Breast Cancer Plasticity after Chemotherapy Highlights the Need for Re-Evaluation of Subtyping in Residual Cancer and Metastatic Tissues
Abstract
1. Introduction
2. Results
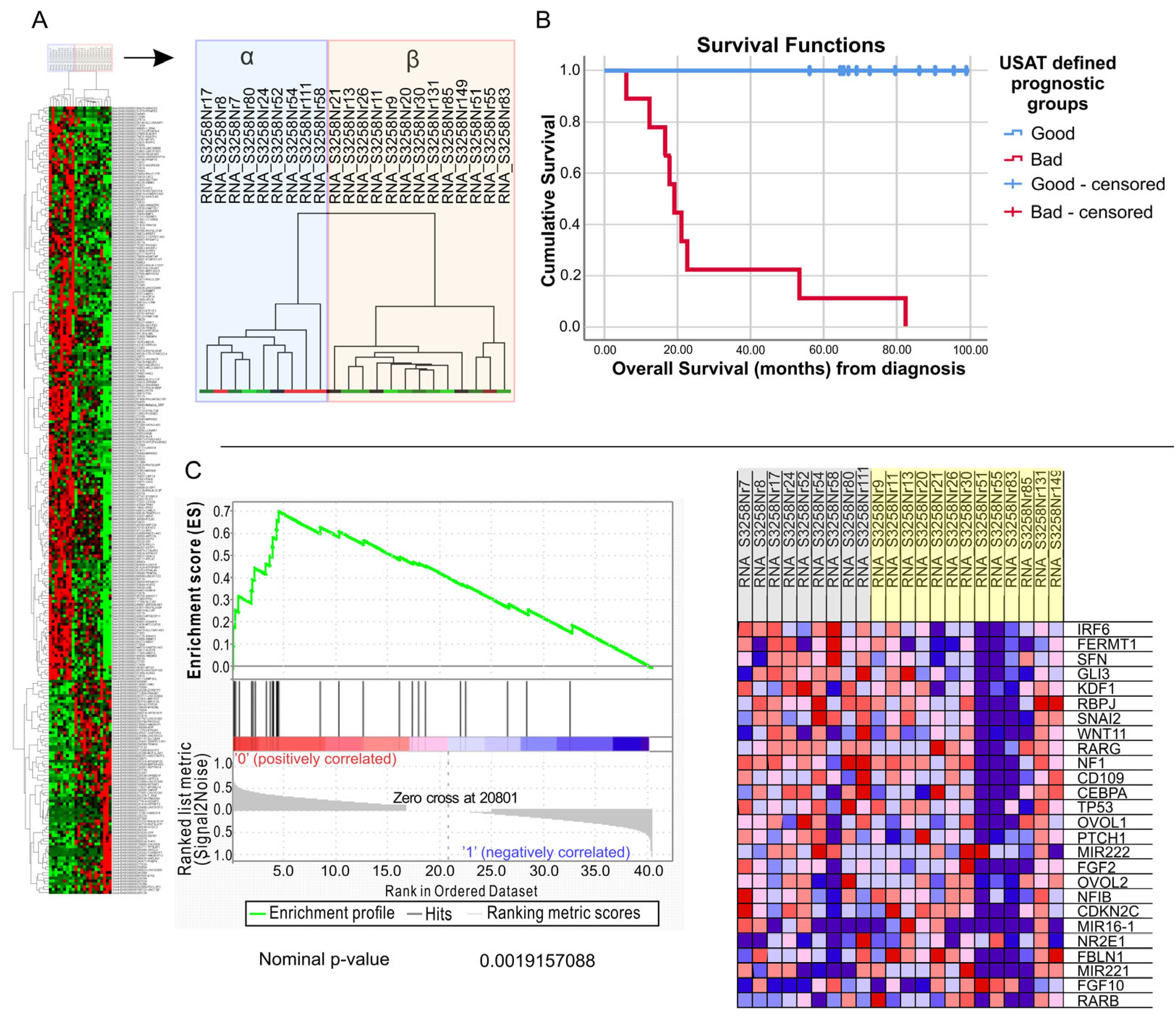
2.1. Identifying a Prognostic Signature for TNBC Patients
2.2. Validating a Previous Signature (Non-Stem Cell-like/EPITHELIAL (NS/E) vs. Stem Cell-like/Mesenchymal (CS/M)) in Our Patient Cohort
2.3. Correlating qPCR to RNAseq Data
2.4. PAM Plasticity
2.5. Identifying Metastatic Markers
3. Discussion
4. Methods and Materials
4.1. Sample Collection
4.2. RNA Isolation and Sequencing
4.3. cDNA Generation and qPCR Analysis
4.4. Analysis of Publicly Available Datasets
4.5. Identifying a Prognostic Signature for TNBC Patients
4.6. Determining Gene Expression Differences in Samples from before and after Chemotherapy
4.7. PAM50 Plasticity
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Global Breast Cancer Initiative Implementation Framework: Assessing, Strengthening and Scaling-Up of Services for the Early Detection and Management of Breast Cancer. Executive Summary; World Health Organisation: Geneva, Switzerland, 2023; ISBN 978-92-4-006713-4.
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics of American 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, M.; Xu, F.; Jiang, S. Wnt Signaling in Breast Cancer: Biological Mechanisms, Challenges and Opportunities. Mol. Cancer 2020, 19, 165. [Google Scholar] [CrossRef] [PubMed]
- Buyuk, B.; Jin, S.; Ye, K. Epithelial-to-Mesenchymal Transition Signaling Pathways Responsible for Breast Cancer Metastasis. Cell. Mol. Bioeng. 2022, 15, 1–13. [Google Scholar] [CrossRef]
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, F.; Senkus, E.; Costa, A.; Papadopoulos, E.; Aapro, M.; André, F.; Harbeck, N.; Aguilar Lopez, B.; Barrios, C.H.; Bergh, J.; et al. 4th ESO-ESMO International Consensus Guidelines for Advanced Breast Cancer (ABC 4). Ann. Oncol. 2018, 29, 1634–1657. [Google Scholar] [CrossRef]
- Canlı, S.D.; Dedeoğlu, E.; Akbar, M.W.; Küçükkaraduman, B.; İşbilen, M.; Erdoğan, Ö.Ş.; Erciyas, S.K.; Yazıcı, H.; Vural, B.; Güre, A.O. A Novel 20-Gene Prognostic Score in Pancreatic Adenocarcinoma. PLoS ONE 2020, 15, e0231835. [Google Scholar] [CrossRef] [PubMed]
- Demirkol, S.; Gomceli, I.; Isbilen, M.; Dayanc, B.E.; Tez, M.; Bostanci, E.B.; Turhan, N.; Akoglu, M.; Ozyerli, E.; Durdu, S.; et al. A Combined ULBP2 and SEMA5A Expression Signature as a Prognostic and Predictive Biomarker for Colon Cancer. J. Cancer 2017, 8, 1113. [Google Scholar] [CrossRef] [PubMed]
- Canli, S.D.; Uner, M.; Kucukkaraduman, B.; Karaoglu, D.A.; Isik, A.; Turhan, N.; Akyol, A.; Gomceli, I.; Gure, A.O. A Novel Gene List Identifies Tumors with a Stromal-Mesenchymal Phenotype and Worse Prognosis in Gastric Cancer. Cancers 2023, 15, 3035. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.W.; Isbilen, M.; Belder, N.; Canli, S.D.; Kucukkaraduman, B.; Turk, C.; Sahin, O.; Gure, A.O. A Stemness and EMT Based Gene Expression Signature Identifies Phenotypic Plasticity and Is A Predictive but Not Prognostic Biomarker for Breast Cancer. J. Cancer 2020, 11, 949. [Google Scholar] [CrossRef] [PubMed]
- Vera-Ramirez, L.; Sanchez-Rovira, P.; Ramirez-Tortosa, C.L.; Quiles, J.L.; Ramirez-Tortosa, M.; Lorente, J.A. Transcriptional Shift Identifies a Set of Genes Driving Breast Cancer Chemoresistance. PLoS ONE 2013, 8, e53983. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Angulo, A.M.; Iwamoto, T.; Liu, S.; Chen, H.; Do, K.A.; Hortobagyi, G.N.; Mills, G.B.; Meric-Bernstam, F.; Symmans, W.F.; Pusztai, L. Gene Expression, Molecular Class Changes and Pathway Analysis after Neoadjuvant Systemic Therapy for Breast Cancer. Clin. Cancer Res. 2012, 18, 1109. [Google Scholar] [CrossRef] [PubMed]
- Creighton, C.J.; Li, X.; Landis, M.; Dixon, J.M.; Neumeister, V.M.; Sjolund, A.; Rimm, D.L.; Wong, H.; Rodriguez, A.; Herschkowitz, J.I.; et al. Residual Breast Cancers after Conventional Therapy Display Mesenchymal as Well as Tumor-Initiating Features. Proc. Natl. Acad. Sci. USA 2009, 106, 13820–13825. [Google Scholar] [CrossRef] [PubMed]
- Vecchi, M.; Confalonieri, S.; Nuciforo, P.; Viganò, M.A.; Capra, M.; Bianchi, M.; Nicosia, D.; Bianchi, F.; Galimberti, V.; Viale, G.; et al. Breast Cancer Metastases Are Molecularly Distinct from Their Primary Tumors. Oncogene 2008, 27, 2148–2158. [Google Scholar] [CrossRef] [PubMed]
- Cejalvo, J.M.; De Dueñas, E.M.; Galván, P.; García-Recio, S.; Gasión, O.B.; Paré, L.; Antolín, S.; Martinello, R.; Blancas, I.; Adamo, B.; et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res. 2017, 77, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Lei, R.; Ding, S.W.; Zhu, S. Skewer: A Fast and Accurate Adapter Trimmer for next-Generation Sequencing Paired-End Reads. BMC Bioinform. 2014, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 April 2024).
- Simon, R.; Lam, A.; Li, M.C.; Ngan, M.; Menenzes, S.; Zhao, Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007, 3, 11. [Google Scholar] [CrossRef]
- de Hoon, M.J.L.; Imoto, S.; Nolan, J.; Miyano, S. Open Source Clustering Software. Bioinformatics 2004, 20, 1453–1454. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, A.J. Java Treeview--Extensible Visualization of Microarray Data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2000; ISBN 9780387987842. [Google Scholar]
- Hothorn, T.; Lausen, B. On the Exact Distribution of Maximally Selected Rank Statistics. Comput. Stat. Data Anal. 2003, 43, 121–137. [Google Scholar] [CrossRef]
- Lausen, B.; Hothorn, T.; Bretz, F.; Schumacher, M. Assessment of Optimal Selected Prognostic Factors. Biom. J. 2004, 46, 364–374. [Google Scholar] [CrossRef]
- R Foundation for Statistical Computing Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Bernard, P.S.; Parker, J.S.; Mullins, M.; Cheung, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160. [Google Scholar] [CrossRef] [PubMed]

| Spearman Rho Values | qPCR Data (-ddCT) | DESeq2 Normalized RNA-seq Data | FPKM of RNA-seq Data | TMM Normalized | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Name | GDSC1 | GDSC2 | CTD | Mean BC | Mean AC | T-Test P | Mean BC | Mean AC | T-Test P | Mean BC | Mean AC | T-Test P | Mean BC | Mean AC | T-Test P | ||||
| ABHD4 | 0.48 | 0.51 | 0.32 | 1 | 6.37 | 2.98 | 0.00 | 0 | 7.67 | 7.98 | 0.08 | 0 | 0.93 | 1.37 | 0.00 | 0 | 7.55 | 8.12 | 0.00 |
| CDYL2 | 0.89 | 0.34 | 0.43 | 1 | 8.35 | 7.39 | 0.45 | 1 | 8.28 | 8.24 | 0.85 | 0 | 0.74 | 0.94 | 0.04 | 0 | 8.15 | 8.38 | 0.27 |
| COPS7A | 0.45 | 0.56 | 0.38 | 1 | 7.72 | 4.23 | 0.00 | 1 | 8.66 | 8.61 | 0.73 | 0 | 1.75 | 2.05 | 0.07 | 0 | 8.53 | 8.75 | 0.25 |
| DPM2 | 0.79 | 0.54 | 0.38 | 1 | 11.64 | 8.75 | 0.00 | 1 | 7.89 | 7.68 | 0.18 | 0 | 1.26 | 1.42 | 0.23 | 0 | 7.75 | 7.82 | 0.77 |
| GSG1 | 0.32 | 0.35 | 0.34 | 1 | 10.74 | 7.88 | 0.01 | 1 | 2.30 | 1.11 | 0.11 | 1 | 0.11 | 0.02 | 0.06 | 1 | 2.24 | 0.98 | 0.05 |
| JMJD7 | −0.35 | −0.47 | −0.32 | 0 | 10.78 | 7.17 | 0.00 | 1 | 5.13 | 5.33 | 0.30 | 1 | 0.43 | 0.59 | 0.00 | 1 | 5.07 | 5.47 | 0.04 |
| KIAA1456 | 0.39 | 0.34 | 0.50 | 1 | 11.04 | 6.33 | 0.00 | 1 | 7.54 | 7.39 | 0.74 | 0 | 0.41 | 0.44 | 0.80 | 0 | 7.41 | 7.53 | 0.78 |
| KRTAP 17-1 | −0.48 | −0.35 | −0.32 | 0 | 12.07 | 6.47 | 0.00 | 0 | 1.05 | 0.36 | 0.13 | 0 | 0.07 | 0.02 | 0.29 | 0 | 0.92 | 0.42 | 0.20 |
| LIN9 | −0.36 | −0.37 | −0.35 | 0 | 9.49 | 7.21 | 0.02 | 0 | 7.83 | 7.40 | 0.09 | 1 | 1.00 | 1.03 | 0.83 | 0 | 7.71 | 7.54 | 0.53 |
| MIGLL | 0.47 | 0.48 | 0.42 | 1 | 6.27 | 4.59 | 0.00 | 0 | 8.67 | 9.09 | 0.10 | 0 | 0.79 | 1.28 | 0.00 | 0 | 8.54 | 9.23 | 0.00 |
| OFD1 | 0.82 | 0.36 | 0.45 | 1 | 6.44 | 4.19 | 0.00 | 0 | 10.64 | 10.76 | 0.50 | 0 | 2.72 | 3.23 | 0.04 | 0 | 10.51 | 10.90 | 0.11 |
| PHKA2 | 0.82 | 0.35 | 0.40 | 1 | 8.25 | 6.05 | 0.01 | 1 | 9.36 | 9.24 | 0.27 | 0 | 1.20 | 1.43 | 0.04 | 0 | 9.23 | 9.38 | 0.26 |
| PRMT7 | 0.82 | 0.36 | 0.48 | 1 | 10.93 | 6.34 | 0.00 | 1 | 9.31 | 9.24 | 0.55 | 0 | 1.12 | 1.38 | 0.01 | 0 | 9.17 | 9.38 | 0.07 |
| ST3GAL4 | 0.37 | 0.34 | 0.31 | 1 | 7.12 | 3.31 | 0.00 | 1 | 8.91 | 8.77 | 0.52 | 0 | 1.57 | 1.80 | 0.21 | 0 | 8.78 | 8.91 | 0.58 |
| TAL2 | −0.37 | −0.44 | −0.34 | 0 | 9.09 | 3.64 | 0.00 | 0 | 1.76 | 0.87 | 0.11 | 0 | 0.15 | 0.08 | 0.15 | 0 | 1.62 | 0.96 | 0.14 |
| TMTM221 | 0.85 | 0.35 | 0.36 | 1 | 7.64 | 3.50 | 0.00 | 1 | 2.35 | 1.53 | 0.14 | 1 | 0.08 | 0.05 | 0.20 | 1 | 2.16 | 1.51 | 0.16 |
| ZDHHC7 | 0.42 | 0.37 | 0.40 | 1 | 6.12 | 3.41 | 0.00 | 0 | 9.35 | 9.51 | 0.27 | 0 | 2.03 | 2.49 | 0.01 | 0 | 9.22 | 9.65 | 0.05 |
| Post-Therapy | ||||||
|---|---|---|---|---|---|---|
| Basal | HER2 | LumA | LumB | Normal | ||
| Pre-therapy | Basal | 2 | 0 | 0 | 0 | 0 |
| HER2 | 1 | 1 | 0 | 2 | 0 | |
| LumA | 0 | 0 | 2 | 3 | 0 | |
| LumB | 0 | 0 | 0 | 1 | 0 | |
| Normal | 3 | 0 | 1 | 1 | 1 | |
| Post-Therapy | ||||||
|---|---|---|---|---|---|---|
| Basal | HER2 | LumA | LumB | Normal | ||
| Pre-therapy | Basal | 2 | 0 | 0 | 0 | 2 |
| HER2 | 1 | 1 | 0 | 1 | 1 | |
| LumA | 0 | 0 | 3 | 0 | 4 | |
| LumB | 1 | 0 | 4 | 3 | 3 | |
| Normal | 0 | 0 | 1 | 0 | 1 | |
| Post-Therapy | ||||||
|---|---|---|---|---|---|---|
| Basal | HER2 | LumA | LumB | Normal | ||
| Pre-therapy | Basal | 5 | 0 | 0 | 0 | 0 |
| HER2 | 0 | 4 | 0 | 0 | 0 | |
| LumA | 0 | 0 | 5 | 1 | 0 | |
| LumB | 0 | 0 | 0 | 1 | 0 | |
| Normal | 2 | 1 | 1 | 0 | 1 | |
| Metastasized Tissue | ||||||
|---|---|---|---|---|---|---|
| Basal | HER2 | LumA | LumB | Normal | ||
| Primary tissue | Basal | 2 | 1 | 0 | 0 | 0 |
| HER2 | 1 | 2 | 0 | 0 | 1 | |
| LumA | 0 | 0 | 4 | 0 | 0 | |
| LumB | 0 | 0 | 0 | 0 | 0 | |
| Normal | 3 | 2 | 0 | 0 | 0 | |
| PAM50 Metastasis | ||||||
|---|---|---|---|---|---|---|
| Sample Name | Primary Tissue | Basal | HER2 | LumA | LumB | Normal |
| A11 | Basal | 5 | 0 | 0 | 0 | 0 |
| A15 | Basal | 2 | 0 | 0 | 0 | 3 |
| A1 | Basal | 4 | 0 | 0 | 0 | 1 |
| A23 | Basal | 5 | 0 | 0 | 0 | 0 |
| A5 | Basal | 2 | 0 | 0 | 0 | 0 |
| A7 | Basal | 5 | 0 | 0 | 0 | 0 |
| A8 | HER2 | 0 | 4 | 0 | 0 | 0 |
| A12 | LumA | 0 | 0 | 2 | 4 | 0 |
| A17 | LumA | 2 | 0 | 0 | 0 | 0 |
| A26 | LumA | 3 | 0 | 0 | 0 | 0 |
| A28 | LumA | 0 | 0 | 3 | 3 | 0 |
| A2 | LumA | 0 | 1 | 0 | 2 | 0 |
| A30 | LumA | 5 | 0 | 0 | 0 | 0 |
| A34 | LumA | 0 | 0 | 4 | 0 | 0 |
| A4 | LumA | 0 | 1 | 1 | 0 | 0 |
| A20 | Normal | 5 | 0 | 0 | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padzińska-Pruszyńska, I.B.; Akbar, M.W.; Isbilen, M.; Górka, E.; Kucukkaraduman, B.; Canlı, S.D.; Dedeoğlu, E.; Azizolli, S.; Cela, I.; Akcay, A.G.; et al. Breast Cancer Plasticity after Chemotherapy Highlights the Need for Re-Evaluation of Subtyping in Residual Cancer and Metastatic Tissues. Int. J. Mol. Sci. 2024, 25, 6054. https://doi.org/10.3390/ijms25116054
Padzińska-Pruszyńska IB, Akbar MW, Isbilen M, Górka E, Kucukkaraduman B, Canlı SD, Dedeoğlu E, Azizolli S, Cela I, Akcay AG, et al. Breast Cancer Plasticity after Chemotherapy Highlights the Need for Re-Evaluation of Subtyping in Residual Cancer and Metastatic Tissues. International Journal of Molecular Sciences. 2024; 25(11):6054. https://doi.org/10.3390/ijms25116054
Chicago/Turabian StylePadzińska-Pruszyńska, Irena Barbara, Muhammad Waqas Akbar, Murat Isbilen, Emilia Górka, Baris Kucukkaraduman, Seçil Demirkol Canlı, Ege Dedeoğlu, Shila Azizolli, Isli Cela, Abbas Guven Akcay, and et al. 2024. "Breast Cancer Plasticity after Chemotherapy Highlights the Need for Re-Evaluation of Subtyping in Residual Cancer and Metastatic Tissues" International Journal of Molecular Sciences 25, no. 11: 6054. https://doi.org/10.3390/ijms25116054
APA StylePadzińska-Pruszyńska, I. B., Akbar, M. W., Isbilen, M., Górka, E., Kucukkaraduman, B., Canlı, S. D., Dedeoğlu, E., Azizolli, S., Cela, I., Akcay, A. G., Hakanoglu, H., Bodnar, L., Cierniak, S., Kozielec, Z., Pruszyński, J. J., Bittel, M., Gure, A. O., Król, M., & Taciak, B. (2024). Breast Cancer Plasticity after Chemotherapy Highlights the Need for Re-Evaluation of Subtyping in Residual Cancer and Metastatic Tissues. International Journal of Molecular Sciences, 25(11), 6054. https://doi.org/10.3390/ijms25116054






