Rna Modulates Expression of Pilus-1 Type in Streptococcus pneumoniae. Microorganisms 2021, 9, 1883. [Google Scholar] [CrossRef]
- Melior, H.; Li, S.; Madhugiri, R.; Stötzel, M.; Azarderakhsh, S.; Barth-Weber, S.; Baumgardt, K.; Ziebuhr, J.; Evguenieva-Hackenberg, E. Transcription Attenuation-Derived Small RNA RnTrpL Regulates Tryptophan Biosynthesis Gene Expression in Trans. Nucleic Acids Res. 2019, 47, 6396–6410. [Google Scholar] [CrossRef]
- Luo, X.; Esberard, M.; Bouloc, P.; Jacq, A. A Small Regulatory RNA Generated from the MalK 59 Untranslated Region Targets Gluconeogenesis in Vibrio Species. Msphere 2021, 6, e0013421. [Google Scholar] [CrossRef]
- Jäger, D.; Pernitzsch, S.R.; Richter, A.S.; Backofen, R.; Sharma, C.M.; Schmitz, R.A. An Archaeal SRNA Targeting Cis-and Trans-Encoded MRNAs via Two Distinct Domains. Nucleic Acids Res. 2012, 40, 10964–10979. [Google Scholar] [CrossRef]
- Matera, G.; Altuvia, Y.; Gerovac, M.; El Mouali, Y.; Margalit, H.; Vogel, J. Global RNA Interactome of Salmonella Discovers a 5′ UTR Sponge for the MicF Small RNA That Connects Membrane Permeability to Transport Capacity. Mol. Cell 2022, 82, 629–644.e4. [Google Scholar] [CrossRef]
- De Lay, N.R.; Garsin, D.A. The Unmasking of “junk” RNA Reveals Novel SRNAs: From Processed RNA Fragments to Marooned Riboswitches. Curr. Opin. Microbiol. 2016, 30, 16–21. [Google Scholar] [CrossRef]
- Adams, P.P.; Baniulyte, G.; Esnault, C.; Chegireddy, K.; Singh, N.; Monge, M.; Dale, R.K.; Storz, G.; Wade, J.T. Regulatory Roles of Escherichia coli 5′ UTR and ORF-Internal RNAs Detected by 3′ End Mapping. Elife 2021, 10, e62438. [Google Scholar] [CrossRef]
- Glaser, P.; Frangeul, L.; Buchrieser, C.; Rusniok, C.; Amend, A.; Baquero, F.; Berche, P.; Bloecker, H.; Brandt, P.; Chakraborty, T.; et al. Comparative Genomics of Listeria Species. Science 2001, 294, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nojima, H.; Okayama, H. High Efficiency Transformation of Escherichia coli with Plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Ivain, L.; Bordeau, V.; Eyraud, A.; Hallier, M.; Dreano, S.; Tattevin, P.; Felden, B.; Chabelskaya, S. An in vivo Reporter Assay for SRNA-Directed Gene Control in Gram-Positive Bacteria: Identifying a Novel SRNA Target in Staphylococcus aureus. Nucleic Acids Res. 2017, 45, 4994–5007. [Google Scholar] [CrossRef] [PubMed]
- Balestrino, D.; Anne Hamon, M.; Dortet, L.; Nahori, M.A.; Pizarro-Cerda, J.; Alignani, D.; Dussurget, O.; Cossart, P.; Toledo-Arana, A. Single-Cell Techniques Using Chromosomally Tagged Fluorescent Bacteria to Study Listeria monocytogenes Infection Processes. Appl. Environ. Microbiol. 2010, 76, 3625–3636. [Google Scholar] [CrossRef]
- Ortega, A.D.; Willers, I.M.; Sala, S.; Cuezva, J.M. Human G3BP1 Interacts with Beta-F1-ATPase MRNA and Inhibits Its Translation. J. Cell Sci. 2010, 123, 2685–2696. [Google Scholar] [CrossRef][Green Version]
- Busch, A.; Richter, A.S.; Backofen, R. IntaRNA: Efficient Prediction of Bacterial SRNA Targets Incorporating Target Site Accessibility and Seed Regions. Bioinformatics 2008, 24, 2849–2856. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morón, Á.; Ortiz-Miravalles, L.; Peñalver, M.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Rli51 Attenuates Transcription of the Listeria Pathogenicity Island 1 Gene mpl and Functions as a Trans-Acting sRNA in Intracellular Bacteria. Int. J. Mol. Sci. 2024, 25, 9380. https://doi.org/10.3390/ijms25179380
Morón Á, Ortiz-Miravalles L, Peñalver M, García-del Portillo F, Pucciarelli MG, Ortega AD. Rli51 Attenuates Transcription of the Listeria Pathogenicity Island 1 Gene mpl and Functions as a Trans-Acting sRNA in Intracellular Bacteria. International Journal of Molecular Sciences. 2024; 25(17):9380. https://doi.org/10.3390/ijms25179380
Chicago/Turabian StyleMorón, Álvaro, Laura Ortiz-Miravalles, Marcos Peñalver, Francisco García-del Portillo, M. Graciela Pucciarelli, and Alvaro Darío Ortega. 2024. "Rli51 Attenuates Transcription of the Listeria Pathogenicity Island 1 Gene mpl and Functions as a Trans-Acting sRNA in Intracellular Bacteria" International Journal of Molecular Sciences 25, no. 17: 9380. https://doi.org/10.3390/ijms25179380
APA StyleMorón, Á., Ortiz-Miravalles, L., Peñalver, M., García-del Portillo, F., Pucciarelli, M. G., & Ortega, A. D. (2024). Rli51 Attenuates Transcription of the Listeria Pathogenicity Island 1 Gene mpl and Functions as a Trans-Acting sRNA in Intracellular Bacteria. International Journal of Molecular Sciences, 25(17), 9380. https://doi.org/10.3390/ijms25179380





