Identification of Two Long Noncoding RNAs, Kcnq1ot1 and Rmst, as Biomarkers in Chronic Liver Diseases in Mice
Abstract
1. Introduction
2. Results
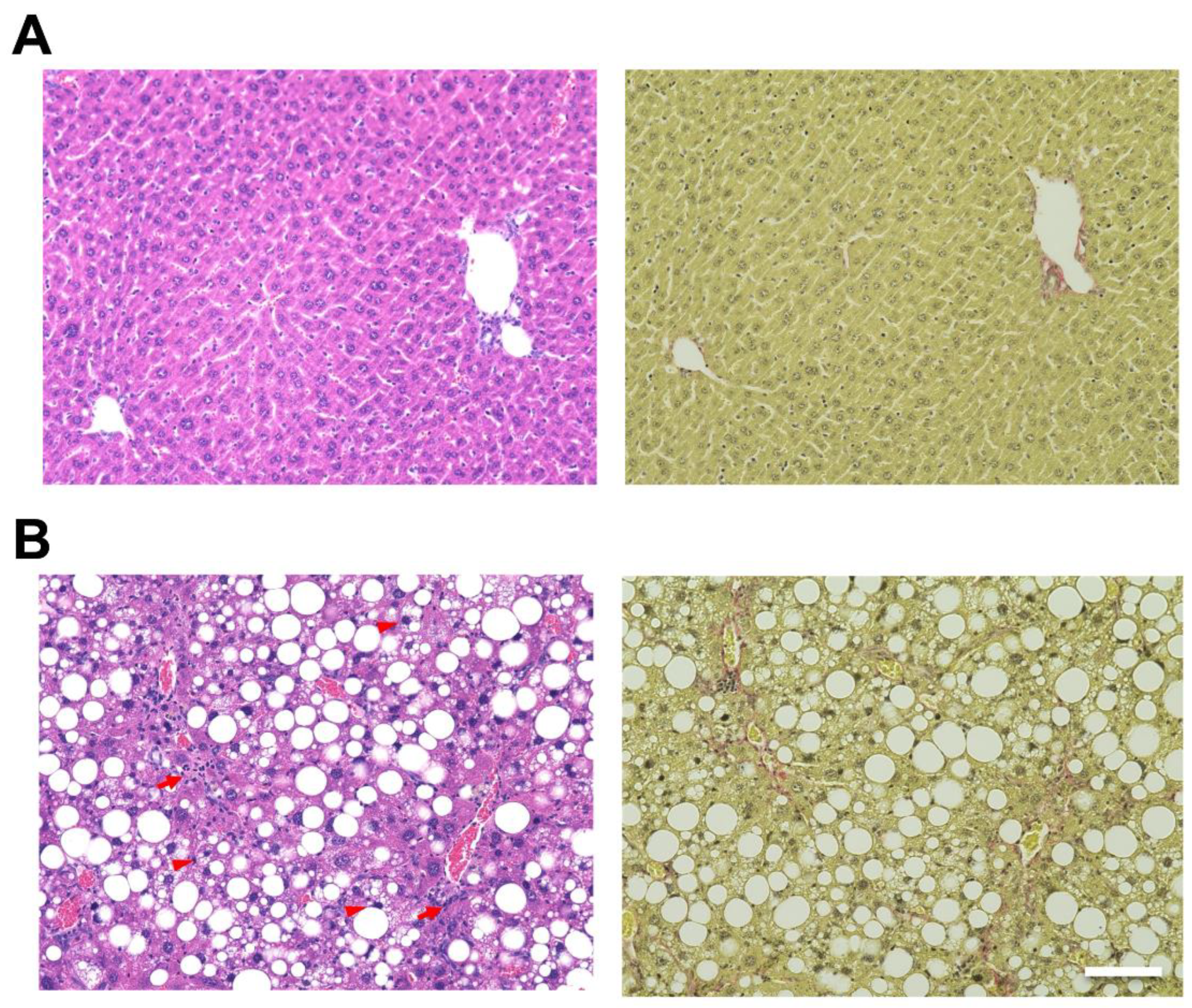
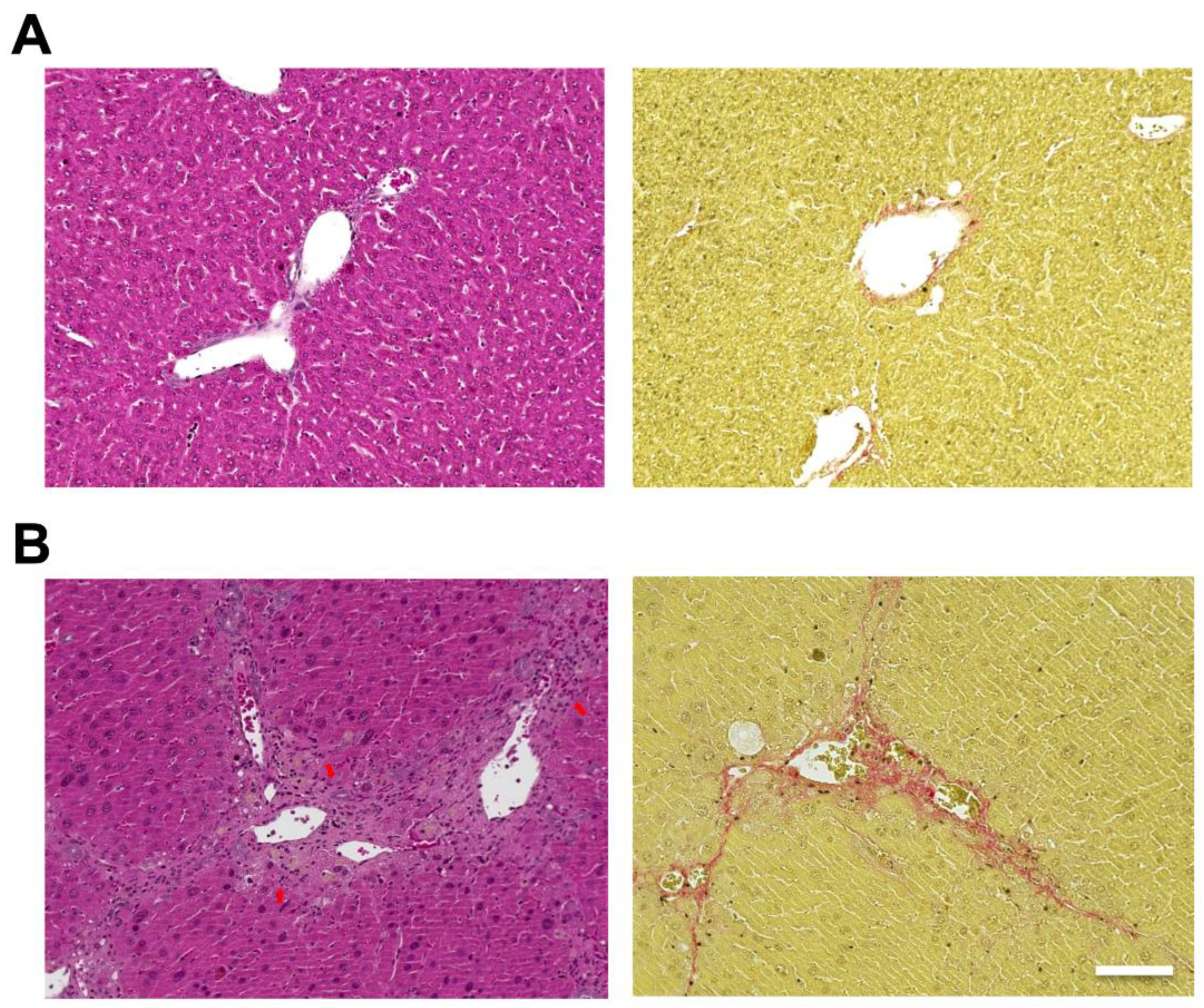
2.1. Evaluation of the Liver in Mice
2.2. LncRNA Expression Changes of the Liver in Mice
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Murine Model
5.2. Histological Analyses
5.3. RNA Isolation
5.4. Quantitative PCR (RT-qPCR)
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muto, H.; Honda, T.; Tanaka, T.; Yokoyama, S.; Yamamoto, K.; Ito, T.; Imai, N.; Ishizu, Y.; Maeda, K.; Ishikawa, T.; et al. Proteomic analysis reveals changes in tight junctions in the small intestinal epithelium of mice fed a high-fat diet. Nutrients 2023, 15, 1473. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ishigami, M.; Zou, B.; Tanaka, T.; Takahashi, H.; Kurosaki, M.; Maeda, M.; Thin, K.N.; Tanaka, K.; Takahashi, Y.; et al. The epidemiology of NAFLD and lean NAFLD in Japan: A meta-analysis with individual and forecasting analysis, 1995–2040. Hepatol. Int. 2021, 15, 366–379. [Google Scholar] [CrossRef]
- Tateishi, R.; Uchino, K.; Fujiwara, N.; Takehara, T.; Okanoue, T.; Seike, M.; Yoshiji, H.; Yatsuhashi, H.; Shimizu, M.; Torimura, T.; et al. A Nationwide Survey on Non-B, Non-C Hepatocellular Carcinoma in Japan: 2011–2015 Update. J. Gastroenterol. 2019, 54, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shen, J.; Sun, T.T.; Zhang, X.; Wong, N. Obesity, insulin resistance, NASH and hepatocellular carcinoma. Semin. Cancer Biol. 2013, 23, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.W.; Adams, L.A. Non-alcoholic fatty liver disease. Crit. Rev. Clin. Lab. Sci. 2011, 48, 97–113. [Google Scholar] [CrossRef]
- Itagaki, H.; Shimizu, K.; Morikawa, S.; Ogawa, K.; Ezaki, T. Morphological and functional characterization of non-alcoholic fatty liver disease induced by a methionine-choline-deficient diet in C57BL/6 mice. Int. J. Clin. Exp. Pathol. 2013, 6, 2683–2696. [Google Scholar]
- Ito, T.; Ishigami, M.; Ishizu, Y.; Kuzuya, T.; Honda, T.; Hayashi, K.; Nishimura, D.; Toyoda, H.; Kumada, T.; Goto, H.; et al. Utility and limitations of noninvasive fibrosis markers for predicting prognosis in biopsy-proven Japanese non-alcoholic fatty liver disease patients. J. Gastroenterol. Hepatol. 2019, 34, 207–214. [Google Scholar] [CrossRef]
- Hagström, H.; Nasr, P.; Ekstedt, M.; Hammar, U.; Stål, P.; Hultcrantz, R.; Kechagias, S. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J. Hepatol. 2017, 67, 1265–1273. [Google Scholar] [CrossRef]
- Angulo, P.; Kleiner, D.E.; Dam-Larsen, S.; Adams, L.A.; Bjornsson, E.S.; Charatcharoenwitthaya, P.; Mills, P.R.; Keach, J.C.; Lafferty, H.D.; Stahler, A.; et al. Liver Fibrosis, but No Other Histologic Features, Is Associated with Long-term Outcomes of Patients with Nonalcoholic Fatty Liver Disease. Gastroenterology 2015, 149, 389–397. [Google Scholar] [CrossRef]
- Charlton, M.; Krishnan, A.; Viker, K.; Sanderson, S.; Cazanave, S.; McConico, A.; Masuoko, H.; Gores, G. Fast food diet mouse: Novel small animal model of NASH with ballooning, progressive fibrosis, and high physiological fidelity to the human condition. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 301, G825–G834. [Google Scholar] [CrossRef]
- Asgharpour, A.; Cazanave, S.C.; Pacana, T.; Seneshaw, M.; Vincent, R.; Banini, B.A.; Kumar, D.P.; Daita, K.; Min, H.K.; Mirshahi, F.; et al. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. J. Hepatol. 2016, 65, 579–588. [Google Scholar] [CrossRef]
- Scholten, D.; Trebicka, J.; Liedtke, C.; Weiskirchen, R. The carbon tetrachloride model in mice. Lab. Anim. 2015, 49, 4–11. [Google Scholar] [CrossRef]
- Kubota, N.; Kado, S.; Kano, M.; Masuoka, N.; Nagata, Y.; Kobayashi, T.; Miyazaki, K.; Ishikawa, F. A high-fat diet and multiple administration of carbon tetrachloride induces liver injury and pathological features associated with non-alcoholic steatohepatitis in mice. Clin. Exp. Pharmacol. Physiol. 2013, 40, 422–430. [Google Scholar] [CrossRef]
- Tsuchida, T.; Lee, Y.A.; Fujiwara, N.; Ybanez, M.; Allen, B.; Martins, S.; Fiel, M.I.; Goossens, N.; Chou, H.I.; Hoshida, Y.; et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 2018, 69, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Obuse, C.; Hirose, T. Functional domains of nuclear long noncoding RNAs: Insights into gene regulation and intracellular architecture. Curr. Opin. Cell Biol. 2023, 85, 102250. [Google Scholar] [CrossRef]
- Onoguchi-Mizutani, R.; Akimitsu, N. Long noncoding RNA and phase separation in cellular stress response. J. Biochem. 2022, 171, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Arakawa, K.; Hirose, T.; Inada, T.; Ito, T.; Kai, T.; Oyama, M.; Tomari, Y.; Yoda, T.; Nakagawa, S. Nondomain biopolymers: Flexible molecular strategies to acquire biological functions. Genes Cells 2023, 28, 539–552. [Google Scholar] [CrossRef]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Mizutani, R.; Salam, K.A.; Tano, K.; Ijiri, K.; Wakamatsu, A.; Isogai, T.; Suzuki, Y.; Akimitsu, N. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012, 22, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 2, ra8. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.T.F.; Ideue, T.; Sano, M.; Mituyama, T.; Hirose, T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc. Natl. Acad. Sci. USA 2009, 24, 2525–2530. [Google Scholar] [CrossRef]
- Abe, R.; Yagi, Y.; Tani, H. Identifying long non-coding RNA as potential indicators of bacterial stress in human cells. BPB Rep. 2023, 6, 226–228. [Google Scholar] [CrossRef]
- Yagi, Y.; Abe, R.; Tani, H. Exploring IDI2-AS1, OIP5-AS1, and LITATS1: Changes in long non-coding RNAs induced by the poly I:C stimulation. Biol. Pharm. Bull. 2024, 47, 1144–1148. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Rolón, A.; Purcell, D.; Rosado, K.; Toro, D.H. A Comparison of Brunt’s Criteria, the Non-Alcoholic Fatty Liver Disease Activity Score (NAS), and a Proposed NAS Scoring that Includes Fibrosis in Non-Alcoholic Fatty Liver Disease Staging. Puerto Rico Health Sci. J. 2015, 34, 189–194. [Google Scholar]
- Cagle, P.; Qi, Q.; Niture, S.; Kumar, D. KCNQ1OT1: An Oncogenic Long Noncoding RNA. Biomolecules 2021, 11, 1602. [Google Scholar] [CrossRef]
- Ng, S.Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef]
- Liu, G.; Shi, L.; Wang, B.; Wu, Z.; Zhao, H.; Zhao, T.; Shi, L. Role of oncogenic long noncoding RNA KCNQ1OT1 in colon cancer. Oncol. Res. 2024, 32, 585–596. [Google Scholar] [CrossRef]
- Wu, Y.; Bi, Q.J.; Han, R.; Zhang, Y. Long noncoding RNA KCNQ1OT1 is correlated with human breast cancer cell development through inverse regulation of hsa-miR-107. Biochem. Cell Biol. 2020, 98, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, X.; Ma, X.; Yuan, Z.; Hu, M. KCNQ1OT1 contributes to sorafenib resistance and programmed death-ligand-1-mediated immune escape via sponging miR-506 in hepatocellular carcinoma cells. Int. J. Mol. Med. 2020, 46, 1794–1804. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.L.; Li, D.J.; Lv, M.Y.; Pei, Y.J.; Zhang, X.J.; Li, L.; Liu, X.Y.; Fan, A.H. LncRNA KCNQ1OT1 regulates the invasion and migration of hepatocellular carcinoma by acting on S1PR1 through miR-149. Cancer Gene Ther. 2021, 28, 850–863. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, H.; Li, Q.; Zhang, L.; Chen, L.; Wang, H.; Chen, Y. Long non-coding RNA KCNQ1OT1 alleviates postmenopausal osteoporosis by modulating miR-421-3p/mTOR axis. Sci. Rep. 2023, 13, 2333. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Li, C.; Wu, Z. lncRNA RMST is associated with the progression and prognosis of gastric cancer via miR-204-5p. Cell Div. 2024, 19, 12. [Google Scholar] [CrossRef] [PubMed]
- De Martino, M.; Pellecchia, S.; Esposito, F.; Liotti, F.; Credendino, S.C.; Prevete, N.; Decaussin-Petrucci, M.; Chieffi, P.; De Vita, G.; Melillo, R.M. The lncRNA RMST is drastically downregulated in anaplastic thyroid carcinomas where exerts a tumor suppressor activity impairing epithelial-mesenchymal transition and stemness. Cell Death Discov. 2023, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ji, L.; Wang, Y.; Zhang, L.; Xu, M.; Su, Y.; Zhang, X. lncRNA RMST suppresses the progression of colorectal cancer by competitively binding to miR-27a-3p/RXRα axis and inactivating Wnt signaling pathway. Acta Biochim. Biophys. Sin. 2023, 55, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Feng, C.; Zhang, X.; Sun, B.; Bian, Y. Abnormal expression of long non-coding RNA rhabdomyosarcoma 2-associated transcript (RMST) participates in the pathological mechanism of atherosclerosis by regulating miR-224-3p. Bioengineered 2022, 13, 2648–2657. [Google Scholar] [CrossRef]
- Zhou, L.; Zhi, Z.; Chen, P.; Du, C.; Wang, B.; Fang, X.; Tang, W.; Li, H. LncRNA-RMST Functions as a Transcriptional Co-regulator of SOX2 to Regulate miR-1251 in the Progression of Hirschsprung’s Disease. Front. Pediatr. 2022, 10, 749107. [Google Scholar] [CrossRef]

| Name | Accession No. | Length (nt) | t1/2 * |
|---|---|---|---|
| Snhg4 | NR_038073 | 1050 | 0.32 |
| Kcnq1ot1 | NR_001461 | 90,612 | 1.60 |
| Rmst | NR_028262 | 2687 | 1.38 |
| Neat1 | NR_003513 | 3190 | 0.01 |
| Gas5 | NR_153812 | 501 | 0.18 |
| Mir17hg | NR_029382 | 2339 | 0.07 |
| Gt(ROSA)26Sor | NR_027010 | 613 | 0.12 |
| Snhg8 | NR_028574 | 785 | 0.14 |
| Snhg17 | NR_015463 | 3175 | 0.48 |
| Peg13 | NR_002864 | 4723 | 0.67 |
| Name | Sence Sequence (5′-3′) | Antisence Sequence (5′-3′) |
|---|---|---|
| Gapdh | TGGTGAAGCAGGCATCTGAG | TGAAGTCGCAGGAGACAACC |
| Actb | CCAGCCATGTACGTAGCCAT | CGGAGTCCATCACAATGCCT |
| Snhg4 | GCATTTTCAGAGTCAACATTCCCA | GAAGCCTCAAGACCTAAACTCCA |
| Kcnq1ot1 | GGCGCAGGTCTGTAATCGTA | CCCAGCAAGGTCCTGAACAT |
| Rmst | GCAGCTAAAGGACAGGAGCA | TTAACTCGCAGTTGGTGGCA |
| Neat1 | AGTGTGAGTCGTAGCAGTGC | TCACTGTGTAGGCGTCAACC |
| Gas5 | ATGGTGGAGTTTGAGGCTGG | CAAGCAAGCCAGCCAAATGA |
| Mir17hg | GCACTTGTTCAGTTCCGCAC | CCACAGCTGTTTTGCCAACA |
| Gt(ROSA)26Sor | GCCACACATGTCCCATTCCA | AACTTTCCAACCCACCACCC |
| Snhg8 | CACAAGGTGGCTATGGTGCT | TCGTCGCGCTAACCTTCAC |
| Snhg17 | TGTCTCCCAAGAGCTCCAGT | ATCCTGCTATGGCCTCCTGA |
| Peg13 | GATTCCACTTGGGTGCCTCA | GACGTCTGCCTGGTCTCTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokoyama, S.; Muto, H.; Honda, T.; Kurokawa, Y.; Ogawa, H.; Nakajima, R.; Kawashima, H.; Tani, H. Identification of Two Long Noncoding RNAs, Kcnq1ot1 and Rmst, as Biomarkers in Chronic Liver Diseases in Mice. Int. J. Mol. Sci. 2024, 25, 8927. https://doi.org/10.3390/ijms25168927
Yokoyama S, Muto H, Honda T, Kurokawa Y, Ogawa H, Nakajima R, Kawashima H, Tani H. Identification of Two Long Noncoding RNAs, Kcnq1ot1 and Rmst, as Biomarkers in Chronic Liver Diseases in Mice. International Journal of Molecular Sciences. 2024; 25(16):8927. https://doi.org/10.3390/ijms25168927
Chicago/Turabian StyleYokoyama, Shinya, Hisanori Muto, Takashi Honda, Yoichi Kurokawa, Hirotaka Ogawa, Riku Nakajima, Hiroki Kawashima, and Hidenori Tani. 2024. "Identification of Two Long Noncoding RNAs, Kcnq1ot1 and Rmst, as Biomarkers in Chronic Liver Diseases in Mice" International Journal of Molecular Sciences 25, no. 16: 8927. https://doi.org/10.3390/ijms25168927
APA StyleYokoyama, S., Muto, H., Honda, T., Kurokawa, Y., Ogawa, H., Nakajima, R., Kawashima, H., & Tani, H. (2024). Identification of Two Long Noncoding RNAs, Kcnq1ot1 and Rmst, as Biomarkers in Chronic Liver Diseases in Mice. International Journal of Molecular Sciences, 25(16), 8927. https://doi.org/10.3390/ijms25168927







