Genome-Wide Identification of the DOF Gene Family in Kiwifruit (Actinidia chinensis) and Functional Validation of AcDOF22 in Response to Drought Stress
Abstract
1. Introduction
2. Results
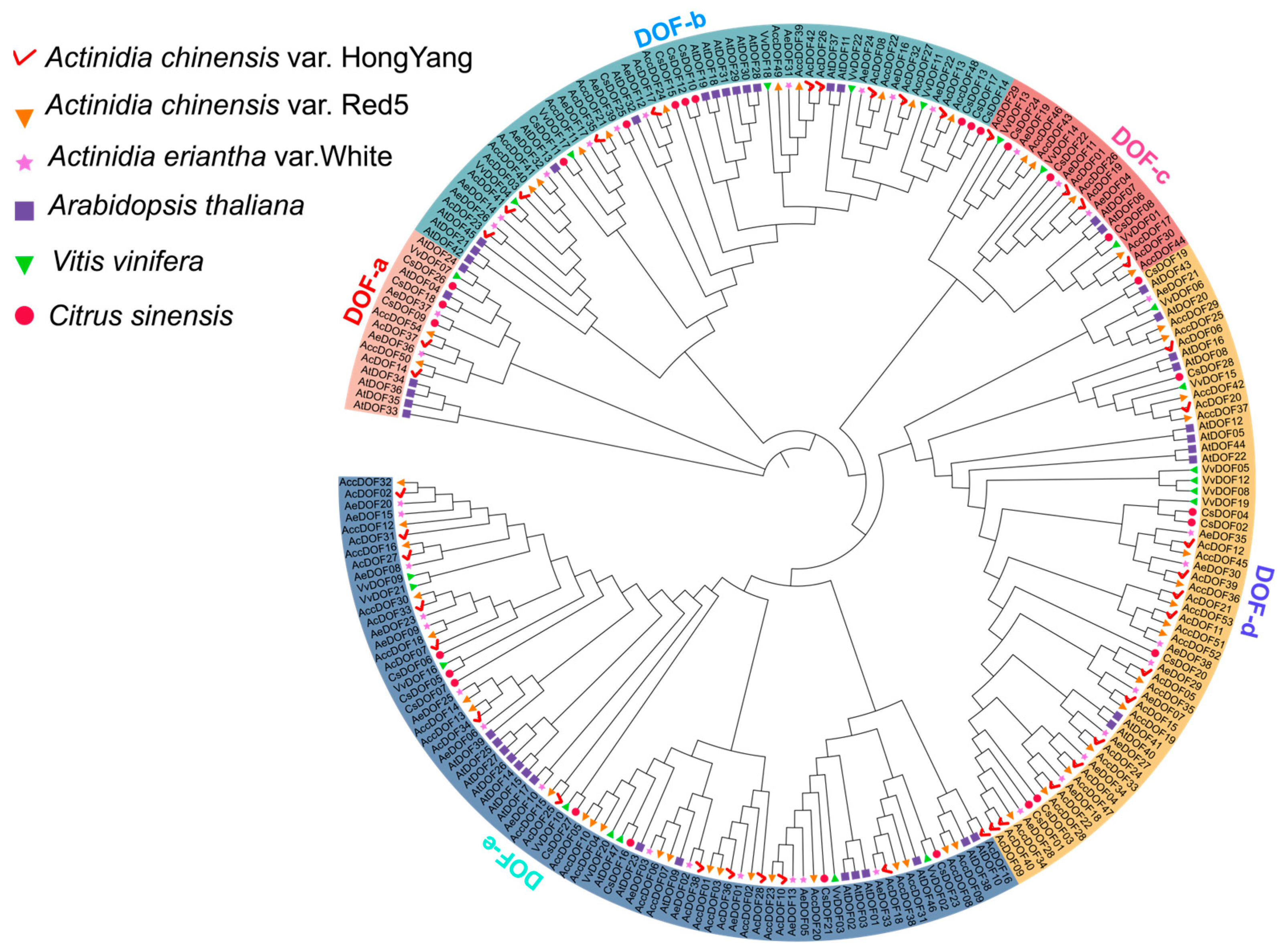
2.1. Identification, Characterization, and Phylogenetic Analysis of the DOF Genes in Kiwifruit
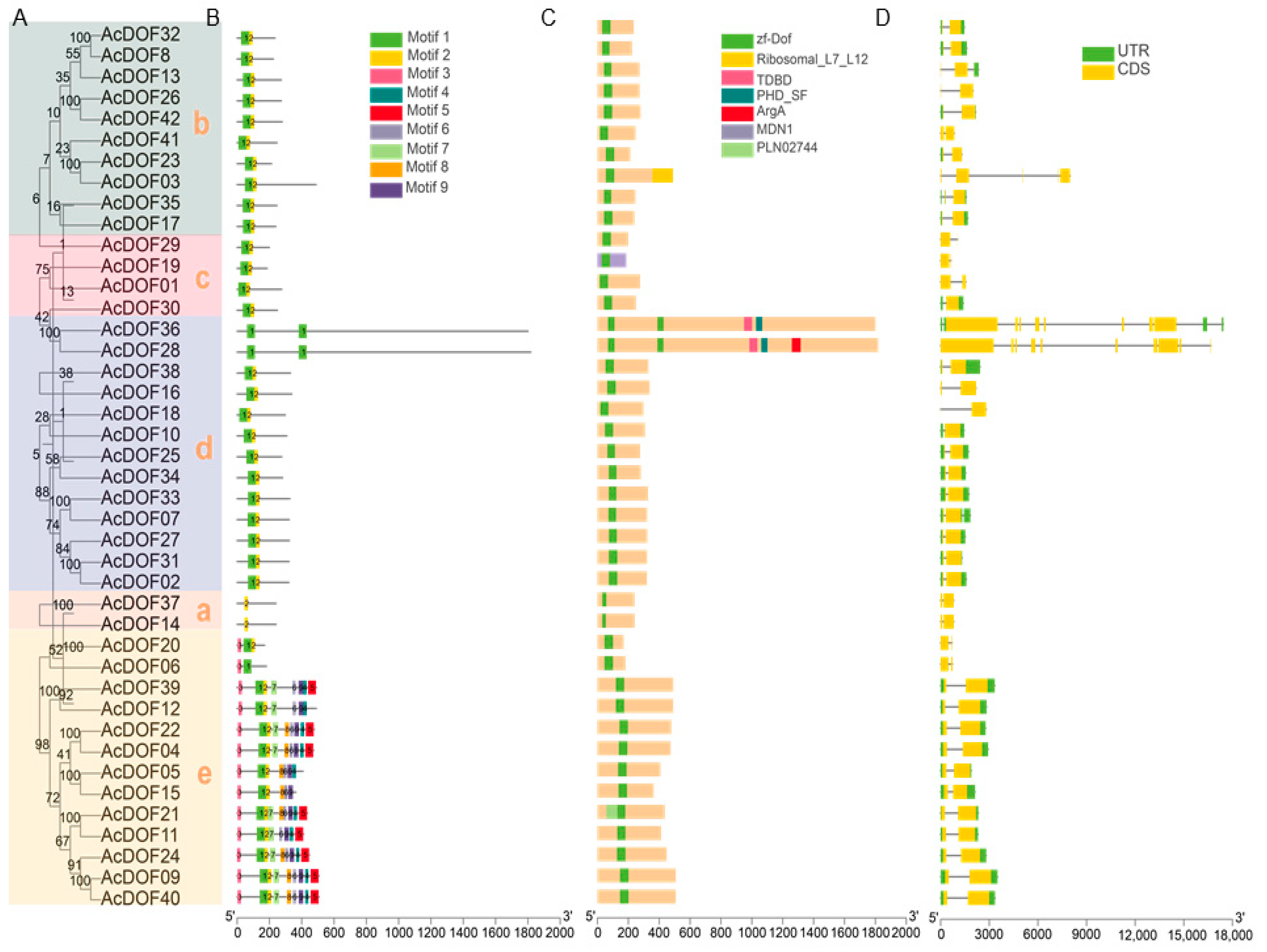
2.2. Evolutionary Analysis, Protein Motifs, Conserved Domains, and AcDOF Gene Structure
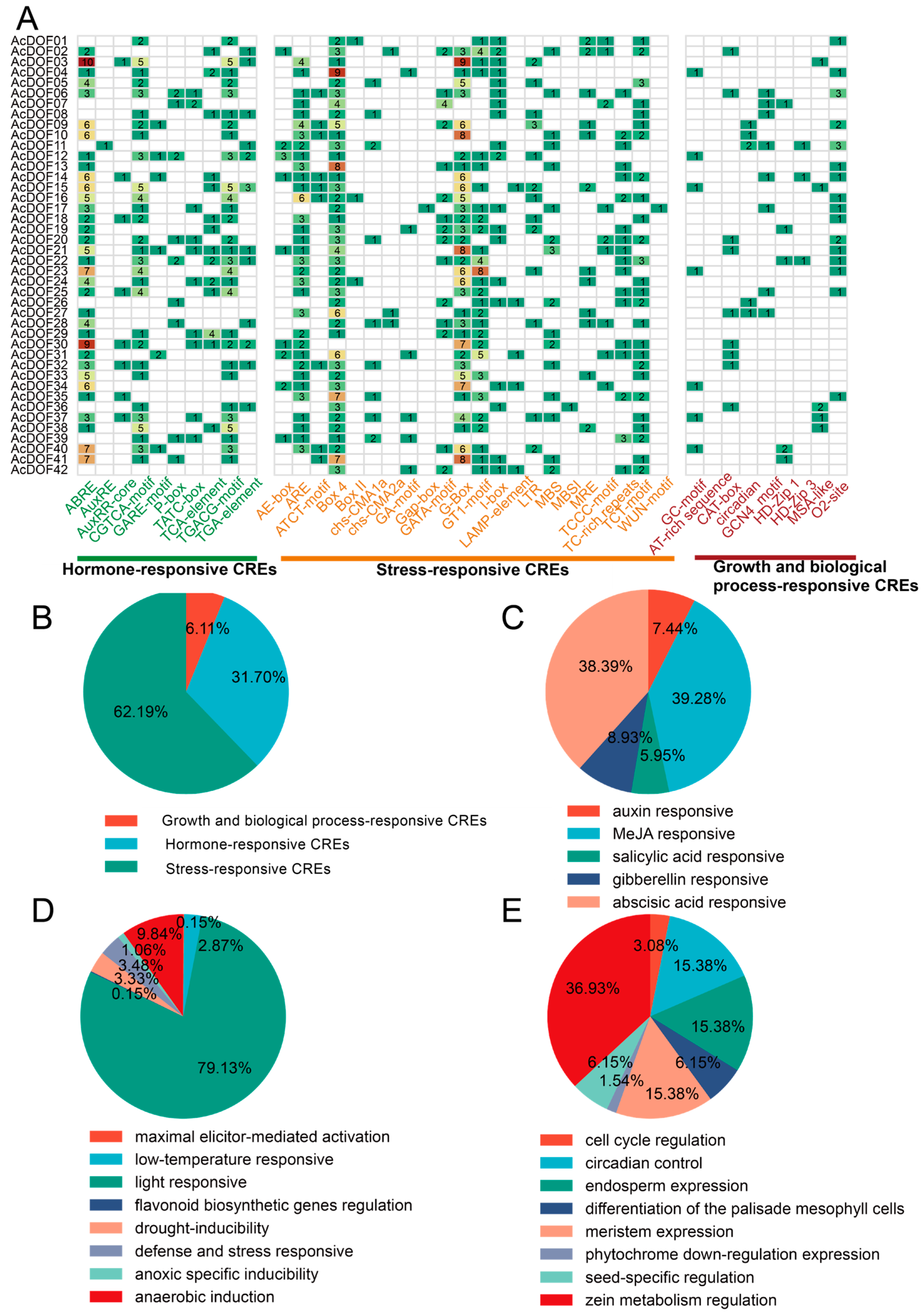
2.3. Analysis of cis-Regulatory Elements of AcDOFs
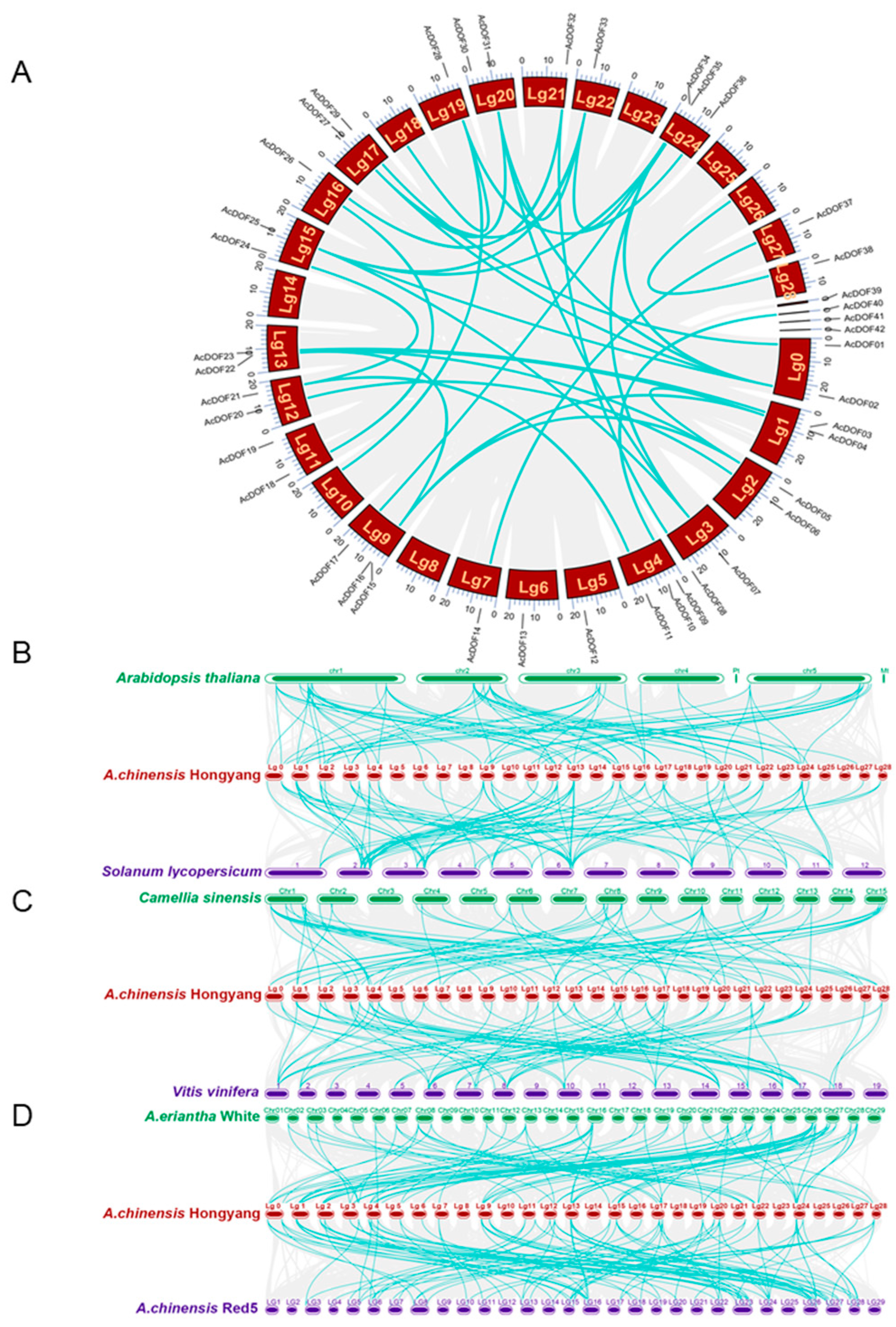
2.4. Collinearity and Synteny Analysis of AcDOFs
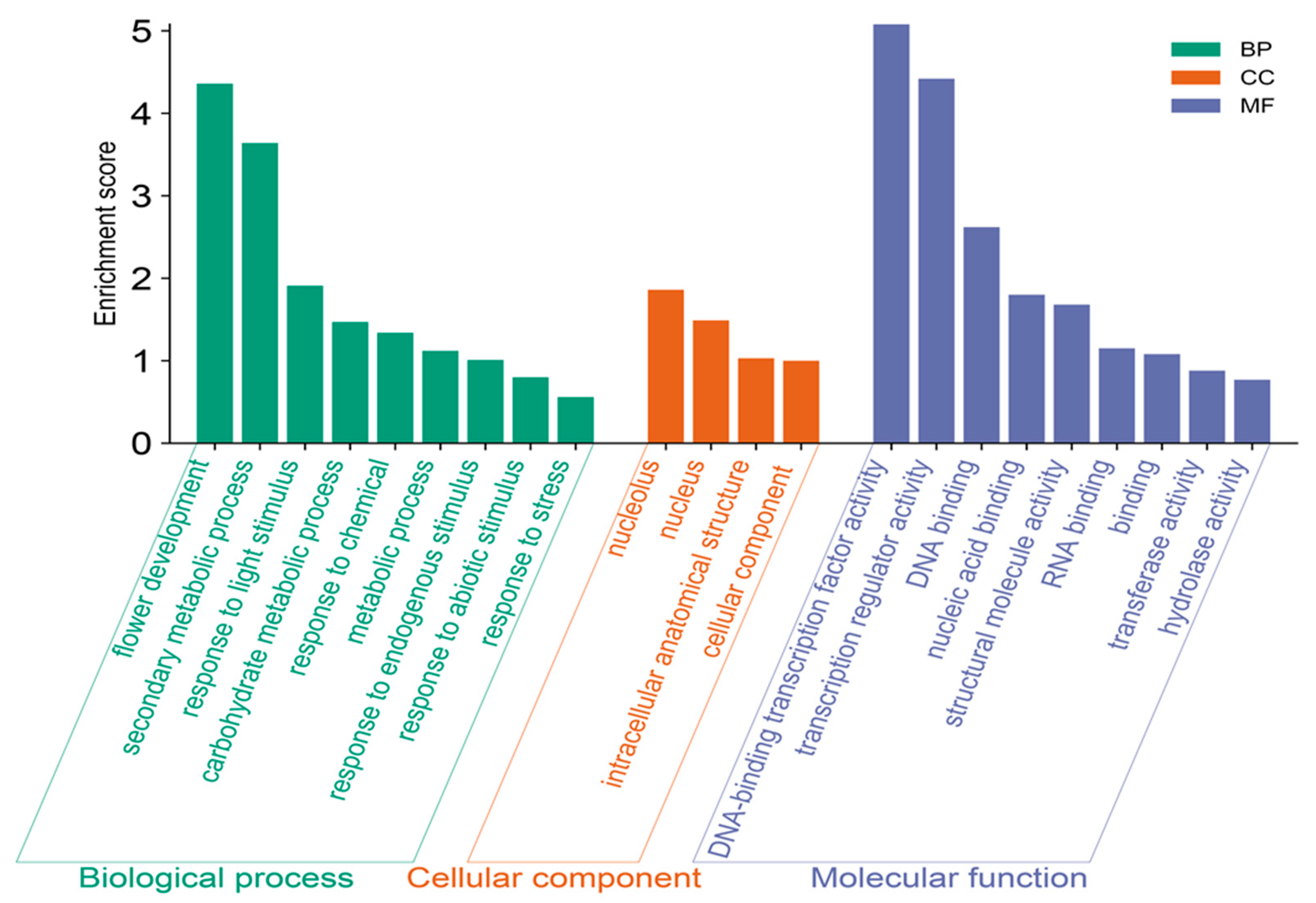
2.5. GO Annotation of the AcDOF Gene Family
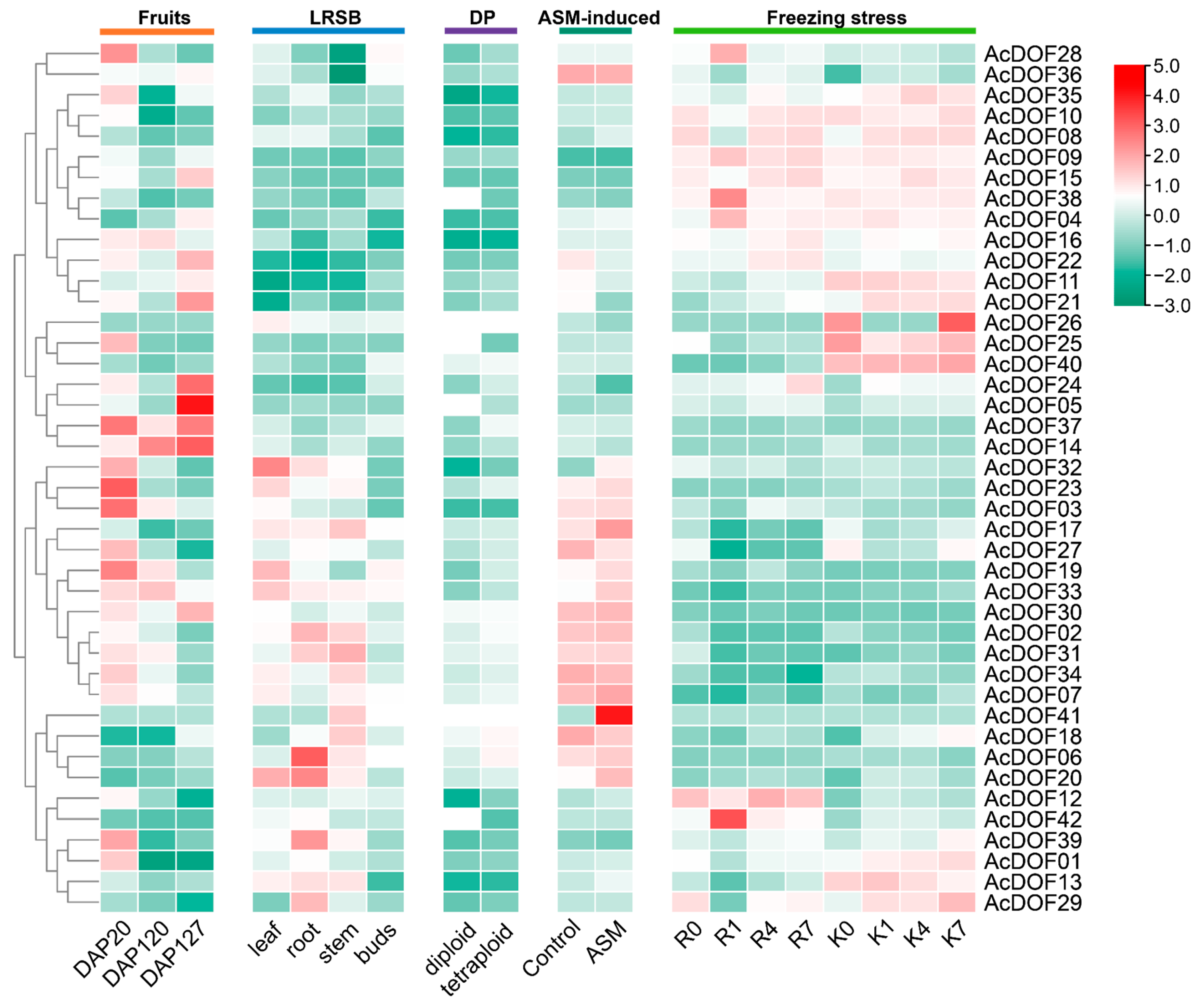
2.6. Expression Patterns of AcDOFs in Kiwifruit
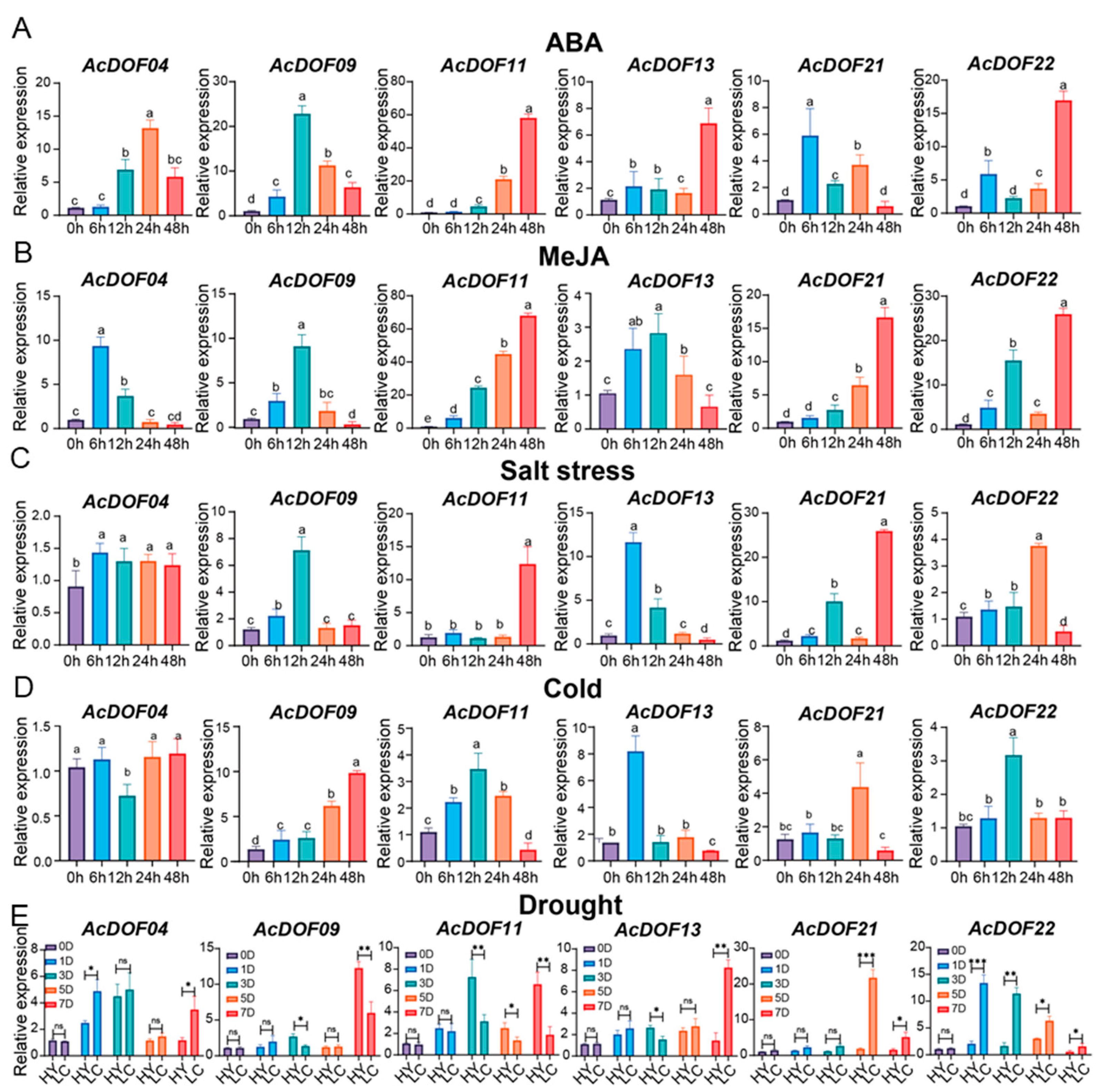
2.7. qRT-PCR Analysis of AcDOFs under Abiotic Stress
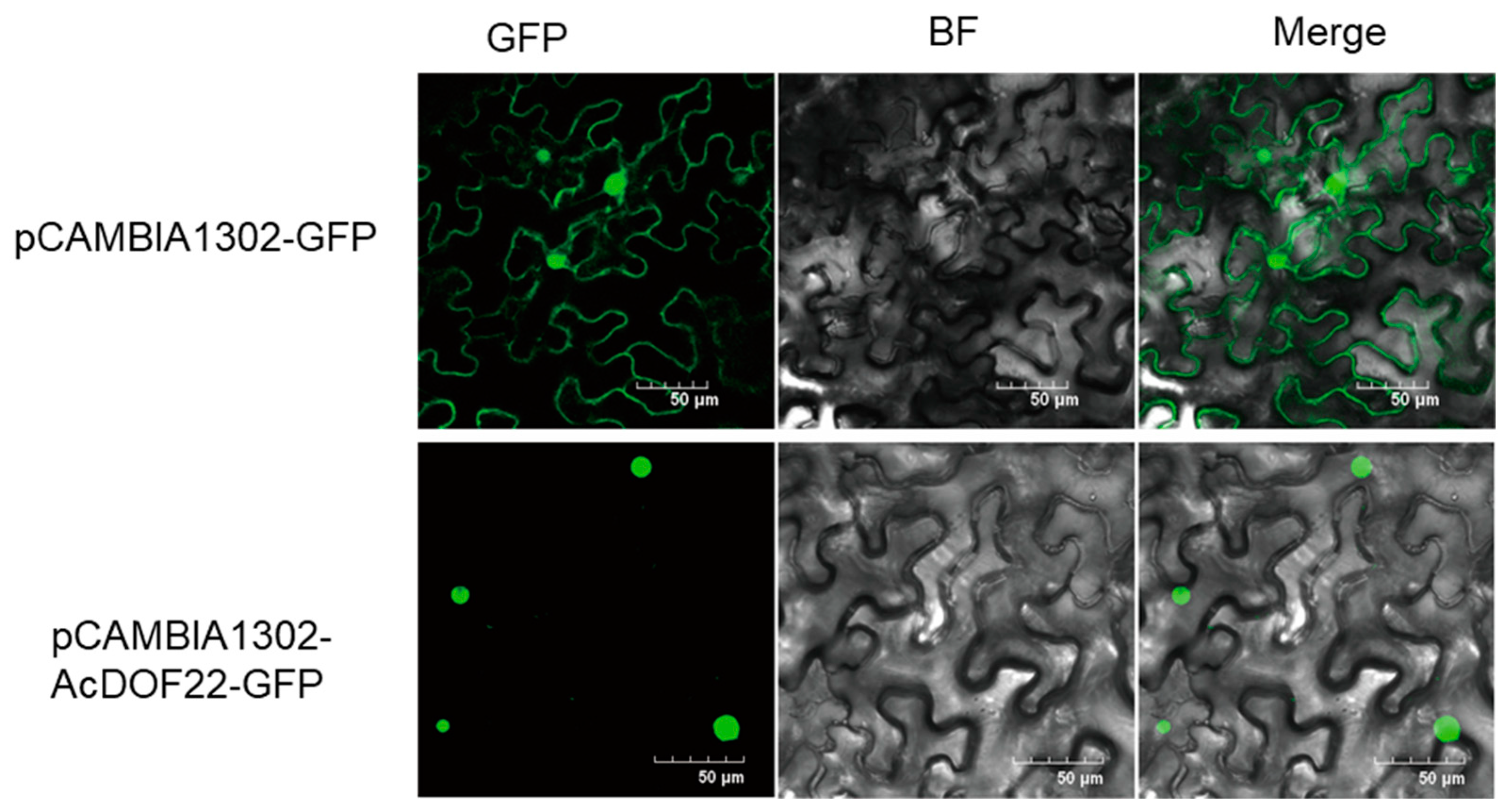
2.8. Subcellular Localization of AcDOF22
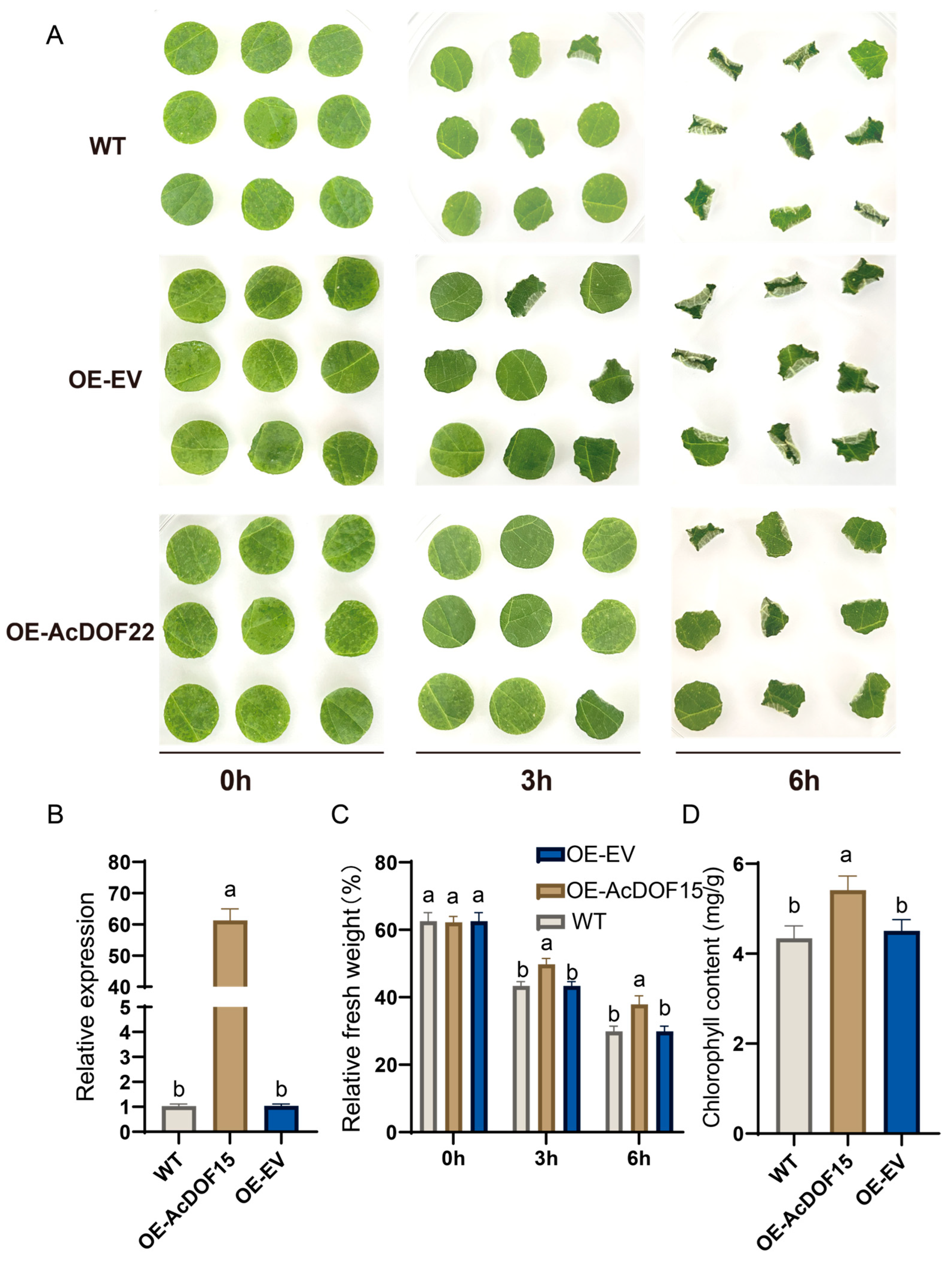
2.9. AcDOF22 Positively Regulates Kiwifruit Drought Resistance
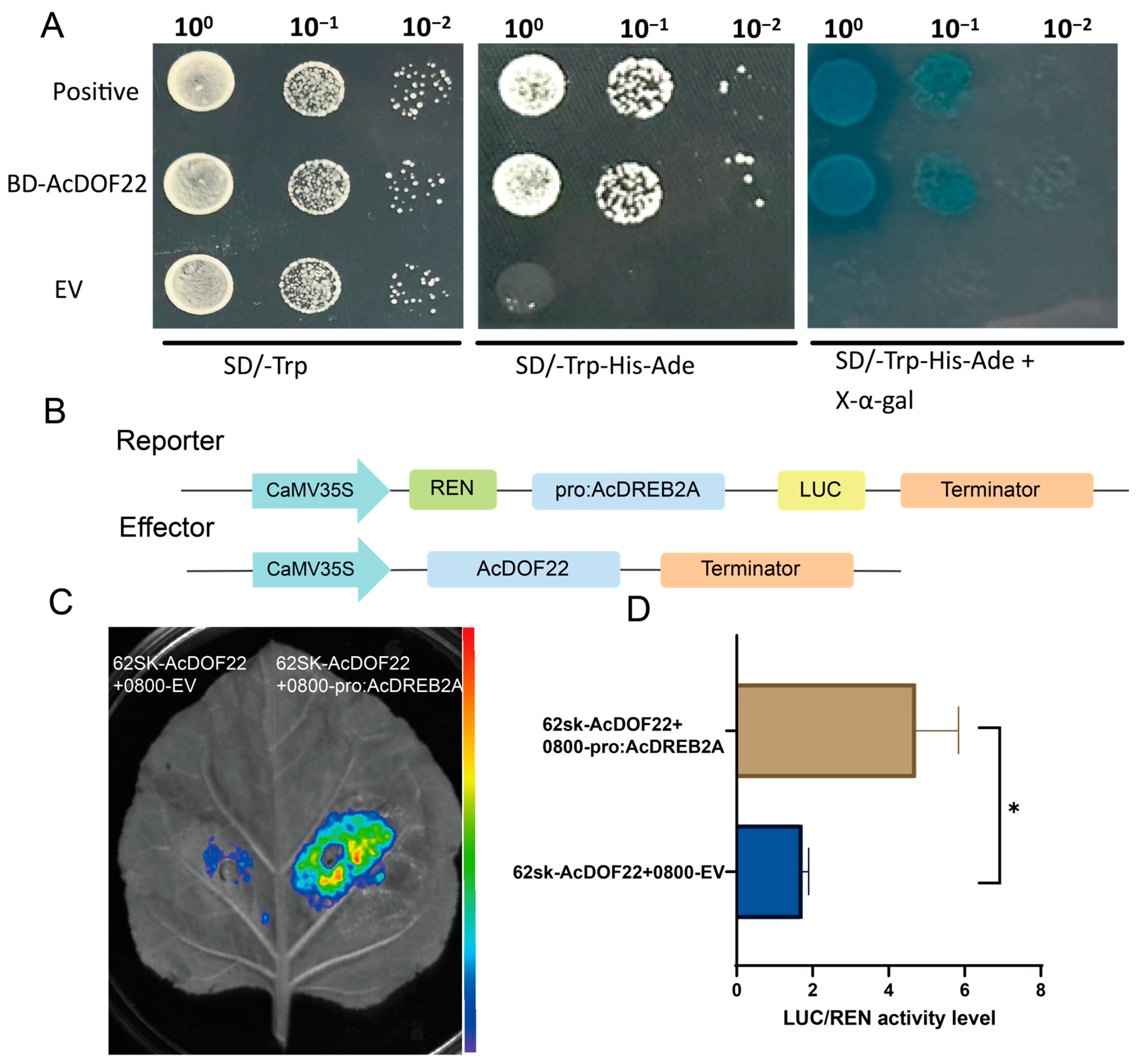
2.10. AcDOF22 Transcriptionally Activates AcDERB2A Expression
3. Discussion
4. Materials and Methods
4.1. Identification of DOF Gene Family Members and Construction of a Phylogenetic Tree
4.2. Analysis of the Structural Domains and Conserved Motifs of the Kiwifruit DOF Gene Family
4.3. CREs Analysis
4.4. Chromosomal Distribution of AcDOF Genes, Duplication, and Synteny Analysis
4.5. GO-Based Annotation of AcDOFs
4.6. Retrieval of RNA-Seq Expression Profile Data
4.7. Kiwifruit Plant Treatment, RNA Extraction, and qRT-PCR Analysis
4.8. Subcellular Localization and Yeast Self-Activation
4.9. Agrobacterium-Mediated Transient Expression in Kiwifruit Leaves
4.10. Dual-Luciferase Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wolber, F.M.; Beck, K.L.; Conlon, C.A.; Kruger, M.C. Kiwifruit and mineral nutrition. Adv. Food Nutr. Res. 2013, 68, 233–256. [Google Scholar] [CrossRef]
- Vissers, M.C.; Carr, A.C.; Pullar, J.M.; Bozonet, S.M. The bioavailability of vitamin C from kiwifruit. Adv. Food Nutr. Res. 2013, 68, 125–147. [Google Scholar] [CrossRef]
- Waswa, E.N.; Ding, S.X.; Wambua, F.M.; Mkala, E.M.; Mutinda, E.S.; Odago, W.O.; Amenu, S.G.; Muthui, S.W.; Linda, E.L.; Katumo, D.M.; et al. The genus Actinidia Lindl. (Actinidiaceae): A comprehensive review on its ethnobotany, phytochemistry, and pharmacological properties. J. Ethnopharmacol. 2024, 319, 117222. [Google Scholar] [CrossRef]
- Yue, J.; Liu, J.; Tang, W.; Wu, Y.Q.; Tang, X.; Li, W.; Yang, Y.; Wang, L.; Huang, S.; Fang, C.; et al. Kiwifruit Genome Database (KGD): A comprehensive resource for kiwifruit genomics. Hortic. Res. 2020, 7, 117. [Google Scholar] [CrossRef]
- Chen, Y.; Su, W.Y.; Ren, C.J.; Lin, Y.L.; Wang, W.Q.; Zhang, H.Q.; Yin, X.R.; Liu, X.F. Restricted responses of AcMYB68 and AcERF74/75 enhanced waterlogging tolerance in kiwifruit. Plant J. 2024, 119, 1059–1072. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, W.; Zhang, Y.; Li, Y.; Ma, C.; Tian, R.; Li, R.; Li, M.; Huang, L. Two transcription factors, AcREM14 and AcC3H1, enhance the resistance of kiwifruit Actinidia chinensis var. chinensis to Pseudomonas syringae pv. actinidiae. Hortic. Res. 2024, 11, uhad242. [Google Scholar] [CrossRef]
- Zhu, L.; Yin, T.; Zhang, M.; Yang, X.; Wu, J.; Cai, H.; Yang, N.; Li, X.; Wen, K.; Chen, D.; et al. Genome-wide identification and expression pattern analysis of the kiwifruit GRAS transcription factor family in response to salt stress. BMC Genom. 2024, 25, 12. [Google Scholar] [CrossRef]
- Strader, L.; Weijers, D.; Wagner, D. Plant transcription factors-being in the right place with the right company. Curr. Opin. Plant Biol. 2022, 65, 102136. [Google Scholar] [CrossRef]
- Zou, X.; Sun, H. DOF transcription factors: Specific regulators of plant biological processes. Front. Plant Sci. 2023, 14, 1044918. [Google Scholar] [CrossRef]
- Khan, I.; Khan, S.; Zhang, Y.; Zhou, J. Genome-wide analysis and functional characterization of the Dof transcription factor family in rice (Oryza sativa L.). Planta 2021, 253, 101. [Google Scholar] [CrossRef]
- Wei, Q.; Wang, W.; Hu, T.; Hu, H.; Mao, W.; Zhu, Q.; Bao, C. Genome-wide identification and characterization of Dof transcription factors in eggplant (Solanum melongena L.). PeerJ 2018, 6, e4481. [Google Scholar] [CrossRef]
- Malviya, N.; Gupta, S.; Singh, V.K.; Yadav, M.K.; Bisht, N.C.; Sarangi, B.K.; Yadav, D. Genome wide in silico characterization of Dof gene families of pigeonpea (Cajanus cajan (L) Millsp.). Mol. Biol. Rep. 2015, 42, 535–552. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Tong, Q.; Xu, G.; Xu, M.; Li, H.; Fan, P.; Li, S.; Liang, Z. Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 2021, 253, 55. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Hu, H.; Xie, J. Genome-wide analysis of the DNA-binding with one zinc finger (Dof) transcription factor family in bananas. Genome 2016, 59, 1085–1100. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, L.; Liu, X.; Chen, X.; Wang, X. Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica. Gene 2018, 639, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Li, C.; Zhang, J.; Tian, Y.; Wang, H.; Zhang, Y.; Zhang, Z.; Xiang, Q.; Han, X.; Zhang, L. Genome-wide identification and expression analysis of the Dof gene family under drought stress in tea (Camellia sinensis). PeerJ 2020, 8, e9269. [Google Scholar] [CrossRef] [PubMed]
- Khaksar, G.; Sangchay, W.; Pinsorn, P.; Sangpong, L.; Sirikantaramas, S. Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent Dof transcription factors. Sci. Rep. 2019, 9, 12109. [Google Scholar] [CrossRef]
- Hou, Q.; Yu, R.; Shang, C.; Deng, H.; Wen, Z.; Qiu, Z.; Qiao, G. Molecular characterization and evolutionary relationships of DOFs in four cherry species and functional analysis in sweet cherry. Int. J. Biol. Macromol. 2024, 263, 130346. [Google Scholar] [CrossRef]
- Liu, X.; Liu, Z.; Hao, Z.; Chen, G.; Qi, K.; Zhang, H.; Jiao, H.; Wu, X.; Zhang, S.; Wu, J.; et al. Characterization of Dof family in Pyrus bretschneideri and role of PbDof9.2 in flowering time regulation. Genomics 2020, 112, 712–720. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur. J. Biochem. 1997, 250, 403–410. [Google Scholar] [CrossRef]
- Manna, M.; Thakur, T.; Chirom, O.; Mandlik, R.; Deshmukh, R.; Salvi, P. Transcription factors as key molecular target to strengthen the drought stress tolerance in plants. Physiol. Plant. 2021, 172, 847–868. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Ye, F.; Fahim, A.M.; Ma, C.; Pang, Y.; Zhang, X.; Zhang, Q.; Lu, X. Transcription factor ZmDof22 enhances drought tolerance by regulating stomatal movement and antioxidant enzymes activities in maize (Zea mays L.). Theor. Appl. Genet. 2024, 137, 132. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, G.L.; Shi, L.; Bergonzi, S.B.; Oortwijn, M.; Franco-Zorrilla, J.M.; Solano-Tavira, R.; Visser, R.; Abelenda, J.A.; Bachem, C. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 2021, 105, 855–869. [Google Scholar] [CrossRef]
- Chen, P.; Yan, M.; Li, L.; He, J.; Zhou, S.; Li, Z.; Niu, C.; Bao, C.; Zhi, F.; Ma, F.; et al. The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 2020, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, T.; Jia, Z.H.; Xuan, J.P.; Pan, D.L.; Guo, Z.R.; Zhang, J.Y. Genome-Wide bioinformatics analysis of MAPK gene family in kiwifruit (Actinidia chinensis). Int. J. Mol. Sci. 2018, 19, 2510. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, C.; Liu, F.; Li, D.; Zhang, A.; Li, L.; Zhang, X. Genome-Wide identification and expression analysis of kiwifruit Leucine-Rich Repeat Receptor-Like proteins reveal their Roles in biotic and abiotic stress responses. Int. J. Mol. Sci. 2024, 25, 4497. [Google Scholar] [CrossRef]
- Wang, W.Q.; Wang, J.; Wu, Y.Y.; Li, D.W.; Allan, A.C.; Yin, X.R. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020, 225, 1618–1634. [Google Scholar] [CrossRef]
- Bao, W.W.; Chen, X.; Li, R.N.; Li, M.; Xie, C.J.; Dou, M.R.; Zhang, K.Z.; Wang, J.; Gao, Z.X.; Liu, Z.D.; et al. Comprehensive assessment of drought resistance and recovery in kiwifruit genotypes using multivariate analysis. Plant J. 2024, 119, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Yang, C.; Liang, Y.; He, Z.; Guo, Y.; Lang, Y.; Wei, J.; Tian, X.; Lin, L.; Deng, H.; et al. Melatonin and arbuscular mycorrhizal fungi synergistically improve drought toleration in kiwifruit seedlings by increasing mycorrhizal colonization and nutrient uptake. Front. Plant Sci. 2022, 13, 1073917. [Google Scholar] [CrossRef]
- Zhang, A.D.; Wang, W.Q.; Tong, Y.; Li, M.J.; Grierson, D.; Ferguson, I.; Chen, K.S.; Yin, X.R. Transcriptome analysis identifies a Zinc Finger protein regulating starch degradation in kiwifruit. Plant Physiol. 2018, 178, 850–863. [Google Scholar] [CrossRef]
- Wu, H.; Ma, T.; Kang, M.; Ai, F.; Zhang, J.; Dong, G.; Liu, J. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic. Res. 2019, 6, 117. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, P.; Logie, C.; Lovering, R.C.; Kuiper, M.; Laegreid, A.; Thomas, P.D. Gene Ontology representation for transcription factor functions. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194752. [Google Scholar] [CrossRef] [PubMed]
- Signor, S.A.; Nuzhdin, S.V. The Evolution of Gene Expression in cis and trans. Trends Genet. 2018, 34, 532–544. [Google Scholar] [CrossRef]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, T.; Yu, J.; Hong, H.; Liu, S.; Guo, F.; Ma, H.; Zhang, J.; Zhu, M.; Meng, X. Genome-wide survey and expression analysis of Dof transcription factor family in sweetpotato shed light on their promising functions in stress tolerance. Front. Plant Sci. 2023, 14, 1140727. [Google Scholar] [CrossRef]
- Song, H.; Ji, X.; Wang, M.; Li, J.; Wang, X.; Meng, L.; Wei, P.; Xu, H.; Niu, T.; Liu, A. Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.). Gene 2024, 910, 148336. [Google Scholar] [CrossRef]
- Cai, K.; Xie, X.; Han, L.; Chen, J.; Zhang, J.; Yuan, H.; Shen, J.; Ren, Y.; Zhao, X. Identification and functional analysis of the DOF gene family in Populus simonii: Implications for development and stress response. Front. Plant Sci. 2024, 15, 1412175. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Jia, H.; Wu, M.; Zhong, W.; Jia, D.; Wang, Z.; Huang, C. Genome-wide identification and characterization of the TIFY gene family in kiwifruit. BMC Genom. 2022, 23, 179. [Google Scholar] [CrossRef]
- Su, L.; Xu, M.; Zhang, J.; Wang, Y.; Lei, Y.; Li, Q. Genome-wide identification of auxin response factor (ARF) family in kiwifruit (Actinidia chinensis) and analysis of their inducible involvements in abiotic stresses. Physiol. Mol. Biol. Plants 2021, 27, 1261–1276. [Google Scholar] [CrossRef]
- Kong, L.; Song, Q.; Wei, H.; Wang, Y.; Lin, M.; Sun, K.; Zhang, Y.; Yang, J.; Li, C.; Luo, K. The AP2/ERF transcription factor PtoERF15 confers drought tolerance via JA-mediated signaling in Populus. New Phytol. 2023, 240, 1848–1867. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef]
- Santopolo, S.; Boccaccini, A.; Lorrai, R.; Ruta, V.; Capauto, D.; Minutello, E.; Serino, G.; Costantino, P.; Vittorioso, P. DOF AFFECTING GERMINATION 2 is a positive regulator of light-mediated seed germination and is repressed by DOF AFFECTING GERMINATION 1. BMC Plant Biol. 2015, 15, 72. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, Z.; Bing, Q.; Duan, X.; Chen, S.; Zeng, M.; Liu, X. Two Dof transcription factors promote flavonoid synthesis in kumquat fruit by activating C-glucosyltransferase. Plant Sci. 2022, 318, 111234. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Chen, M.; Zhou, W.; Dong, Q.; Jiang, H.; Cheng, B. The DOF-Domain transcription factor ZmDOF36 positively regulates starch synthesis in transgenic maize. Front. Plant Sci. 2019, 10, 465. [Google Scholar] [CrossRef]
- Xu, P.; Chen, H.; Ying, L.; Cai, W. AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana. Sci. Rep. 2016, 6, 27705. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, Z.; Ai, Q.; Li, X.; Yang, J.; Zhang, N.; Si, H. DNA-Binding with One Finger (Dof) transcription factor gene family study reveals differential stress-responsive transcription factors in contrasting drought tolerance potato species. Int. J. Mol. Sci. 2024, 25, 3488. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Wei, J.T.; Zhao, S.P.; Zhang, H.Y.; Jin, L.G.; Yu, T.F.; Zheng, L.; Ma, J.; Chen, J.; Zhou, Y.B.; Chen, M.; et al. GmDof41 regulated by the DREB1-type protein improves drought and salt tolerance by regulating the DREB2-type protein in soybean. Int. J. Biol. Macromol. 2023, 230, 123255. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Lin, M.; Qi, X.; Chen, J.; Gu, H.; Zhong, Y.; Sun, L.; Muhammad, A.; Bai, D.; Hu, C.; et al. Full-length transcriptome profiling reveals insight into the cold response of two kiwifruit genotypes (A. arguta) with contrasting freezing tolerances. BMC Plant Biol. 2021, 21, 365. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhao, C.; Liu, L.; Huang, D.; Ma, C.; Li, R.; Huang, L. Genome-wide identification of the TGA gene family in kiwifruit (Actinidia chinensis spp.) and revealing its roles in response to Pseudomonas syringae pv. actinidiae (Psa) infection. Int. J. Biol. Macromol. 2022, 222, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhong, S.; Dong, Q.; Yang, H.; Yang, H.; Tan, F.; Chen, C.; Ren, T.; Shen, J.; Cao, G.; et al. Identification of Photoperiod- and Phytohormone-Responsive DNA-Binding One Zinc Finger (Dof) transcription factors in Akebia trifoliata via Genome-Wide expression analysis. Int. J. Mol. Sci. 2023, 24, 4973. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Kokkirala, V.R.; Yonggang, P.; Abbagani, S.; Zhu, Z.; Umate, P. Subcellular localization of proteins of Oryza sativa L. in the model tobacco and tomato plants. Plant Signal. Behav. 2010, 5, 1336–1341. [Google Scholar] [CrossRef]
- Luan, Y.; Chen, Z.; Fang, Z.; Meng, J.; Tao, J.; Zhao, D. PoWRKY69-PoVQ11 module positively regulates drought tolerance by accumulating fructose in Paeonia ostii. Plant J. 2024, 119, 1782–1799. [Google Scholar] [CrossRef]
- Geng, L.; Yu, S.; Zhang, Y.; Su, L.; Lu, W.; Zhu, H.; Jiang, X. Transcription factor RcNAC091 enhances rose drought tolerance through the abscisic acid-dependent pathway. Plant Physiol. 2023, 193, 1695–1712. [Google Scholar] [CrossRef]
- McNabb, D.S.; Reed, R.; Marciniak, R.A. Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 2005, 4, 1539–1549. [Google Scholar] [CrossRef]
- Ramongolalaina, C. Dual-luciferase assay and siRNA silencing for nodD1 to study the competitiveness of Bradyrhizobium diazoefficiens USDA110 in soybean nodulation. Microbiol. Res. 2020, 237, 126488. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Bai, H.; Li, C.; Pang, Z.; Xuan, L.; Lv, D.; Niu, S. Genome-Wide Identification of the DOF Gene Family in Kiwifruit (Actinidia chinensis) and Functional Validation of AcDOF22 in Response to Drought Stress. Int. J. Mol. Sci. 2024, 25, 9103. https://doi.org/10.3390/ijms25169103
Zhao C, Bai H, Li C, Pang Z, Xuan L, Lv D, Niu S. Genome-Wide Identification of the DOF Gene Family in Kiwifruit (Actinidia chinensis) and Functional Validation of AcDOF22 in Response to Drought Stress. International Journal of Molecular Sciences. 2024; 25(16):9103. https://doi.org/10.3390/ijms25169103
Chicago/Turabian StyleZhao, Chao, Hao Bai, Chaoshuo Li, Zhaojin Pang, Lifeng Xuan, Dezhi Lv, and Shuaike Niu. 2024. "Genome-Wide Identification of the DOF Gene Family in Kiwifruit (Actinidia chinensis) and Functional Validation of AcDOF22 in Response to Drought Stress" International Journal of Molecular Sciences 25, no. 16: 9103. https://doi.org/10.3390/ijms25169103
APA StyleZhao, C., Bai, H., Li, C., Pang, Z., Xuan, L., Lv, D., & Niu, S. (2024). Genome-Wide Identification of the DOF Gene Family in Kiwifruit (Actinidia chinensis) and Functional Validation of AcDOF22 in Response to Drought Stress. International Journal of Molecular Sciences, 25(16), 9103. https://doi.org/10.3390/ijms25169103





